ABSTRACT
This study investigated the antioxidant defense system involved in the tolerance of soybean (Glycine max) to aluminum (Al) stress. Physiological assays showed that the amount of superoxide free radicals (O2−), hydrogen peroxide (H2O2), and malondialdehyde (MDA) were significantly lower in an Al-resistant soybean cultivar (cv. PI416937) than in an Al-sensitive soybean cultivar (cv. Huachun18). Comparative analysis of microarray data from both genotypes following Al-stress treatment revealed that the expression of a series of antioxidant enzymes genes was induced in the Al-resistant cultivar. The quantitative real time-PCR (qRT-PCR) assay showed that the transcript levels of genes encoding antioxidant enzymes, including GmCAT1, GmPOD1, GmGST1, GmAPX, GmGSH1, and GmSOD, were higher in the Al-resistant cultivar than in the Al-sensitive cultivar in Al-stress conditions. Furthermore, GmCAT1-overexpressing Arabidopsis plants had improved tolerance to Al-stress and lower O2−, H2O2, and MDA contents than wild-type plants. Therefore, providing evidence that the antioxidant defense system is essential for Al tolerance in soybean.
Abbreviations
Al: aluminum; O2−: superoxide free radicals; ROS: reactive oxygen species; H2O2: hydrogen peroxide; MDA: malondialdehyde; qRT-PCR: quantitative reverse transcription polymerase chain reaction; GO: gene ontology; WT: wild type; MS medium: Murashige and Skoog medium
GRAPHICAL ABSTRACT
Antioxidant defense system play essential roles in soybean Al tolerance. The results are of great significance in understanding the molecular mechanism of Al stress responses in soybean.
Aluminum (Al) is mainly found in the roots of plants and is an essential element for growth and development [Citation1,Citation2]. Even low concentrations of Al3+ (< 0.1 mg/L) can promote plant growth. However, concentrations from 0.1–1.0 mg/L can be toxic to Al3+-sensitive plants. Furthermore, concentrations of Al3+ that are greater than 1 mg/L are toxic to most plants. Under natural conditions, Al is mainly present in the soil as aluminosilicates, which have little bioavailability [Citation3]. However, in recent years, the use of agricultural fertilizers and effects of acid rain have increased the dissolution of Al owing to soil acidification [Citation3,Citation4].
Previous studies have shown that Al affects plants by limiting the absorption and accumulation of important mineral elements, such as calcium, phosphorus, potassium, magnesium, iron, manganese, copper, and zinc [Citation5–Citation7]. For example, after enrichment of Al, intracellular calcium-regulated proteins, which control calcium balance and metabolism, are in an inactive state [Citation6,Citation8]. The non-activated state can affect various physiological and biochemical processes, such as plant meristem DNA synthesis, orderly synthesis and fragmentation of plant cell wall pectin chains, damage or rupture of cell membranes, and dysregulation of cAMP as a second messenger, which damages plant growth and development [Citation6]. In addition, binding of excess Al to the phosphate in the cell wall of root surfaces may prevent phosphorus from entering the cytoplasm; therefore, further affecting the absorption and metabolism of phosphorus [Citation5,Citation7]. Al can also destroy cells and cause death by dehydrating the plant cell protoplasm, which combines with pectin in the cell wall to strengthen the cross-linked structure of pectin, leading to the disordered absorption of water and nutrients [Citation9].
To survive, plants have evolved sophisticated strategies to respond to stress through changes at the physiological and molecular levels [Citation10]. These processes are modulated by a complicated signaling network that regulates the expression of stress-responsive genes [Citation11,Citation12]. Stress-responsive genes are usually classified into two groups: effector genes and regulatory genes [Citation12]. Effector genes encode enzymes that function as osmoprotectants, late embryogenesis abundant proteins, chaperone, and detoxification enzymes that are involved in protecting cell membrane integrity, controlling ion balances, and scavenging reactive oxygen species (ROS) [Citation9,Citation13]. Regulatory genes encode protein kinases and transcription factors, which play essential roles in signal perception and signal transduction [Citation9,Citation13].
Since the 1960s, researchers have given considerable attention to understanding the mechanisms underlying Al toxicity in plants and screening and cultivating Al-resistant varieties. As a major economic crop, soybean cultivation has a huge production potential worldwide. However, acid-Al stress that occurs in acidic red soil areas is a serious problem that has become a major limitation for crop growth [Citation2,Citation4,Citation14]. Despite this, few studies have investigated acid-Al stress-responsive mechanisms in soybean. Therefore, it is important to measure the expression of soybean Al-responsive genes. In this study, physiological and molecular assays were used to understand the mechanisms of Al-tolerance in soybean under Al stress conditions. In particular, we show that several antioxidant enzymes, especially GmCAT1, have an important role in Al stress responses. These findings may provide a theoretical basis for screening and breeding Al-resistant soybean varieties.
Materials and methods
Plant materials and growth conditions
Seeds of an Al-resistant soybean cultivar (Glycine max cv. PI416937) and Al-sensitive cultivar (Glycine max cv. Huachun 18) were obtained from the Zhejiang Academy of Agricultural Sciences (Zhejiang, China). Soybean seeds were surface sterilized in 1.0% sodium hypochlorite (ν/ν) for 5 min, rinsed thoroughly with distilled deionized water, and subsequently germinated in an incubator for 72 h at 25°C. Seedlings with uniform growth were transplanted to black-painted pots that were filled with a nutrient solution that contained 0.5 mM CaCl2 (pH = 4.5) for 2 days. Next, seedlings were subjected to a 0.5 mM CaCl2 solution (pH = 4.5) without (control group) or with 50 µM AlCl3 (experimental group) for 72 h. Roots were harvested 72 h after the treatment, frozen in liquid nitrogen, and stored at −80°C until RNA isolation.
Data sources
We searched the ArrayExpress database to establish the Al stress-responsive mechanism. Two keywords (“Al stress” and “soybean root”) were used to blast search microarray data from Al stress-related root tissue from soybean. In this study, according to the sample size and experimental content, E-GEOD-18517 was used to identify Al stress-responsive genes. The microarray data was uploaded by Dongquan Chen [Citation15] and the GPL4592 [Soybean] Affymetrix Soybean Genome Array platform was used.
Screening and clustering of differently expressed genes
Differentially expressed genes (DEGs) between the Al-resistant group and Al-sensitive group in different treatments at different times were screened by limma Version 3.34.9 in R. The threshold was set at the false discovery rate FDR (P < 0.05 and |log2FC| > 1). For screened DEGs in each group, expression clustering was performed for all samples using the pheatmap Version 1.0.10 package in R3.4.3.
Functional enrichment analysis
Gene Ontology (GO) analysis was performed using the platform PlantRegMap (Plant Transcriptional Regulatory Map). PlantRegMap provides a comprehensive, high-quality resource of plant transcription factors (TFs), regulatory elements, and interactions between them. Glycine max was used as the selected species. The gene list of each group was used as an input and an enrichment analysis was performed for Biological Process (P), Molecular Function (F), and Cellular Component (C). The threshold was p-value < 0.05.
RNA extraction and quantitative real-time (qRT)-PCR
Total RNA was extracted from root tissue samples using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Total RNA was purified using an RNase-free DNase and cDNA was synthesized from RNA using a Reverse Transcription Kit (TaKaRa) according to the manufacturer’s protocol. qRT-PCR was performed using Fast SYBR green master mix on the Fast Real-Time PCR system (Applied Biosystems). The primer sequences used in this study are provided in Table S1. The thermal cycling parameters used for qRT-PCR were as follows: 95°C for 5 min, 45 cycles of amplification at 95°C for 10 s, 55°C for 20 s, and 72°C for 20 s; melting at 95°C for 1 min, 65°C for 1 min, and 95°C continuous; and cooling at 40°C for 30 s. Relative fold changes of gene expression were evaluated and normalized by the 2−ΔΔCTmethod. GmTubulin (Glyma.08G014200.1) was used as a reference gene for normalization of soybean transcripts and AtActin2 (At3G18780) was used for Arabidopsis data. Each experiment was performed independently at least three times.
Generation of transgenic Arabidopsis plants
The GmCAT1-overexpression vector was generated by inserting the coding sequence of GmCAT1 into the pBI121 vector with expression driven by a CaMV 35S promoter. This vector was transformed into wild-type (WT) Arabidopsis (Arabidopsis thaliana Col-0) using the vacuum infiltration method [Citation16] to produce GmCAT1-overexpressing plants. Homozygous T4 transgenic lines were used for phenotypic analyses.
Measurement of H2O2, O2−, and MDA contents
For physiological trait assays, 7-day-old GmCAT1-overexpressing and WT Arabidopsis seedlings were grown for 2 days on Murashige and Skoog (MS) (Caisson Labs, USA) medium that contained 0.5 mM CaCl2 (pH = 4.5). Next, these seedlings were grown for 2 weeks on MS medium that contained 0.5 mM CaCl2 supplemented with 0 or 25 µM AlCl3 (pH = 4.5). Approximately 0.2 g of leaves were used to measure physiological parameters. A Varioskan LUX Multimode Microplate Reader (ThermoFisher Scientific, USA) was used for absorbance measurements. The O2− and H2O2 contents were determined as previously described [Citation12]. MDA contents were also measured as previously described2
Statistical analysis
All data are shown as the mean ± SD of three biological replicates. A Student’s t-test (*P < 0.05 and **P < 0.01) was used to identify statistically significant differences between cultivars.
Results
The Al-resistant soybean cultivar PI416937 has better Al stress-induced oxidative stress tolerance than Al-sensitive soybean cultivar Huachun 18
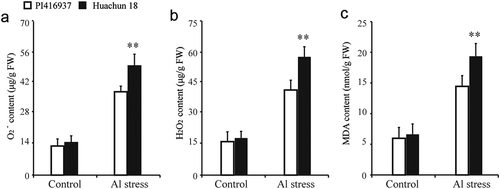
Al stress causes various biochemical and physiological changes in soybean. To identify Al stress-tolerance-related physiological processes in soybean, we analyzed the levels of O2−, H2O2, and MDA in Al-resistant (cv. PI416937) and Al-sensitive (cv. Huachun 18) soybean cultivars. There were no significant differences between the Al-sensitive and Al-resistant soybean cultivars in control conditions (). However, Al stress treatment induced remarkable physiological differences between the cultivars. In particular, the Al-resistant soybean cultivar had significantly lower O2−, H2O2, and MDA contents than the Al-sensitive cultivar during Al stress (–)).
Multiple stress related genes were activated in by Al stress in the Al-resistant soybean cultivar (cv. PI416937)
To further investigate the Al stress-response mechanism, we analyzed previously published microarray data to identify differentially expressed genes (DEGs) between the Al-sensitive (cv. Huachun18) and Al-resistant (cv. PI416937) cultivars following Al stress. A 2-fold change threshold and a Student’s t-test significance cutoff of P < 0.05 were used to identify significant DEGs. Microarray data obtained from the two cultivars grown under normal conditions were compared. Overall, 380 up-regulated genes (115 in control condition; 265 in control and Al-stress condition) and 222 down-regulated genes (94 in control condition; 128 in control and Al-stress condition) genes were identified in the Al-sensitive soybean cultivar (cv. Huachun 18). After Al stress treatment, 398 up-regulated genes (133 in Al-stress condition; 265 in control condition and Al-stress condition) and 227 down-regulated genes (99 in Al stress condition; 128 in control and Al stress condition) were identified in the in Al-resistant cultivar (cv. PI416937) ()). These results reveal differential gene expression in response to Al between Al-resistant and Al-sensitive cultivars of soybean.
Figure 2. Analysis of differentially expressed genes (DEGs) between an Al-resistant (cv. PI416937) and Al-sensitive (cv. Huachun18) soybean cultivars under Al stress conditions.
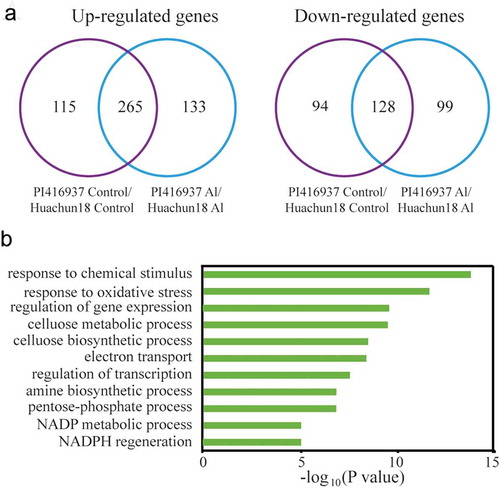
The Gene Ontology (GO) analysis showed that DEGs between the Al-sensitive and Al-resistant soybean cultivars after Al stress treatment were significantly enriched in biological process categories including “response to chemical stimulus”, “response to oxidative stress”, “regulation of gene expression”, and “regulation of metabolic and biosynthetic processes”()).
The expression of genes encoding antioxidant enzymes was induced in an Al-resistant soybean cultivar (cv. PI416937) following Al stress treatment
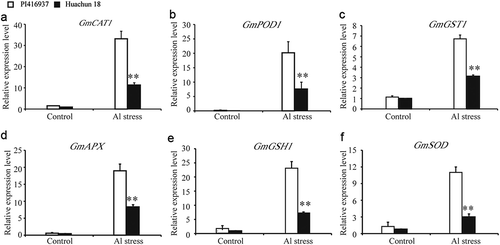
The GO analysis results show that the expression of many genes encoding antioxidant enzymes was induced in the Al-resistant soybean cultivar and not the Al-sensitive cultivar. Interestingly, we noted that genes encoding a range of well-known antioxidant enzyme were among the up-regulated DEGs for the Al stress-treated Al-resistant soybean cultivar. To confirm the reliability of our bioinformatics data, genes encoding 6 antioxidant enzymes were selected to measure their pattern of expression during Al stress conditions using qRT-PCR experiments. These experiments confirmed the up-regulated expression trends for antioxidant enzyme genes, including GmCAT1, GmPOD1, GmGST1, GmAPX, GmGSH1, and GmSOD. In control conditions, no significant differences were identified between the two soybean cultivars (–)). Furthermore, the expression of GmCAT1, GmPOD1, GmGST1, GmAPX, GmGSH1, and GmSOD was significantly higher in the Al-resistant cultivar than in the A1-sensitive cultivar following Al stress treatment (–)).
GmCAT1 overexpression confers Al tolerance to transgenic Arabidopsis
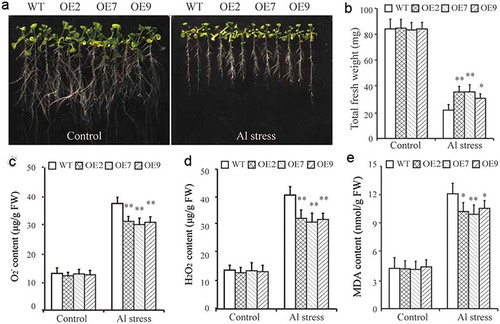
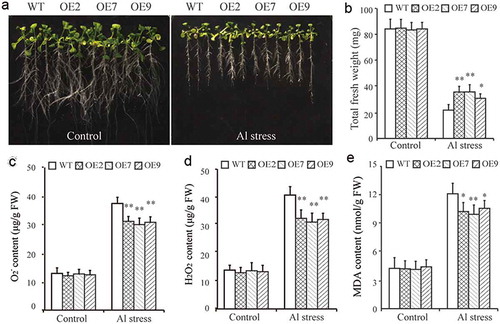
The results of our qRT-PCR assay showed that the expression of GmCAT1 was induced the most in response to Al stress. To further analyze the function of GmCAT1 under Al stress, we generated transgenic Arabidopsis plants that were constitutively overexpressing GmCAT1. Three independent homozygous T4 transgenic lines (lines OE2, OE7, and OE9) that had relatively high expression of GmCAT1 were chosen for phenotypic analysis (Figure S1). To assess the tolerance of GmCAT1-overexpressing lines to Al stress, 7-day-old GmCAT1-overexpressing and WT seedlings were grown for 2 days on MS medium that contained 0.5 mM CaCl2 (pH = 4.5). Next, these seedlings were transferred into MS medium that contained 0.5 mM CaCl2 supplemented with 0 or 25 µM AlCl3 (pH = 4.5). The morphology of the roots and fresh weight of GmCAT1-overexpressing and WT plants were similar in control conditions ()). However, Al stress treatment impaired the growth of both lines. The fresh weight of GmCAT1-overexpressing plants was significantly greater than WT plants in response to Al stress ()). GmCAT1-overexpressing lines had improved tolerance to Al stress owing to physiological and biochemical changes. The O2−, H2O2, and MDA contents of GmCAT1-overexpressing and WT lines were similar in control conditions (–)) but significantly lower in GmCAT1-overexpressing lines than WT plants under Al stress conditions (–)).
Discussion
Al toxicity acts as a major environmental stimulus that affects plant growth and development; therefore, posing a great challenge to agricultural production [Citation4,Citation17,Citation18]. In the present study, a basis for breeding of Al-resistant soybean varieties was established using physiological and bioinformatic assays to measure differences in the physiology and gene expression between an Al-resistant (cv. PI416937) and Al-sensitive (cv. Huachun18) soybean cultivar during Al stress. The Al-resistant cultivar had significantly lower O2−, H2O2, and MDA contents than the A1-sensitive cultivar under Al stress conditions. Previous studies have shown that abiotic stress resulted in the accumulation of ROS [Citation12,Citation18,Citation19], which have dual roles in stress responses [Citation19,Citation20]. Excessive accumulation of ROS can cause oxidative stress, damage the cell structure, and cause cell death [Citation18,Citation21]. Furthermore, an excessive accumulation of ROS also increases MDA [Citation22]. The amount of MDA reflects the stability of the cell membrane, which is negatively related to environmental stress in plants [Citation12]. Therefore, cellular redox homeostasis has an important function in the response of plants to abiotic stress.
GO analysis revealed that the expression of genes that encode a series of antioxidant enzyme genes were activated in the Al-resistant cultivar under Al-stress, which is consistent with reduced ROS contents in this variety. Furthermore, qRT-PCR assays showed that the transcript levels of genes encoding antioxidant enzymes, including GmCAT1, GmPOD1, GmGST1, GmAPX, GmGSH1, and GmSOD, were significantly higher than in the Al-sensitive cultivar. These antioxidant enzyme genes encode detoxification enzymes that play essential roles in maintaining cellular redox homeostasis and protecting organisms against oxidative stress caused by Al stress [Citation18,Citation23]. These results suggest that antioxidant enzymes have important roles in alleviating Al stress-induced oxidative stress.
CATs function as an efficient ROS scavenging system to alleviate abiotic stress-induced oxidative damage. Previous studies have illustrated that IbCAT2 expression was induced by drought and salt stress in Ipomoea batatas. Furthermore, over-expression of IbCAT2 conferred salt and drought tolerance in Escherichia coli and Saccharomyces cerevisiae [Citation24]. The peroxisomal MeCAT1 has been shown to positively regulate cold and drought stress in cassava (Manihot esculenta) by scavenging ROS [Citation25]. In the present study, the expression of GmCAT1 was induced by Al stress and the presence of H2O2. Over-expression of GmCAT1 increased Al stress tolerance and reduced the contents of O2−, H2O2, and MDA. Therefore, these results suggest that CATs could have an important role in Al stress tolerance by scavenging ROS.
In conclusion, this study has identified upregulated genes that are likely involved in Al-stress resistance in soybean. Furthermore, we have shown that overexpression of GmCAT1 improves stress tolerance in transgenic Arabidopsis. These findings contribute to efforts to identify targets and develop Al resistant soybean varieties that have improved yields on acidic soils.
Author contribution
WL and YS designed experiments and wrote the manuscript; BW, HX and JW analyzed sequencing data and experimental results; ZN carried out experiments.
Supplemental_data.pdf
Download PDF (52.7 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Ahn SJ, Matsumoto H. The role of the plasma membrane in the response of plant roots to aluminum toxicity. Plant Signal Behav. 2006 Mar;1(2):37–45.
- Bojorquez-Quintal E, Escalante-Magana C, Echevarria-Machado I, et al. Aluminum, a friend or foe of higher plants in acid soils. Front Plant Sci. 2017;8:1767.
- Kopittke PM, Moore KL, Lombi E, et al. Identification of the primary lesion of toxic aluminum in plant roots. Plant Physiol. 2015 Apr;167(4):1402–1411.
- Carcamo MP, Reyes-Diaz M, Rengel Z, et al. Aluminum stress differentially affects physiological performance and metabolic compounds in cultivars of highbush blueberry. Sci Rep. 2019 Aug 2;9(1):11275.
- Miller JB, Pratap A, Miyahara A, et al. Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell. 2013 Dec;25(12):5053–5066.
- Bernardino KC, Pastina MM, Menezes CB, et al. The genetic architecture of phosphorus efficiency in sorghum involves pleiotropic QTL for root morphology and grain yield under low phosphorus availability in the soil. BMC Plant Biol. 2019 Feb 28;19(1):87.
- Chen J, Duan RX, Hu WJ, et al. Unravelling calcium-alleviated aluminium toxicity in Arabidopsis thaliana: insights into regulatory mechanisms using proteomics. J Proteomics. 2019 May;15(199):15–30.
- Kochian LV. Cellular mechanisms of aluminum toxicity and resistance in plants. Ann Rev Plant Physiol Plant Mol Biol. 1995 Jun;46(1):237–260.
- Huang XS, Wang W, Zhang Q, et al. A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 2013 Jun;162(2):1178–1194.
- Qi J, Song CP, Wang B, et al. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol. 2018 Sep;60(9):805–826.
- Wu Q, Wang M, Shen J, et al. ZmOST1 mediates abscisic acid regulation of guard cell ion channels and drought stress responses. J Integr Plant Biol. 2019 Apr;61(4):478–491.
- Cui XY, Gao Y, Guo J, et al. BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 2019 May;180(1):605–620.
- Liu W, Tai H, Li S, et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014 Mar;201(4):1192–11204.
- Hamed SM, Hassan SH, Selim S, et al. Physiological and biochemical responses to aluminum-induced oxidative stress in two cyanobacterial species. Environ Pollut. 2019 Aug;251:961–969.
- Duressa D, Soliman K, Chen D. Identification of aluminum responsive genes in Al-tolerant soybean line PI 416937. Int J Plant Genomics. 2010;2010:1–13.
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998 Jun;82:259–266.
- Jaskowiak J, Kwasniewska J, Milewska-Hendel A, et al. Aluminum alters the histology and pectin cell wall composition of barley roots. Int J Mol Sci. 2019 Jun 21;20(12):12.
- Awasthi JP, Saha B, Panigrahi J, et al. Redox balance, metabolic fingerprint and physiological characterization in contrasting North East Indian rice for aluminum stress tolerance. Sci Rep. 2019 Jun 18;9(1):8681.
- Wu Z, Yin X, Banuelos GS, et al. Indications of selenium protection against cadmium and lead toxicity in oilseed rape (Brassica napus L.). Front Plant Sci. 2016;7:1875.
- Liu S, Liu S, Wang M, et al. A wheat SIMILAR TO RCD-ONE gene enhances seedling growth and abiotic stress resistance by modulating redox homeostasis and maintaining genomic integrity. Plant Cell. 2014 Jan;26(1):164–180.
- Chichiricco G, Poma A. Penetration and toxicity of nanomaterials in higher plants. Nanomaterials (Basel). 2015 May 26;5(2):851–873.
- Farmer EE, Mueller MJ. ROS-mediated lipid peroxidation and RES-activated signaling. Annu Rev Plant Biol. 2013 Feb;64(1):429–450.
- Sharma P, Dubey RS. Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep. 2007 Nov;26(11):2027–2038.
- Yong B, Wang X, Xu P, et al. Isolation and abiotic stress resistance analyses of a catalase gene from Ipomoea batatas (L.) lam. BioMed Res int. 2017May;2017:6847532
- Xu J, Duan X, Yang J, et al. Coupled expression of Cu/Zn-superoxide dismutase and catalase in cassava improves tolerance against cold and drought stresses. Plant Signal Behav. 2013 Apr;8(6):e24525.