ABSTRACT
Here, we describe a procedure to fluorescently contrast the nuclear boundary using the lipophilic carbocyanine dye DiI in cultured human cells. Our procedure is simple and is applicable to detect nuclear boundary defects, which may be relevant to studies on nuclear envelope dynamics, micronuclei formation and cancer biology.
Abbreviations
DiI: 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; DiO: 3,3ʹ-dioctadecyloxacarbocyanine perchlorate; NE: nuclear envelope; RanBP2: Ran-binding protein 2/Nucleoporin 358
Fluorescence visualization of nuclear morphology is an important technique in not only basic cell biology, but also in applied medical sciences, because many physiological and pathological features of cells appear to be accompanied by alterations of the nuclear size, number, and shape [Citation1–Citation3]. In nearly all eukaryotic cells, the boundary of the nucleus is the nuclear envelope (NE). NEs comprise a double membrane sheet that includes two closely juxtaposed lipid membrane bilayers referred to as the inner and outer nuclear membranes, as well as proteins that interact with the inner and/or outer membranes, including nuclear pore proteins [Citation2]. While multiple methods to fluorescently outline the NE structure have been well developed, including immunostaining using antibodies against NE-localizing proteins, little attention has been focused on a method(s) that fluorescently contrasts the nuclear boundary via the visualization of the NE lipid membrane structure.
Here, we tested the lipophilic carbocyanine dye DiI [also known as DiIC18(3); 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate] to fluorescently visualize the nuclear boundary of human cervical cancer HeLa cells. DiI is a synthetic compound with two 18-alkyl chains, which is incorporated efficiently into the lipid membrane layer. The dye is weakly fluorescent in aqueous solutions, but emits characteristic bright red fluorescence when its alkyl chains are incorporated into membranes. It is generally used to fluorescently visualize membrane structures in cells, including the endoplasmic reticulum, Golgi apparatus, and plasma membrane, and monitor intracellular membrane structures induced by lipidated proteins [Citation4–Citation7]. Therefore, we expected that the nuclear boundary of HeLa cells could be detected fluorescently, possibly via visualization the NE lipid membrane layer, under the appropriate experimental settings.
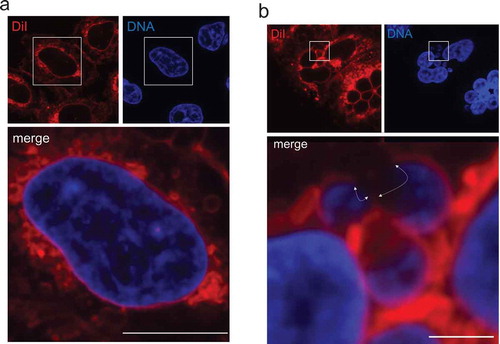
As shown in ), when exponentially growing HeLa cells were exposed to 50 μg/mL DiI for 30 min at 37°C, followed by fixation using 4% paraformaldehyde for 10 min at room temperature, accumulation of a significant amount fluorescent signals was observed not only in the surface, but also in the inside area of cells. Costaining with 1 μg/mL Hoechst 33,342 allowed us to visualize the nuclear boundary which was outlined by the DiI-stained layer. Similar results were obtained when we tested two other cultured cell lines, HuH-7 human hepatocytes and C2C12 mouse myocytes (Figure S1). We suggest that 30–60 min of DiI exposure to cells is optimal for fluorescence detection of the nuclear boundary, because incubation for less than 30 min appears to result in little fluorescence intensity around the nucleus, exhibiting a weakly outlined nuclear boundary (data not shown). We also tested DiO [also known as DiOC18(3); 3,3ʹ-dioctadecyloxacarbocyanine perchlorate], another lipophilic dye which emits green fluorescence, and found that the nuclear boundary could be visualized (Figure S2). While the molecular mechanism by which DiI/DiO is accumulated at the membrane surrounding the nucleus over other ER subdomains remains to be elucidated, our results indicate that multiple lipophilic dyes can be used for visualization of the nuclear boundary in cultured cells. The detailed procedure and reagents used for the staining method are described in the Supplementary information.
Figure 1. Fluorescence visualization of the nuclear boundary using the lipophilic dye DiI. (a) HeLa cells were incubated with DiI, followed by fixation in paraformaldehyde and DNA staining by Hoechst 33,342. DiI and DNA staining patterns are represented at the upper-left and upper-right, respectively. A merged image is shown at the bottom. Bar: 10 μm. (b) After MN were induced pharmacologically in HeLa cells, DiI staining was performed (upper-left). Nuclear DNA was visualized with Hoechst 33,342 (upper-right). A merged image is shown at the bottom. The area exhibiting discontinuous DiI signals around MN are indicated by white arrows. Bar: 2.5 μm. In A and B, all fluorescence images were taken by an FV1200 laser scanning confocal microscope imaging system.
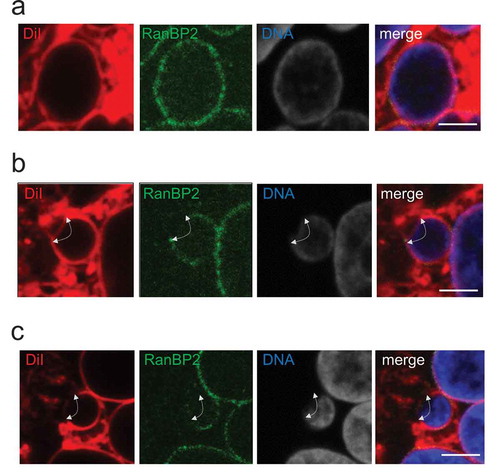
To verify that the nuclear boundary signal outlined by DiI staining possibly represented the NE lipid membrane layer, we performed a colocalization experiment using an antibody recognizing RanBP2/Nup358 that has been demonstrated to be associated with the nuclear pore complex embedded in the NE [Citation8]. As shown in Figure S3, RanBP2 signals appeared to merge with DiI signals, at least in part, along with the Hoechst 33,342-staied nuclear border, predicting that the DiI signal detected around the nuclear boundary may overlap the area including the NE lipid membrane layer. The detailed DiI and antibody costaining procedure is described in the Supplementary information.
To elucidate potential applications of the DiI staining as well as the DiI and antibody costaining procedures, we investigated the nuclear boundary of micronuclei (MN). Because MN are small nuclei sheltering extranuclear chromatin bodies in cells and nuclear boundary defects, such as NE rupture, associated with MN is suspected to trigger chromothripsis, leading to development of cancerous cells [Citation9–Citation12] and references therein], we expected that the DiI staining patterns obtained by our procedures would provide beneficial information to researchers who are interested in studying the linkage between nuclear boundary defects and MN fate, which may be relevant to cancer biology.
When DiI staining was performed using cells in which MN were induced by mitotic slippage inducers paclitaxel and reversine [Citation13,Citation14] and see Supplementary procedures], we detected regions deficient for DiI staining in many MN ()), suggesting the existence of nuclear boundary defects, which might represent discontinued NE due to membrane rupture or altered lipid membrane composition in the local vicinity. Intriguingly, when the DiI and antibody costaining method was carried out in the MN-induced cells, we observed several different patterns of the DiI and RanBP2 signals: i) nuclear boundary, in which both DiI and RanBP2 signals were continuously detected () and S4a), ii) nuclear boundary containing a region deficient for RanBP2 signals, but detectable by DiI staining () and S4b), and iii) nuclear boundary containing a region deficient for both DiI and RanBP2 signals () and S4 c). These results suggested that the DiI staining and the costaining procedures have potential to provide additional information concerning defects associated with MN boundary, possibly representing NE assembly defects.
Figure 2. After MN were induced pharmacologically in HeLa cells, the nuclear boundary was visualized by the DiI and antibody costaining method. Representative images of micronucleus with seemingly intact nuclear boundary, which showed continuous DiI and RanBP2 signals (a and see also S4a), micronucleus with the nuclear boundary region exhibiting continuous DiI signals, but deficient for RanBP2 signals (b and see also S4b), and micronucleus with the nuclear boundary region deficient for both DiI and RanBP2 signals (c and see also S4 c) are shown. In a, b and c, DiI, anti-RanBP2 antibody and DNA staining patterns are represented at the left, middle-left and middle-right, respectively. A merged image is shown at the right. White arrows in b indicate the area exhibiting continuous DiI signals, but discontinuous RanBP2 signals, around the micronucleus. White arrows in c indicate the area deficient for both DiI and RanBP2 signals around the micronucleus. All fluorescence images were taken by an FV1200 laser scanning confocal microscope imaging system. Bar: 2.5 μm.
In summary, we describe a procedure to fluorescently contrast the nuclear boundary using lipophilic dyes, such as DiI and DiO. This procedure is very simple and its combined use with DNA and antibody staining provides additional information concerning defects associated with NE, nuclear boundary and MN.
Author’s contributions
K. Miyazaki and H. Saitoh conceptualized the project, designed experiments. K. Miyazaki performed and analyzed experiments. K.Yano helped microscopic imaging analyses. H. Saitoh wrote the manuscript. All authors helped editing the manuscript.
0414SuppleInfoMiyazaki.pdf
Download PDF (663.4 KB)Acknowledgments
We thank all members of the Saitoh Laboratory for helpful discussions. This work was supported by JP17H01878 and The Novartis Foundation Japan for the Promotion of Science to H.S.
Disclosure statement
The authors declare no conflicts of interest.
Supplementary material
Supplementary data for this article can be accessed here
Additional information
Funding
References
- Edens LJ, White KH, Jevtic P, et al. Nuclear size regulation: from single cells to development and diseasel. Trends Cell Bio. 2013;23:151–159.
- Ungricht R, Kutay U. Mechanisms and functions of nuclear envelope remodeling. Nat Rev Mol Cell Biol. 2017;18:229–245.
- Ibrahim DM, Mundlos S. Three-dimensional chromatin in disease: what holds us together and what drives us apart? Curr Opin Cell Biol. 2019;64:1–9.
- Honig MG, Hume RI. Fluorescent carbocyanine dyes allow living neurons of identified origin to be studied in long-term cultures. J Cell Biol. 1986;103:171–187.
- Terasaki M, Jaffe LA. Organization of the sea urchin egg endoplasmic reticulum and its reorganization at fertilization. J Cell Biol. 1991;114:929–940.
- Haugland RP, Spence MTZ, Johnson ID. Handbook of fluorescent probes and research chemicals. Eugene OR, USA (4849 Pitchford Ave., Eugene 97402): Molecular Probes; 1996.
- Linde N, Stick R. Intranuclear membranes induced by lipidated proteins are derived from the nuclear envelope. Nucleus. 2010;1:343–353.
- Yokoyama N, Hayashi N, Seki T, et al. A giant nucleopore protein that binds Tan/TC4. Nature. 1995;376:184–188.
- Hatch ET, Fischer AH, Deerinck T,J, et al. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60.
- Shah P, Wolf K, Lammerding J. Bursting the bubble – nuclear envelope rupture as a path to genomic instability? Trends Cell Biol. 2017;27:546–555.
- Liu S, Kwon M, Mannino M, et al. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature. 2018;561:551–555.
- Willan J, Cleasby AJ, Flores-Rodriguez N, et al. ESCRT-III is necessary for the integrity of the nuclear envelope in micronuclei but is aberrant at ruptured micronuclear envelopes generating damage. Oncogenesis. 2019;8:29.
- Samwer M, Schneider MWG, Hoefler R, et al. DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell. 2017;170:956–972.
- Miyazaki K, Ichikawa Y, Saitoh N, et al. Three types of nuclear envelope assemblies associated with micronuclei. CellBio. 2020;9:14–28.
