ABSTRACT
The purpose of our article was to probe the influence of GRINA on rectal cancer and how GRINA is regulated in rectal cancer. Based on the public data, we found that GRINA was highly expressed in rectal cancer tissues and related to worse prognosis in rectal cancer patients. MiR-296 was predicted as an upstream regulatory miRNA of GRINA, which was further verified by dual-luciferase reporter assay. Moreover, we revealed that up-regulation/down-regulation of GRINA facilitated/suppressed SW1463/SW837 cell proliferation, migration, and invasion. Rescue assays indicated that the facilitating impact of GRINA on SW1463 cell proliferation and motility was abolished by miR-296 over-expression whilst the suppressing influence of GRINA on SW837 cell proliferation, migration, and invasion was reversed by miR-296 depletion. These consequences indicated that GRINA, which might be regulated by miR-296, acted stimulative important impact on rectal cancer cells, insinuating that GRINA might be a novel potential target for rectal cancer therapy.
GRAPHICAL ABSTRACT
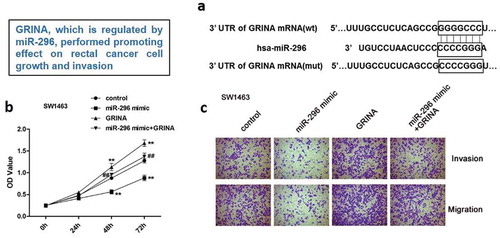
GRINA, which is regulated by miR-296, performed promoting effect on rectal cancer cell growth and invasion.
Rectal cancer is relatively common in men and older persons around the world and is one of the most common causes of death [Citation1,Citation2]. Neoadjuvant chemoradiation is a gold standard treatment for rectal cancer, which followed by total mesorectal excision to ameliorate resectability, anal sphincter reservation, and portion of control [Citation3]. Improvement in early diagnosis, surgical techniques, and radiation therapy can distinctly fortify the survival rate of rectal cancer patients [Citation4]. Due to the peculiar anatomical location of the rectum and the lack of serosa around it, leading to the unsatisfactory of rectal cancer radical resection, and the risk of postoperative recurrence is higher [Citation5]. In the absence of breakthroughs in these therapies for rectal cancer, potential new therapeutic targets need to be identified.
According to some studies, Glutamate Receptor, Ionotropic, N-Methyl D-Aspartate-Associated Protein 1 (GRINA), belongs to the N-Methyl D-Aspartate (NMDA) receptors which are closely related to cancer progression, is located at chromosome 8q24.3 [Citation6–Citation8]. Blockade of NMDA receptors could accelerate the apoptosis of breast cancer [Citation9] or small cell lung cancer cells [Citation10]. Moreover, NMDA receptors are considered as latent therapeutic targets for ovarian cancer [Citation11]. A previous study has revealed that GRINA facilitated cell growth in gastric cancer [Citation6]. However, there is few research about the impact of GRINA on rectal cancer. Besides, the upstream mechanism of how GRINA is regulated is still unclear, which all need further exploration.
During carcinogenesis, researchers have focused on microRNAs (miRNAs) as momentous epigenetic regulators of gene expression through binding to its 3ʹ UTR [Citation12,Citation13]. Emerging studies displayed that dysfunction and abnormal expression of miRNAs acted vital roles in human malignancies (including rectal cancer) pathogenesis and tumorigenicity [Citation14,Citation15]. MiRNAs are a class of small non-coding RNA with the nucleotides length of 18–25, which have been concerned with various cellular processes containing cell proliferation, differentiation, apoptosis, motility, and so on [Citation16,Citation17]. Interestingly, miR-296 was discovered to play crucial influences in multifarious human tumors containing lung cancer [Citation18], colorectal cancer [Citation19], and so on. By targeting S100A4, miR-296 restrains colorectal cancer metastasis [Citation19]. However, the direct role of miR-296 in rectal cancer is not much, and further research is needed.
Herein, we noticed that GRINA was up-regulated in rectal cancer tissues and cell lines. Moreover, the high level of GRINA was related to poor prognosis of patients with rectal cancer. Our data demonstrated that GRINA facilitated rectal cancer cell proliferation, invasion, and migration, which might be modulated by miR-296.
Materials and methods
Analyses of the expression of GRINA and miR-296 utilizing public databases
First, on the basis of the data gained from TCGA database (https://cancergenome.nih.gov/), the expression levels of GRINA were explored, which containing 167 rectal cancer tissues and 10 normal samples. Furthermore, the data acquired from ONCOMINE database (www.oncomine.org), including 65 tumor tissues and 65 normal samples, were employed to ulteriorly analyze the expression of GRINA. Then, overall survival analyses of GRINA were implemented for rectal cancer patients based on TCGA datasets. Basing on the median value of GRINA expression level, patients were fell into high and low expression groups.
Culture of rectal cancer cell lines
Rectal cancer cell lines SW837, SW1463, and the control CRC cell line FHC were acquired from American Type Culture Collection (ATCC, USA) and cultivated in RPMI-1640 supplemented with 10% fetal bovine serum (FBS), 0.1 mg/mL streptomycin, and 100 U/mL penicillin under normal conditions.
Cell transfection
si-con: 5ʹ-CGAACUCACUGGUCUGACC-3ʹ, si-GRINA-1: 5ʹ-GTGCTCTTCATCTTCGCCAT-3ʹ, si-GRINA-2: 5ʹ-CCTGTCTACTCATTGTTGCA-3ʹ, pcDNA3.1, pcDNA3.1-GRINA; miR-296 mimic/inhibitor and their corresponding negative control (NC) were synthesized by GenePharma (Shanghai, China). Then, they were transfected into rectal cancer cells utilizing Lipofectamine 2000 transfection reagent (Invitrogen, USA) according to the manual. After incubation for 24 h, the relative mRNA expression levels were detected by qRT-PCR.
qRT-PCR
Total RNA from rectal cancer cells was extracted utilizing TRIzol reagent (Invitrogen). PrimeScript RT Reagent Kit (Takara) was utilized to synthesize cDNA. Then, the mRNA expression of GRINA was analyzed by qPCR, which was conducted applying an SYBR Premix Ex Taq II (TaKaRa) kit on an ABI 7300 fast real-time PCR system (Thermo Fisher Scientific). GAPDH was regarded as an internal reference. All the primers utilized for qPCR were listed in .
Table 1. The primers used in qRT-PCR.
Western blotting assay
Proteins were isolated by a lysis buffer (containing protease suppressor). The concentration of the proteins was tested by a BCA method. Then, the protein lysates were separated by SDS-PAGE and transferred onto PVDF membranes. After being blocked with 5% defatted milk, the membrane was incubated with primary antibodies overnight at 4°C. Subsequently, the membrane was blotted with peroxidase-conjugated secondary antibodies at room temperature for 1 hour. The signals were tested utilizing an ECL detection kit (GE Healthcare). The quantitative analysis was normalized to GAPDH. All experiments were executed in 3 times.
Cell Counting Kit 8 (CCK8)
To measure cell proliferation, the number of viable cells at 0, 24, 48, 72 h after transfection was evaluated by CCK8 (Dojindo Laboratories, Kumamoto, Japan) following the instruction. The OD450 was tested utilizing a microplate reader. Cell proliferation curve was drawn by Graphpad Prism 7.0.
Transwell assay
Rectal cancer cell invasion and migration capacity were detected by transwell assay (24-well chambers) pre-coating with or without Matrigel (BD Biosciences). The top chamber was seeded with 2 times 105 cells in 500 μL of serum-free medium and the lower chamber was added with the complete medium with 10% FBS. Next day, the non-invading or non-migrating cells on the surface of the top chamber were shifted with a cotton swab. Invaded or migrated cells were fixed with 4% polyoxymethylene for 30 min and stained with 0.1% crystal violet for 10 min at 37°C. Finally, the outcomes were visualized utilizing a light microscope at magnification × 200. Five fields were randomly selected under the microscope, and the number of cells was counted. Each assay was executed in triplicate.
Target miRNAs prediction and survival analysis of miR-296
The miRNA target gene prediction websites miRanda, miRWalk, and TargetScan were employed to predict the upstream regulatory miRNA of GRINA. The common miRNAs were acquired from the intersection miRNAs predicted by three databases and the down-regulated miRNAs in rectal cancer that gained on the basis of TCGA database. The miR-296 expression data (including 162 rectal cancer samples) were acquired from the TCGA database to assess its correlation with clinical outcome. Then, the overall survival analysis of miR-296 was implemented for rectal cancer patients based on TCGA dataset.
Dual-Glo luciferase assay
Cells (1 × 104/well) were plated in 96-well plate and continued to culture for 24 h. To inquiry whether miR-296 could interact with the 3ʹ-UTRs of GRINA, wild type (WT) 3ʹ-UTR of GRINA predicted to interact with miR-296 or the mutant (Mut) GRINA 3ʹ-UTR was amplified and cloned into plasmids. MiR-296 mimic/inhibitor/NC and PGL3-3ʹUTR-GRINA-WT or PGL3-3ʹUTR-GRINA-Mut were co-transfected into rectal cancer cells utilizing Lipofectamine 2000 (Invitrogen). After 24 h transfection, the luciferase activity was evaluated by utilizing the Dual-Glo® Luciferase Assay Kit (Promega, Corporation).
Statistical analysis
Kaplan-Meier approach was utilized to construct survival curves relying on the expression level of GRINA and miR-296, and then log-rank test was employed to examine the survival differences.
All statistical data were analyzed with SPSS 22.0 software and GraphPad Prism 7.0 software (San Diego, CA, USA). Comparison between two groups was determined by Student’s t-test. One-way analysis of variance (ANOVA) followed by a Tukey’s post hoc test was utilized for comparison of 3 or more than 3 groups. Data were presented as the Mean ± SD. P < 0.01 was considered as a statistically significant difference.
Results
High expression of GRINA is discovered in rectal cancer tissues
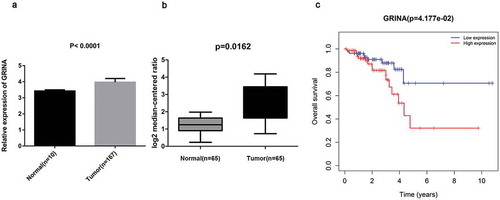
As displayed in ), basing on TCGA database, GRINA expression level was visibly increased in rectal cancer tissues (n = 167), compared with normal tissues (n = 10) (P < 0.0001). Analogously, GRINA expression level was over-expressed in rectal cancer tissues (n = 65) relative to normal tissues (n = 65) (P = 0.0162, )) on the basis of ONCOMINE database. Moreover, high expression of GRINA was related to worse survival (P = 0.04177), indicating that GRINA might act as a prognostic biomarker in rectal cancer patients ()).
Figure 1. GRINA is up-regulated in rectal cancer tissues. (a) Based on TCGA database, GRINA expression level in rectal cancer tissues (n = 167) and normal tissues (n = 10) was analyzed. (b) Based on ONCOMINE database, the expression level of GRINA in rectal cancer tissues (n = 65) and normal tissues (n = 65). (c) Based on TCGA database, the overall survival of GRINA was analyzed by Kaplan–Meier.
Detection of GRINA expression in rectal cancer cell lines
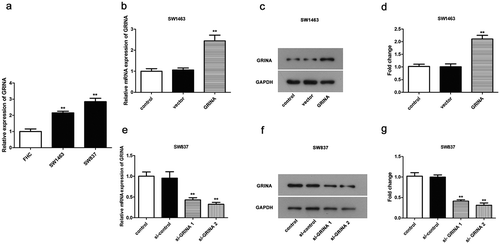
On the purpose of inquiry the expression of GRINA in rectal cancer cell lines, SW1463, SW837 (two rectal cancer cell lines), and FHC (a human rectal normal cell line) were elected. The outcomes revealed that GRINA expression level in rectal cancer cell lines was obviously increased compared to that in FHC cell line (P < 0.01, )). Among these rectal cancer cell lines, SW1463 cell line displayed the lower GRINA expression level than SW837. Hence, cell line SW1463 was selected to over-express GRINA for subsequent assays. Nevertheless, GRINA expression level was higher in SW837 cell line; thus, it was chosen to down-regulate GRINA expression in the following assays. So, pcDNA3.1 and pcDNA3.1-GRINA were transfected into SW1463 cells and si-control, si-GRINA-1, si-GRINA-2 were transfected into SW837 cells. Next, qRT-PCR was carried out to detect the relative mRNA expression levels. Compared to vector, the mRNA and protein expression levels of GRINA were distinctly elevated by pcDNA3.1-GRINA in SW1463 cells (), whilst si-GRINA 1 and si-GRINA 2 weakened the GRINA mRNA and protein level in SW837 cells (). Besides, si-GRINA 2 presented a lower GRINA expression than si-GRINA 1; thus, si-GRINA 2 was utilized in the follow-up experiments.
Figure 2. GRINA is over-expressed in rectal cancer cell lines. (a) GRINA expression in rectal cancer cell lines SW1463, SW837 and normal cell line FHC was detected by qPCR. **P < 0.01 vs. FHC. (b) The mRNA expression of GRINA in SW1463 cells. **P < 0.01 vs. vector. (c, d) The protein levels of GRINA in SW1463 cells. **P < 0.01 vs. vector. (e) The mRNA expression of GRINA in SW837 cells. **P < 0.01 vs. si-control. (f, g) The protein levels of GRINA in SW837 cells. **P < 0.01 vs. si-control.
MiR-296 is an upstream regulatory miRNA of GRINA and is low expressed in human rectal cancer tissues
To probe the mechanism by which GRINA is regulated in rectal cancer, we predicted its potential upstream regulatory miRNAs by miRanda, miRWalk, and TargetScan. The common miRNA miR-296 was acquired from the intersection miRNAs predicted by three databases (61 miRNAs) and the down-regulated miRNAs (1 miRNA) in rectal cancer that gained on the basis of TCGA database ()). Thus, miR-296 was chosen as an upstream regulatory miRNA of GRINA for further research. As illustrated in ), GRINA expression was negatively concerned with miR-296 expression (r = −0.1707, P = 0.0415). The software analysis manifested that miR-296 may identify as a potential upstream regulatory miRNA for the regulation of GRINA ()). Next, the luciferase assay was carried out to further corroborate this result. –e) elucidated that the introduction of miR-296 mimic distinctly weakened whereas miR-296 inhibitor enhanced the luciferase activity of cells containing GRINA 3ʹ-UTR-WT instead of GRINA 3ʹ-UTR-MUT, compared to NC group.
Figure 3. MiR-296 is an upstream regulatory miRNA of GRINA and is low expressed in human rectal cancer tissues. (a) Venn diagram for common miRNAs in miRNAs predict websites miRanda, miRWalk, TargetScan, miRDB (the blue round on the left) and the down-regulated miRNAs in rectal cancer acquired on the basis of TCGA database (the red round on the right). (b) In rectal cancer tissue samples, GRINA expression was negatively related to miR-296 expression. (c) The 3ʹUTR of GRINA including the wide type (WT) or mutant (Mut) binding sites between GRINA and miR-296. The relative luciferase activity of GRINA-WT was reduced/elevated in miR-296 mimic/inhibitor group relative to NC group in SW1463 cells (d) and SW837 cells. (e) **P < 0.01 vs. NC.
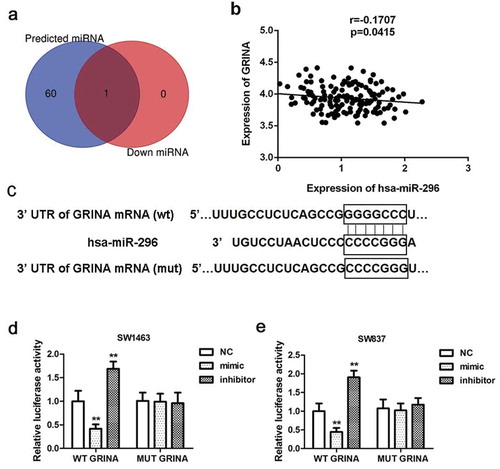
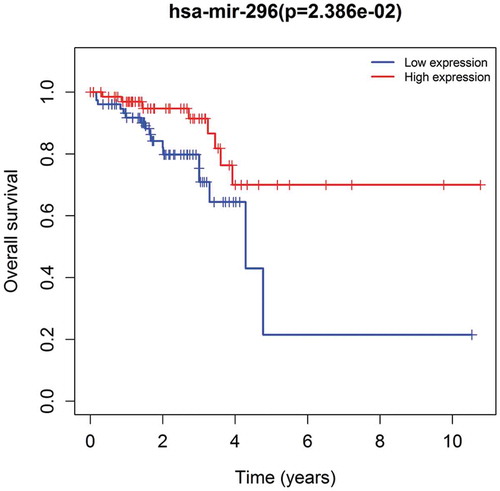
We analyzed the correlation between miR-296 and the clinical prognosis of patients with rectal cancer using the dataset from TCGA database which includes 162 samples. The results showed that low expression of miR-296 was related to poor survival (P = 0.02386), manifesting that miR-296 played a suppressive influence in rectal cancer development ()).
GRINA is negatively modulated by miR-296 in rectal cancer cells
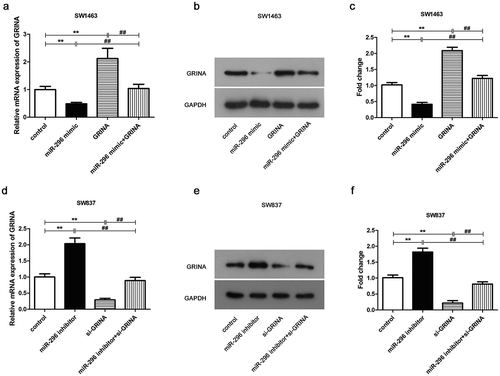
To investigate the effect of miR-296 on GRINA expression, changes in endogenous GRINA expression were inspected in SW1463 and SW837 cells with up or down-regulated miR-296. qPCR and Western blotting analyses disclosed a decline in GRINA expression both at mRNA and protein expression levels in miR-296-overexpressing SW1463 cells, whereas enhanced mRNA and protein expression of GRINA were shown in miR-296-depleted SW837 cells (). These data proofed that GRINA expression was negatively modulated by miR-296 in rectal cancer cells.
Figure 5. GRINA expression was negatively modulated by miR-296 in rectal cancer cell lines. As evaluated by qRT-PCR, mRNA levels of GRINA in miR-296-overexpressing and miR-296 knockout SW1463 (a) and SW837 (d) cells. The protein levels of GRINA in miR-296-overexpressing and miR-296 knockdown SW1463 (b) and SW837 (e) cells. (c) Quantification of (b). (f) Quantification of (e). **P < 0.01 vs. control group. ##P < 0.01 vs. miR-296 mimic + GRINA or miR-296 inhibitor + si-GRINA.
MiR-296/GRINA axis regulates cell growth, migration, and invasion in rectal cancer
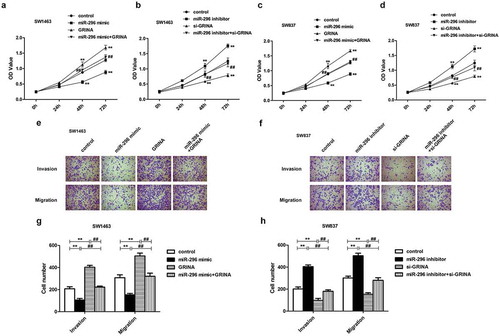
To ulteriorly probe whether miR-296/GRINA axis could modulate rectal cancer cell proliferation, migration, and invasion, we executed CCK8 and transwell assays. We observed that the facilitative impacts of GRINA over-expression on SW1463 cell proliferation, migration, and invasion were partly countervailed by high-expression of miR-296 (). Moreover, the impacts of miR-296/GRINA over-expression on SW837 cell proliferation were also measured using CCK8 assay. Homoplastically, miR-296 mimic can be attenuated the promoting effects of GRINA over-expression on the SW837 cell ()). On the contrary, the suppressive influences of GRINA ablation on SW837 cell proliferation, migration, and invasion were partially reversed by miR-296 depletion (). Besides, miR-296 inhibitor restored the anti-proliferative effects of si-GRINA on SW1463 cell ()). In summary, the above data showed that GRINA, which might be modulated by miR-296, could affect rectal cancer cell proliferation, migration, and invasion.
Figure 6. The influence of GRINA on rectal cancer cell proliferative, invasive and migratory activities was modulated by miR-296. GRINA-overexpression accelerated the proliferative ability of SW1463 (a) and SW837 cells (c), which were weakened by miR-296 mimic. GRINA-knockdown reduced the proliferative ability of SW1463 (b) and SW837 cells (d), which were reversed by miR-296 inhibitor. (e, f) Transwell assays demonstrated that miR-296/GRINA affected the invasive and migratory capacity of SW1463 cells (e) and SW837 cells (f). (g) Quantification of (e). (h) Quantification of (f). **P < 0.01 vs. control group. ##P < 0.01 vs. miR-296 mimic + GRINA or miR-296 inhibitor + si-GRINA.
Discussion
In the present study, we exhibited that GRINA level was over-expressed in rectal cancer tissues and cell lines. Moreover, the high expression of GRINA was concerned with poor prognosis in rectal cancer patients. The accelerating influences of GRINA over-expression on rectal cancer cell proliferation, migration, and invasion were lessened by miR-296 mimic whilst the repressive impacts of GRINA ablation on rectal cancer cell proliferation, migration, and invasion were fortified by miR-296 inhibitor. Our work is the first to report that GRINA may be a tumor accelerator gene in rectal cancer cells which is modulated by miR-296. Overall, miR-296/GRINA may serve as a potential therapeutic target for treating rectal cancer.
GRINA is one member of the NMDA receptors [Citation20]. Studies have demonstrated that NMDA receptors are closely related to cancer progression [Citation21,Citation22]. As reported, ketamine bated colorectal cancer cell migration, which might be through NMDA receptor blockage [Citation23]. In small cell lung cancer, the combination of ifenprodil and NMDA receptor blockers would visibly improve the prognosis of patients [Citation10]. Based on the above reports, we deduced that GRINA may be closely concerned with tumor progression. Xu et al. revealed that GRINA facilitated the proliferation, invasion, and migration of gastric cancer cells [Citation6]. Consistent with previous reports, our study discovered that GRINA was highly expressed in rectal cancer tissues and cell lines. With transfection technology, we verified the effects of GRINA on the phenotypes of rectal cancer cells from both positive and negative aspects. Rectal cancer cell lines SW1463 and SW837 were, respectively, selected to carry out gain- and loss-of-function of GRINA assays, due to their relatively low and high GRINA expression. We considered that this design might better explain the effect of GRINA on rectal cancer cells, and the results indicated that GRINA could accelerate rectal cancer cell proliferation, invasion, and migration.
Through binding to 3ʹ UTR of target genes, miRNAs act as negative modulators of gene expression at the post-transcriptional level [Citation24]. MicroRNAs (miRNAs) are a category of highly conserved non-coding single-stranded RNAs in organisms that have displayed to be positively associated with tumorigenesis, containing cell proliferation, motility, apoptosis, and so on [Citation16,Citation25,Citation26]. Thereinto, miR-296, located at human 20q13.32 gene locus, was found to act important impacts on various human cancers containing glioblastoma [Citation27], lung cancer [Citation18], and colorectal cancer [Citation19]. The study of glioblastoma manifested that miR-296 acted a prohibitive impact by restraining glioblastoma cell stemness [Citation28]. Besides, miR-296 targeted RABL3 to inhibit lung cancer cell proliferation and migration [Citation29]. Through targeting ARRB1, miR-296 repressed cell growth whilst facilitated cell apoptosis of colorectal cancer cells [Citation30]. All these above studies manifested its potential as a cancer-suppressor miRNA in rectal cancer. In our work, we also found that patients with a low level of miR-296 had a worse clinical outcome. Moreover, to better determine the effects of miR-296/GRINA on the rectal cancer cell proliferation, miR-296/GRINA-knockdown and -overexpression experiments were implemented in both SW837 and SW1463 cells. As expected, GRINA-overexpression promoted the two kinds of cell proliferation which can be abated by miR-296 mimic, whereas GRINA-knockdown suppressed these two kinds of cells which can be reversed by miR-296 inhibitor. For the invasion and migration, we also found that the promotive effect of GRINA-overexpression on the SW1463 cells could be weakened by miR-296 mimic, whereas the suppressive effect of GRINA-knockdown on SW837 cells could be reversed by miR-296 inhibitor. All of these findings hinted that GRINA, which might be modulated by miR-296, facilitated rectal cancer cell proliferation, migration, and invasion.
In short, our research is the first study about the impact of GRINA in rectal cancer. Briefly, GRINA was over-expressed in rectal cancer tissues and cell lines. Moreover, high expression of GRINA was associated with poor prognosis in rectal cancer patients. Furthermore, GRINA, which might be regulated by miR-296, accelerated proliferation, invasion, and migration of rectal cancer cells. These consequences offered GRINA might regard as a novel prognostic target for rectal cancer treatment.
Author contributions
Huan Ma and Xianyu Zhang performed the experiment. Na Li and Xiurong Lu analyzed and interpreted the data. Yulei Wei and Na Yuan drafted the article. Guiying Tian designed the study and Shuguang Li critically revised the manuscript. All authors have read and approved the final manuscript.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Herrinton LJ, Altschuler A, McMullen CK, et al. Conversations for providers caring for patients with rectal cancer: comparison of long-term patient-centered outcomes for patients with low rectal cancer facing ostomy or sphincter-sparing surgery. CA Cancer J Clin. 2016;66:387–397.
- Grosek J, Kosir JA, Novak J, et al. Validation of the Slovenian version of the low anterior resection syndrome score for rectal cancer patients after surgery. Zdr Varst. 2019;58:148–154.
- Dayde D, Tanaka I, Jain R, et al. Predictive and prognostic molecular biomarkers for response to neoadjuvant chemoradiation in rectal cancer. Int J Mol Sci. 2017;18(3):573.
- Grosek J, Velenik V, Edhemovic I, et al. The influence of the distal resection margin length on local recurrence and long- term survival in patients with rectal cancer after chemoradiotherapy and sphincter- preserving rectal resection. Radiol Oncol. 2017;51:169–177.
- Zhang YX, Liu HY, Jiang B, et al. Stepwise neoadjuvant chemoradiotherapy in the management of mid-low locally advanced rectal cancer. Eur J Surg Oncol. 2020;46(3):410–414. .
- Xu DH, Li Q, Hu H, et al. Transmembrane protein GRINA modulates aerobic glycolysis and promotes tumor progression in gastric cancer. J Exp Clin Cancer Res. 2018;37:308.
- Ahmed H, Haider A, Varisco J, et al. Structure-affinity relationships of 2,3,4,5-tetrahydro-1H-3-benzazepine and 6,7,8,9-tetrahydro-5H-benzo[7]annulen-7-amine Analogues and the Discovery of a Radiofluorinated 2,3,4,5-tetrahydro-1H-3-benzazepine congener for imaging GluN2B subunit-containing N-methyl-d-aspartate receptors. J Med Chem. 2019;62:9450–9470.
- Mahadeen A, Mullaguri N, George P, et al. Anti-N-methyl-D-aspartate encephalitis concomitantly with tall-cell variant papillary thyroid carcinoma. Cureus. 2019;11:e5415.
- Morelli MB, Amantini C, Nabissi M, et al. Role of the NMDA receptor in the antitumor activity of chiral 1,4-dioxane ligands in MCF-7 and SKBR3 breast cancer cells. ACS Med Chem Lett. 2019;10:511–516.
- North WG, Liu F, Dragnev KH, et al. Small-cell lung cancer growth inhibition: synergism between NMDA receptor blockade and chemotherapy. Clin Pharmacol. 2019;11:15–23.
- North WG, Liu F, Tian R, et al. NMDA receptors are expressed in human ovarian cancer tissues and human ovarian cancer cell lines. Clin Pharmacol. 2015;7:111–117.
- Ma Y, Ma M, Ma L, et al. Downregulation of miR-552 in hepatocellular carcinoma inhibits cell migration and invasion, and promotes cell apoptosis via RUNX3. Exp Ther Med. 2019;18(5):3829–3836. .
- Xu L, Wang P, Feng X, et al. SETD3 is regulated by a couple of microRNAs and plays opposing roles in proliferation and metastasis of hepatocellular carcinoma. Clin Sci. 2019;133:2085–2105.
- Machackova T, Prochazka V, Kala Z, et al. Translational potential of MicroRNAs for preoperative staging and prediction of chemoradiotherapy response in rectal cancer. Cancers (Basel). 2019;11:1545.
- Bjornetro T, Redalen KR, Meltzer S, et al. An experimental strategy unveiling exosomal microRNAs 486-5p, 181a-5p and 30d-5p from hypoxic tumour cells as circulating indicators of high-risk rectal cancer. J Extracell Vesicles. 2019;8:1567219.
- Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15(1):1–19.
- Zhang C, Wan J, Long F, et al. Identification and validation of microRNAs and their targets expressed in osteosarcoma. Oncol Lett. 2019;18:5628–5636.
- Dong Y, Xu T, Zhong S, et al. Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296-5p. Life Sci. 2019;239:116984.
- He Z, Yu L, Luo S, et al. miR-296 inhibits the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100A4. BMC Cancer. 2017;17:140.
- Cihakova D, Eaton WW, Talor MV, et al. Gut permeability and mimicry of the Glutamate Ionotropic Receptor NMDA type Subunit Associated with protein 1 (GRINA) as potential mechanisms related to a subgroup of people with schizophrenia with elevated antigliadin antibodies (AGA IgG). Schizophr Res. 2019;208:414–419.
- Deutsch SI, Tang AH, Burket JA, et al. NMDA receptors on the surface of cancer cells: target for chemotherapy? Biomed Pharmacother. 2014;68:493–496.
- Dai WL, Yan B, Jiang N, et al. Simultaneous inhibition of NMDA and mGlu1/5 receptors by levo-corydalmine in rat spinal cord attenuates bone cancer pain. Int J Cancer. 2017;141:805–815.
- Duan W, Hu J, Liu Y. Ketamine inhibits colorectal cancer cells malignant potential via blockage of NMDA receptor. Exp Mol Pathol. 2019;107:171–178.
- Tyagi S, Sharma S, Ganie SA, et al. Plant microRNAs: biogenesis, gene silencing, web-based analysis tools and their use as molecular markers. 3 Biotech. 2019;9(11):413. .
- Wang Y, Mu L, Huang M. MicroRNA‑195 suppresses rectal cancer growth and metastasis via regulation of the PI3K/AKT signaling pathway. Mol Med Rep. 2019;20:4449–4458.
- Shirmohamadi M, Eghbali E, Najjary S, et al. Regulatory mechanisms of microRNAs in colorectal cancer and colorectal cancer stem cells. J Cell Physiol. 2020;235:776–789.
- Lee H, Hwang SJ, Kim HR, et al. Neurofibromatosis 2 (NF2) controls the invasiveness of glioblastoma through YAP-dependent expression of CYR61/CCN1 and miR-296-3p. Biochim Biophys Acta. 2016;1859:599–611.
- Lopez-Bertoni H, Lal B, Michelson N, et al. Epigenetic modulation of a miR-296-5p: HMGA1axis regulates Sox2 expression and glioblastoma stem cells. Oncogene. 2016;35:4903–4913.
- Ge T, Wu HC, Zhou YY, et al. MiR-296-3p may affect the proliferation and migration of non-small cell lung cancer cells via regulating RABL3. Eur Rev Med Pharmacol Sci. 2019;23(13):5823–5830. .
- Zhang Z, Zhong X, Xiao Y, et al. MicroRNA-296 inhibits colorectal cancer cell growth and enhances apoptosis by targeting ARRB1-mediated AKT activation. Oncol Rep. 2019;41:619–629.