ABSTRACT
Melanin metabolism disorders may cause severe impacts on the psychological and social activities of patients. Different from the other two steps of melanin metabolism, namely synthesis and transport, little has been known about the mechanism of melanin degradation. Isoimperatorin (ISO) suppressed the activity of tyrosinase, an essential enzyme in melanin biosynthesis, hence, we investigated the effects and mechanism of ISO in melanin reduction. ISO stimulation significantly reduces the melanin contents and PMEL 17 protein levels; meanwhile, the activity and the protein levels of two critical lysosomal enzymes, Cathepsin B and Cathepsin D, can be significantly increased by ISO treatment. MiR-3619 inhibited the expression of CSTB and CSTD, therefore affecting ISO-induced degradation of melanin. In summary, ISO reduces the melanin content via miR-3619/CSTB and miR-3619/CSTD axes. ISO could be a potent skin-whitening agent, which needs further in vivo and clinical investigation.
Graphical Abstract
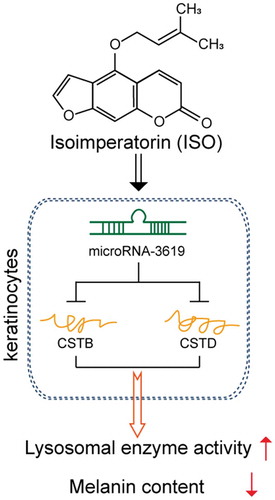
ISO reduces the melanin content and increase lysosomal enzye activity via miR-3619/CSTB and miR-3619/CSTD axes.
Pigmented diseases are a common type of skin disease caused by melanin metabolism disorders, which has a considerable impact on the psychological and social activities of patients [Citation1–Citation8]. Therefore, exploring the pathogenesis and prevention measures of such diseases has become a research hotspot. The metabolic process of melanin includes three significant steps: synthesis, transport, and degradation of melanin. During the past decades, studies of melanin metabolism have been largely limited to synthesis and transport steps, especially synthesis [Citation9–Citation13]; in the meantime, little research has been done on the mechanism of melanin degradation.
The substances in the cells are mainly degraded via two pathways: protease degradation and autophagy degradation [Citation14,Citation15]. Studies have found that when keratinocytes reach the stratum corneum, the structure of melanosomes also disappears [Citation16], suggesting that melanosomes may be degraded by specific proteases in keratinocytes. As the keratinocytes continue to move up the epidermis to complete the final differentiation process, the cytoplasmic melanin bodies in the cytoplasm are also continuously degraded by lysosomes in the keratinocytes, and finally, excreted with the exfoliation of epidermal cells [Citation17,Citation18]. These all suggest that lysosomes and lysosomal enzymes in keratinocytes exert a critical effect on melanin degradation.
The lysosome is one of single-layer membrane vesicular organelles that is ubiquitous in various types of cells and contains more than 60 hydrolases; these hydrolases could degrade or digest nucleic acids and polysaccharides [Citation19–Citation22]. Among them, acid phosphatase (ACP) is ubiquitously present in lysosomes and can be displayed by electron microscopy enzyme cytochemistry, thus is generally used as a marker lysosomal enzyme. Other studies have found differential expression of epidermal lysosomal hydrolase at the upper base, including B3GTL, Cathepsin B, Cathepsin L2, Cathepsin D, ACPP, and β-GLU. These differences in expression lead to different degradation of melanin in keratinocytes from different tissues [Citation23,Citation24].
Isoimperatorin (ISO) is an abundant dietary flavonoid and is considered to be a key chemical component of Angelica dahurica. Reportedly, ISO can suppress the activity [Citation25] or synthesis [Citation26] of tyrosinase, an essential enzyme within the biosynthesis of melanin [Citation27]. The role of ISO in the activity of tyrosinase might be associated with its inhibitory effect on the formation of melanin within melanocytes [Citation26]. Previously, isoliquiritigenin, another main chemical component of Angelica dahurica, has been reported to decrease the levels of melanin and PMEL 17 protein within the keratinocytes of the human epidermis; in the meantime, autophagy was involved in isoliquiritigenin-induced melanin degradation; eventually, PI3 K/AKT/mTOR signaling contributed to the isoliquiritigenin-induced melanin degradation [Citation15]. Autophagy activators have been proved to reduce the melanin content in three-dimensional human skin substitutes and tissue-cultured human skin [Citation28]. Moreover, some lysosomal enzymes activation depends upon Atg5 and Atg7 mediated autophagosome formation [Citation29]. Based on the critical roles of ISO in melanin content reduction, herein, we speculated that ISO might exert its effects via similar mechanisms and further examined whether ISO could affect melanin content in a lysosome and lysosomal enzymes-related manner by regulating one or more lysosomal enzymes mentioned above.
Reportedly, microRNAs (miRNAs) exert potent effects on regulating the generation of melanin in mammals. Liu et al. [Citation30] reported that short tandem target mimic (STTM)-miR-508-3p could effectively block miR-508-3p, therefore enhancing the generation of melanin. The microphthalmia-associated transcription factor (MITF) is known as a critical regulatory factor of melanogenic enzymes for the production of melanin; miR-218 can suppress the production of melanin via direct inhibition of MITF expression [Citation31]. miR-21a-5p modulates the production of melanin via binding to Sox5 within the melanocytes of mouse skin [Citation32]. Thus, we hypothesize miRNAs might be involved in ISO functions in melanin reduction by regulating lysosomal enzymes.
Herein, the involvements of different doses of ISO treatment in the cell viability and the melanin degradation were examined in HaCaT cells; next, the specific role of ISO treatment in the activity and protein levels of four vital lysosomal enzymes, ACP, Cathepsin B, Cathepsin D, and Cathepsin L2, could be detected. Since Cathepsin B and Cathepsin D could be significantly altered by ISO treatment, next, the effects of CSTB or CSTD knockdown on ISO-induced melanin reduction were evaluated. Regarding the molecular mechanism, we used an online tool to identify miRNAs that might target both CSTB and CSTD to inhibit their expression; among 17 miRNAs, miR-3619 was selected and examined for its effect on ISO-induced melanin degradation and its binding with CSTB and CSTD. These data indicate that we might provide a new mechanism for ISO from the perspective of inducing melanin degradation in HaCaT cells lysosome- and lysosomal-enzyme-dependently.
Materials and methods
Cell culture and cell transfection
Human immortalized fibrocytes HaCaT cells were obtained from CLS Cell Lines Service (300,493/p800_HaCaT, CLS Cell Lines Service, Eppelheim, Germany) and were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 4.5 g/L glucose, 2 mM L-glutamine, and 10% fetal bovine serum. Cells were incubated at 37°C in 5% CO2.
For CSTB/CSTD knockdown, specific short hairpin RNA (sh-CSTB/CSTD) and negative control (sh-NC) were purchased from Genepharma (Shanghai, China) and transfected into HaCaT cells. For miR-3619 expression, cells were transfected with miR-3619 mimics or inhibitor (Genepharma). Cells were harvested after 48 h for further experiments.
MTT assay
MTT analysis was performed to detect the cell viability following the methods described before [Citation15]. The OD values were measured at 490 nm. The viability of cells in each group was calculated, respectively, from the viability of the non-treated cells (control group).
Melanin determination in keratinocytes
The levels of melanin in HaCaT cells were determined following the methods described before [Citation15,Citation33]. Relative melanin content was determined by absorbance at 405 nm in an ELISA plate reader.
Immunoblotting analyses
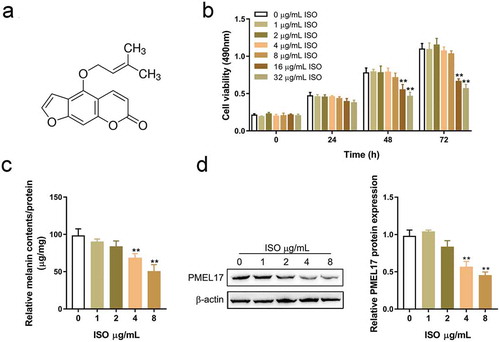
Before PMEL17 protein level determination, HaCaT cells were incubated with melanosomes (isolated from A375 melanocytes) for 48 h, cultured in mediums containing different concentrations of ISO (0, 1, 2, 4, 8 μg/mL), and cultured for 24 h. Afterward, the protein level of PMEL17 was determined.
For ACP, Cathepsin B, Cathepsin D, and Cathepsin L2 protein levels, the protein concentrations were first determined using the BCA protein assay kit (Pierce). After that, the proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA, USA) and determined for the protein levels using the following antibodies: anti-ACP (MBS2016306, MyBioSource, Inc., San Diego, CA, USA), anti-Cathepsin B (ab58802, Abcam, Cambridge, MA, USA), anti-Cathepsin D (ab75852, Abcam), and Cathepsin L2 (ab166894, Abcam). Signals were detected by enhanced chemiluminescence (ECL) and visualized with the ChemiDOC XRS system (Bio-Rad).
Determination of the activity of Cathepsin B/D/L2 and ACP
The activity of Cathepsin B/D/L2 and ACP was determined using Cathepsin B Activity Assay Kit (Fluorometric) (ab65300, Abcam), Cathepsin D Activity Assay Kit (Fluorometric) (ab65302, Abcam), Cathepsin L Activity Assay Kit (Fluorometric) (ab65306, Abcam), and Acid Phosphatase (ACP) Assay Kit (BC2135, Solarbio, Beijing, China), respectively, following the protocols.
Statistical analysis
Data from at least three independent experiments were processed using SPSS 17.0 statistical software (SPSS, Chicago, IL, USA) and then exhibited as a mean ± SD. The difference among the groups was estimated by Student’s t-test (two groups) or one-way ANOVA (more than two groups). P values of <0.05 were considered statistically significant.
Results
ISO reduces melanin within HaCaT cells
ISO molecular structure schematic diagram was shown in . To confirm the effects of ISO on the degradation of melanin, we co-incubated HaCaT cells with melanosomes for 48 h, followed by the treatment with ISO (0, 1, 2, 4, 8, 16, 32 μg/mL) for 0 h, 24 h, 48 h, and 72 h. Then cell viability, melanin contents, and PMEL 17 protein levels, a critical factor in melanin synthesis, were examined [Citation34]. As shown in , 16 and 32 μg/mL ISO treatment caused a significant inhibition on the cell viability of HaCaT cells; thus, HaCaT cells were then stimulated with 0 μg/mL, 1 μg/mL, 2 μg/mL, 4 μg/mL, and 8 μg/mL ISO before detecting the melanin contents. As shown in , melanin produced during the co-incubation with melanosomes could be significantly degraded with 4 μg/mL or 8 μg/mL ISO stimulation, as manifested via the significant decreases in melanin contents in response to 4 μg/mL or 8 μg/mL ISO stimulation. Furthermore, 4 μg/mL or 8 μg/mL ISO stimulation also induced a significant decrease in PMEL 17 protein levels, indicating that 4 and 8 μg/mL ISO could significantly reduce the levels of melanin within HaCaT cells.
The involvement of ISO in lysosomal enzyme activity and related proteins
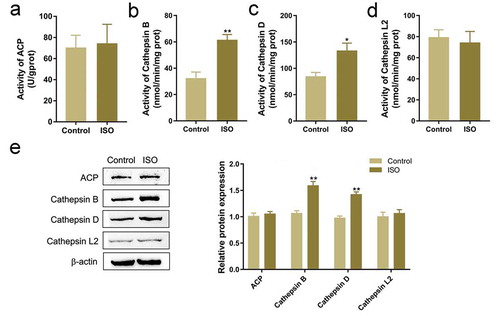
Based on the critical roles of lysosome in melanin degradation [Citation35,Citation36], the effects of ISO on related lysosomal enzymes were examined. We co-incubated HaCaT cells with melanosomes for 48 h, followed by 8 μg/mL ISO treatment for 24 h. Then we evaluated the activity and the protein levels of ACP, Cathepsin B, Cathepsin D, and Cathepsin L2, four representative lysosomal enzymes [Citation23,Citation24]. showed that ISO stimulation significantly enhanced the activity and the protein levels of Cathepsin B, and Cathepsin D but not ACP and Cathepsin L2, indicating that ISO exerts its effects via promoting lysosomal enzyme-dependent melanin degradation; Cathepsin B and Cathepsin D, but not Cathepsin L2, are involved in ISO functions.
Cathepsin B or Cathepsin D silence blocked ISO-induced melanin reduction
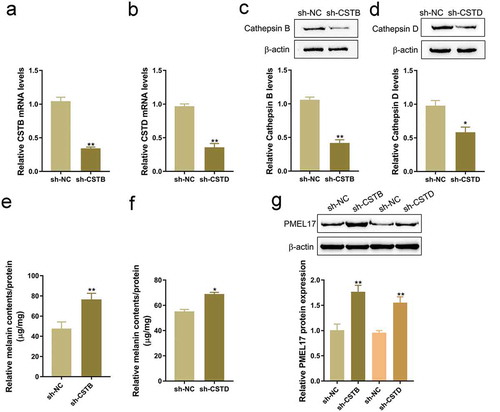
To further investigate whether Cathepsin B and Cathepsin D contribute to ISO functions, next, we transfected sh-CSTB or sh-CSTD to conduct Cathepsin B or Cathepsin D knockdown in HaCaT cells, and performed real-time PCR and Immunoblotting to verify the transfection efficiency (-d). We co-incubated the transfected HaCaT cells with melanosomes for 48 h, and examined melanin contents and PMEL 17 protein levels in HaCaT cells. -f showed that CSTB or CSTD knockdown significantly increased the contents of the melanin in HaCaT cells. Furthermore, CSTB or CSTD knockdown also considerably increased the protein levels of PMEL 17 (), indicating that Cathepsin B and Cathepsin D are involved in ISO-induced melanin reduction.
Selection of miRNAs that might target CSTB and CSTD
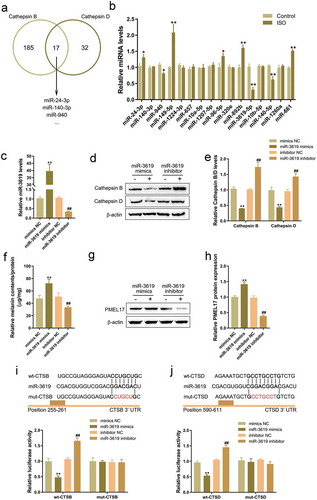
As we have mentioned, miRNAs participate in the melanogenesis by targeting different downstream targets [Citation31,Citation32,Citation37,Citation38]. Here, we employed an online tool to predict miRNAs that might target both CSTB and CSTD; as shown in , a total of 17 miRNAs might target both CSTB and CSTD. Next, we stimulated HaCaT cells with 8 μg/mL ISO for 24 h, then performed real-time PCR to examine 17 miRNA expression. showed that miR-940, miR-3619, and miR-140 expression could be significantly inhibited by ISO treatment, miR-3619 more downregulated. Thus miR-3619 was selected for further experiments.
miR-3619 modulates ISO-induced melanin reduction via targeting CSTB and CSTD
To confirm the function and mechanism of miR-3619 within ISO-induced melanin degradation, we transfected miR-3619 mimics or miR-3619 inhibitor to conduct miR-3619 expression in HaCaT cells, and performed real-time PCR to verify the transfection efficiency (). It has been demonstrated by the online tool that miR-3619 negatively regulated Cathepsin B and Cathepsin D protein levels (-e). Then, we co-incubated miR-3619 mimics- or inhibitor-transfected HaCaT cells with melanosomes for 48 h, followed by the treatment with 8 μg/mL ISO for 24 h. Then we evaluated melanin contents and PMEL17 protein levels. As shown in -g, miR-3619 overexpression significantly increased, while miR-3619 inhibition decreased both melanin contents () and PMEL 17 protein levels (-h), suggesting that miR-3619 could modulate the content of melanin, possibly via targeting CSTB and CSTD.
To validate the predicted bindings between miR-3619 and CSTB/CSTD, the luciferase reporter assays were performed. We constructed wild- and mutant-type CTSB and CTSD 3ʹ-UTR luciferase reporter vectors; 5 or 7 bp within the putative miR-3619 binding site could be mutated to remove the miR-3619 complementarity (-j). We co-transfected these vectors in 293 T cells with miR-3619 mimics or miR-3619 inhibitor, then examined the luciferase activity. Fig.5i-j showed that wild-type reporter vector luciferase activity could be significantly downregulated via the overexpression of miR-3619 whereas upregulated via the inhibition of miR-3619; mutating the putative miR-3619 binding sites could abolish the alterations in the luciferase activity, suggesting that miR-3619 can target CSTB and CSTD in their 3ʹ-UTR to negatively regulate their expression.
Discussion
Herein, we revealed that ISO stimulation significantly reduces the melanin contents and PMEL 17 protein levels; meanwhile, the activity and the protein levels of two critical lysosomal enzymes, Cathepsin B and Cathepsin D, can be significantly increased by ISO treatment. The ISO-induced decrease in the contents of melanin and the protein levels of PMEL 17 are significantly attenuated via CSTB or CSTD knockdown. As predicted by an online tool and confirmed by luciferase reporter assays, miR-3619 can inhibit the expression of CSTB and CSTD via direct binding to their 3ʹ-UTR, therefore affecting ISO-induced reduction of melanin.
Studies have found that ISO significantly affects the proliferation of melanocyte and cancer cells. ISO caused G2/M arrest in B16F10 cells under UVA irradiation, as well as significantly reduced tumor growth and final tumor weight in UVA-irradiated B16F10 melanoma-bearing HRM mice [Citation39]. A series of six furanocoumarins, including ISO, could significantly inhibit the proliferation of A549 (non small cell lung), SK-OV-3 (ovary), SK-MEL-2 (melanoma), XF498 (central nervous system) and HCT-15 (colon) in vitro [Citation40]. More importantly, ISO has been reported to inhibit melanocyte migration, melanin synthesis, and tyrosinase activity [Citation39–Citation42]. Herein, we observed similar inducible effects of ISO upon melanin degradation in HaCaT cells; in the meantime, ISO treatment also induces changes in lysosomal enzyme activity and protein levels, including Cathepsin B and Cathepsin D, indicating that ISO might exert its effects on melanin degradation via a lysosome and lysosomal enzyme-dependent manner.
Lysosomes contain a variety of enzymes that break down biomolecules eaten by a cell. As recently reported, the expression of Cathepsin V (CTSV, also known as Cathepsin L2), a cysteine protease in lysosomes, is greater within light-colored skin compared to that within dark skin [Citation43]. Besides, the degradation of melanosome could be reduced within CTSV knockdown cells [Citation44]. It has been reported that lysosomal proteinases cathepsin B and L activation depends upon Atg5 and Atg7 mediated autophagosome formation [Citation29]. Murase et al also reported that the autophagic pathway, from cytoplasm to lysosomes, plays a crucial role in the melanosomes degradation in keratinocytes [Citation28]. In the present study, the activity and protein levels of Cathepsin B and Cathepsin D, but not ACP and Cathepsin L2 (Cathepsin V) could be significantly altered by ISO treatment. To further confirm the involvement of Cathepsin B and Cathepsin D in ISO treatment on melanin reduction, we conducted CTSB or CTSD knockdown in HaCaT cells to investigate the detailed effects of CTSB or CTSD knockdown on ISO-induced melanin degradation. Similar to that of CTSV knockdown reported previously [Citation44], CTSB or CTSD knockdown increased melanin contents and PMEL 17 protein levels, indicating that CTSB or CTSD knockdown also suppressed ISO-induced melanin reduction. However, the role of autophagy in ISO-induced melanin reduction needs further investigation.
As we have mentioned, miRNAs exert an essential effect on melanin synthesis and degradation. Based on the potent roles of Cathepsin B and Cathepsin D in ISO-induced melanin reduction, we employed online tool to identify miRNAs that might target both CTSB and CTSD to inhibit their expression, therefore affecting ISO-induced melanin reduction; among 17 candidate miRNAs, miR-3619 expression was the most inhibited by ISO treatment. As previously reported, miR-3619 could contribute to autophagy within THP-1 derived macrophages by controlling the expression of Cathepsin S [Citation45]. Herein, miR-3619 can decrease the levels of Cathepsin B and Cathepsin D via direct binding to CSTB and CSTD in their 3ʹ-UTR. Consistent with its suppression on CSTB and CSTD, miR-3619 overexpression significantly increased the protein levels of PMEL 17 and the contents of melanin, indicating that miR-3619 participate in ISO-induced melanin reduction via targeting CSTB and CSTD in a lysosome and lysosomal enzymes-related manner.
In conclusion, we demonstrate a novel, lysosomal enzymes-related mechanism by which ISO induces the reduction of melanin. ISO could be a potent skin-whitening agent, which needs further in vivo and clinical investigation, especially the usage safety
Author Contribution Statement
Bijun Zeng, Kai Li, and Zhibo Yang contributed to experimental design and supervising the whole experimental process; Haizhen Wang and Chang Wang were involved in the experimental conducting; Pan Huang and Yi Pan contributed to the data analysis and manuscript preparation. All the authors read, revised and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Roy S. Melanin, melanogenesis, and vitiligo. Fortschr Chem Org Naturst. 2007;88:131–185. PubMed PMID: 17302180.
- Baraboi VA. Melanin: structure, biosynthesis, biological functions. Ukr Biokhim Zh (1999). 1999 Jul-Aug;71(4):5–14. PubMed PMID: 10791050.
- Griffond B, Baker BI. Cell and molecular cell biology of melanin-concentrating hormone. Int Rev Cytol. 2002;213:233–277. PubMed PMID: 11837894.
- Plonka PM, Grabacka M. Melanin synthesis in microorganisms–biotechnological and medical aspects. Acta Biochim Pol. 2006;53(3):429–443. PubMed PMID: 16951740.
- Jian D, Jiang D, Su J, et al. Diethylstilbestrol enhances melanogenesis via cAMP-PKA-mediating up-regulation of tyrosinase and MITF in mouse B16 melanoma cells. Steroids. 2011 Nov;76(12):1297–1304. PubMed PMID: 21745488.
- Zeng Q, Wang Q, Chen X, et al. Analysis of lncRNAs expression in UVB-induced stress responses of melanocytes. J Dermatol Sci. 2016 Jan;81(1):53–60. PubMed PMID: 26596215.
- Wang X, Liu Y, Chen H, et al. LEF-1 regulates tyrosinase gene transcription in vitro. PLoS One. 2015;10(11):e0143142. PubMed PMID: 26580798; PubMed Central PMCID: PMCPMC4651308.
- Zhang N, Liu ZJ, Li G, et al. Clinical and pathologic features of melanocytic lesion of the central nervous system. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2007 Aug;32(4):713–717. PubMed PMID: 17767073.
- Van Den Bossche K, Naeyaert JM, Lambert J. The quest for the mechanism of melanin transfer. Traffic. 2006 Jul;7(7):769–778. PubMed PMID: 16787393.
- Yang YS, Li N, Deng XM, et al. MC1R–the key gene in mammalian melanin synthesis. Yi Chuan. 2004 Jul;26(4):544–550. PubMed PMID: 15640059.
- Hearing VJ. Determination of melanin synthetic pathways. J Invest Dermatol. 2011 Nov 17;131(E1):E8–E11. PubMed PMID: 22094404.
- Jiang L, Huang J, Lu J, et al. Ganoderma lucidum polysaccharide reduces melanogenesis by inhibiting the paracrine effects of keratinocytes and fibroblasts via IL-6/STAT3/FGF2 pathway. J Cell Physiol. 2019 May 21;234(12):22799–22808. PubMed PMID: 31115052.
- Hu S, Huang J, Pei S, et al. Ganoderma lucidum polysaccharide inhibits UVB-induced melanogenesis by antagonizing cAMP/PKA and ROS/MAPK signaling pathways. J Cell Physiol. 2019 May;234(5):7330–7340. PubMed PMID: 30362532.
- Schraermeyer U, Peters S, Thumann G, et al. Melanin granules of retinal pigment epithelium are connected with the lysosomal degradation pathway. Exp Eye Res. 1999 Feb;68(2):237–245. PubMed PMID: 10068489.
- Yang Z, Zeng B, Pan Y, et al. Autophagy participates in isoliquiritigenin-induced melanin degradation in human epidermal keratinocytes through PI3K/AKT/mTOR signaling. Biomed Pharmacother. 2018 Jan;97:248–254. PubMed PMID: 29091873.
- Boissy RE. Melanosome transfer to and translocation in the keratinocyte. Exp Dermatol. 2003;12(Suppl 2):5–12. PubMed PMID: 14756517.
- Hu DN. Methodology for evaluation of melanin content and production of pigment cells in vitro. Photochem Photobiol. 2008 May-Jun;84(3):645–649. PubMed PMID: 18435617.
- Mammone T, Marenus K, Muizzuddin N, et al. Evidence and utility of melanin degrading enzymes. J Cosmet Sci. 2004 Jan-Feb;55(1):116–117. PubMed PMID: 15037924.
- Ciechanover A. Intracellular protein degradation: from a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Medicina (B Aires). 2010;70(2):105–119. PubMed PMID: 20447892.
- Ferguson SM. Beyond indigestion: emerging roles for lysosome-based signaling in human disease. Curr Opin Cell Biol. 2015 Aug;35:59–68. PubMed PMID: 25950843; PubMed Central PMCID: PMCPMC4529762.
- Lettau M, Kabelitz D, Janssen O. Lysosome-related effector vesicles in T lymphocytes and NK cells. Scand J Immunol. 2015 Sep;82(3):235–243. PubMed PMID: 26118957.
- Hasanagic M, Waheed A, Eissenberg JC. Different pathways to the lysosome: sorting out alternatives. Int Rev Cell Mol Biol. 2015;320:75–101. PubMed PMID: 26614872.
- Ebanks JP, Koshoffer A, Wickett RR, et al. Epidermal keratinocytes from light vs. dark skin exhibit differential degradation of melanosomes. J Invest Dermatol. 2011 Jun;131(6):1226–1233. PubMed PMID: 21326292.
- Ebanks JP, Koshoffer A, Wickett RR, et al. Hydrolytic enzymes of the interfollicular epidermis differ in expression and correlate with the phenotypic difference observed between light and dark skin. J Dermatol. 2013 Jan;40(1):27–33. PubMed PMID: 23088390.
- Wang J, Peng L, Shi M, et al. Spectrum effect relationship and component knock-out in angelica dahurica radix by high performance liquid chromatography-q exactive hybrid quadrupole-orbitrap mass spectrometer. Molecules. 2017 Jul 21;22(7):1231. PubMed PMID: 28754032; PubMed Central PMCID: PMCPMC6152310.
- Cho YH, Kim JH, Park SM, et al. New cosmetic agents for skin whitening from Angelica dahurica. J Cosmet Sci. 2006 Jan-Feb;57(1):11–21. PubMed PMID: 16676120.
- Nerya O, Vaya J, Musa R, et al. Glabrene and isoliquiritigenin as tyrosinase inhibitors from licorice roots. J Agric Food Chem. 2003 Feb 26;51(5):1201–1207. PubMed PMID: 12590456.
- Murase D, Hachiya A,Takano K, et al. Autophagy has a significant role in determining skin color by regulating melanosome degradation in keratinocytes. J Investi Dermatol. 2003;133(10):2416–2424.
- Zhou J, Tan SH, Nicolas V, et al. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res. 2013 Apr;23(4):508–523. PubMed PMID: 23337583; PubMed Central PMCID: PMCPMC3616426. eng.
- Liu B, Zhang J, Yang S, et al. Effect of silencing microRNA-508 by STTM on melanogenesis in alpaca (Vicugna pacos). Gene. 2018 Dec 15;678:343–348. PubMed PMID: 30098430.
- Guo J, Zhang JF, Wang WM, et al. MicroRNA-218 inhibits melanogenesis by directly suppressing microphthalmia-associated transcription factor expression. RNA Biol. 2014;11(6):732–741. PubMed PMID: 24824743; PubMed Central PMCID: PMCPMC4156504.
- Wang P, Zhao Y, Fan R, et al. MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting Sox5 in mouse skin melanocytes. Int J Mol Sci. 2016 Jun 24;17(7):959. PubMed PMID: 27347933; PubMed Central PMCID: PMCPMC4964364.
- Alesiani D, Cicconi R, Mattei M, et al. Cell cycle arrest and differentiation induction by 5,7-dimethoxycoumarin in melanoma cell lines. Int J Oncol. 2008 Feb;32(2):425–434. PubMed PMID: 18202765; eng.
- Du J, Miller AJ, Widlund HR, et al. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am J Pathol. 2003 Jul;163(1):333–343. PubMed PMID: 12819038; PubMed Central PMCID: PMCPMC1868174.
- Park DJ, Sekhon SS, Yoon J, et al. Color reduction of melanin by lysosomal and peroxisomal enzymes isolated from mammalian cells. Mol Cell Biochem. 2016 Feb;413(1–2):119–125. 10.1007/s11010-015-2645-2. PubMed PMID: 26738491.
- Fujita H, Motokawa T, Katagiri T, et al. Inulavosin, a melanogenesis inhibitor, leads to mistargeting of tyrosinase to lysosomes and accelerates its degradation. J Invest Dermatol. 2009 Jun;129(6):1489–1499. PubMed PMID: 19110539.
- Dai X, Rao C, Li H, et al. Regulation of pigmentation by microRNAs: MITF-dependent microRNA-211 targets TGF-beta receptor 2. Pigment Cell Melanoma Res. 2015 Mar;28(2):217–222. PubMed PMID: 25444235.
- Zhao Y, Wang P, Meng J, et al. MicroRNA-27a-3p inhibits melanogenesis in mouse skin melanocytes by targeting Wnt3a. Int J Mol Sci. 2015 May 14;16(5):10921–10933. PubMed PMID: 26006230; PubMed Central PMCID: PMCPMC4463683.
- Kimura Y, Sumiyoshi M, Sakanaka M, et al. In vitro and in vivo antiproliferative effect of a combination of ultraviolet-A and alkoxy furocoumarins isolated from Umbelliferae medicinal plants, in melanoma cells. Photochem Photobiol. 2013 Sep-Oct;89(5):1216–1225. PubMed PMID: 23802687.
- Kim YK, Kim YS, Ryu SY. Antiproliferative effect of furanocoumarins from the root of Angelica dahurica on cultured human tumor cell lines. Phytother Res. 2007 Mar;21(3):288–290. PubMed PMID: 17143927.
- Hultman KA, Scott AW, Johnson SL. Small molecule modifier screen for kit-dependent functions in zebrafish embryonic melanocytes. Zebrafish. 2008 Dec;5(4):279–287. 10.1089/zeb.2008.0542. PubMed PMID: 19133826; PubMed Central PMCID: PMCPMC2757780.
- Fujioka T, Furumi K, Fujii H, et al. Antiproliferative constituents from umbelliferae plants. V. A new furanocoumarin and falcarindiol furanocoumarin ethers from the root of Angelica japonica. Chem Pharm Bull (Tokyo). 1999 Jan;47(1):96–100. PubMed PMID: 9987830.
- Chen N, Seiberg M, Lin CB. Cathepsin L2 levels inversely correlate with skin color. J Invest Dermatol. 2006 Oct;126(10):2345–2347. PubMed PMID: 16728970.
- Homma T, Kageyama S, Nishikawa A, et al. Melanosome degradation in epidermal keratinocytes related to lysosomal protease cathepsin V. Biochem Biophys Res Commun. 2018 Jun 2;500(2):339–343. PubMed PMID: 29654760.
- Pawar K, Sharbati J, Einspanier R, et al. Mycobacterium bovis BCG interferes with miR-3619-5p control of Cathepsin S in the process of autophagy. Front Cell Infect Microbiol. 2016;6:27. PubMed PMID: 27014637; PubMed Central PMCID: PMCPMC4783571.