?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Feature selection is a data processing method which aims to select effective feature subsets from original features. Feature selection based on evolutionary computation (EC) algorithms can often achieve better classification performance because of their global search ability. However, feature selection methods using EC cannot get rid of invalid features effectively. A small number of invalid features still exist till the termination of the algorithms. In this paper, a feature selection method using genetic programming (GP) combined with feature ranking (FRFS) is proposed. It is assumed that the more the original features appear in the GP individuals' terminal nodes, the more valuable these features are. To further decrease the number of selected features, FRFS using a multi-criteria fitness function which is named as MFRFS is investigated. Experiments on 15 datasets show that FRFS can obtain higher classification performance with smaller number of features compared with the feature selection method without feature ranking. MFRFS further reduces the number of features while maintaining the classification performance compared with FRFS. Comparisons with five benchmark techniques show that MFRFS can achieve better classification performance.
1. Introduction
Feature selection (Sreeja, Citation2019; Too & Abdullah, Citation2020) is to select effective feature subsets from high-dimensional original features, which is one of the key issues for machine learning. High-quality features play a key role in building an efficient model, and irrelevant or redundant features may cause difficulties (Xue et al., Citation2013). Therefore, feature selection methods have been extensively applied to solve practical classification tasks (Espejo et al., Citation2010; Liang et al., Citation2017; Loughran et al., Citation2017; Mei et al., Citation2017; Patel & Upadhyay, Citation2020). Recently, Gaining Sharing Knowledge based Algorithm (GSK) (Mohamed et al., Citation2020) was proposed for feature selection and achieved good performance (Agrawal, Ganesh, & Mohamed, Citation2021a, Citation2021b; Agrawal, Ganesh, Oliva, et al., Citation2022). Evolutionary computation (EC) algorithms (Al-Sahaf et al., Citation2019; Hancer, Citation2019) have been widely used for feature selection due to their global search ability (Ma & Gao, Citation2020b; Xue et al., Citation2014). EC algorithms are population based, which need to initialise individuals randomly. Sometimes, the genetic operators of genetic algorithm (GA) and genetic programming (GP), or the updating strategy of particle swarm optimisation (PSO) (Nagra et al., Citation2020; Too et al., Citation2021), are not sufficient to delete invalid features. So, a small number of invalid features still exist till the termination of the algorithms.
GP can be used for feature selection due to its global search ability and is proved to achieve good classification performance. However, with the evolution of GP, a large number of individuals with the same best fitness are generated. GP only outputs the best individual, which may lose some good individuals. Moreover, the generated features in the output individual may still contain redundant features.
Feature ranking (Ahmed et al., Citation2014; Friedlander et al., Citation2011; Neshatian, Citation2010) is the ranking of original features based on specific evaluation criteria, which is usually a step of feature selection. It is employed to find out which features or feature sets are more important. So, this paper proposes a GP-based feature selection method combined with feature ranking (FRFS), which considers the large number of best individuals generated during the evolution of GP instead of only one best individual. A correlation-based evaluation criterion is used as the fitness function. It is assumed that the more the original features appear in the GP individuals' terminal nodes, the more valuable these features are. Therefore, the individuals with the best fitness are stored. The occurrence times of features appearing in the individuals' terminal nodes are counted and the top β features are selected as the candidate feature subset. To decrease the number of selected features while maintaining the classification performance, another feature selection method that combines FRFS with a multi-criteria fitness function is investigated. The motivation of this algorithm is to set higher fitness values for the individuals with the same correlation value in the case of smaller feature number.
The overall objective of this paper is to propose a feature selection algorithm that combines feature ranking with a multi-criteria fitness function and verify the improvement of the algorithm by feature ranking and the dimensionality reduction effect of multi-criteria fitness function. To achieve the overall objective, the following three objectives will be investigated.
Objective 1. Propose a feature selection method combined with feature ranking using GP (FRFS) and verify whether it can achieve better classification performance with smaller number of features than using feature selection only (FS).
Objective 2. Investigate a feature selection method that combines FRFS with a multi-criteria fitness function which is named as MFRFS, and verify whether the multi-criteria fitness function can reduce the number of features of FRFS while maintaining the classification performance.
Objective 3. Investigate whether our proposed MFRFS can achieve better classification performance than five benchmark techniques, including ReliefF feature selection method (Kira & Rendell, Citation1992), linear forward feature selection method (LFS) (Gutlein et al., Citation2009) and three GSK related methods (Agrawal, Ganesh, & Mohamed, Citation2021b; Agrawal, Ganesh, Oliva, et al., Citation2022).
The rest of this paper is arranged as follows. Section 2 outlines the background information of this paper. Section 3 describes in detail the three GP-based feature selection methods. Section 4 shows the experimental design. Section 5 presents the experimental results and discussion. Section 6 is the conclusion and future work.
2. Related works
2.1. Genetic programming (GP)
GP is one of the evolutionary computation algorithms (Koza, Citation1992; Koza et al., Citation1999). It is very similar to genetic algorithm (GA). The main difference between GP and GA is the representation of individuals. Due to GP's flexible representation methods, GP can be used to construct high-level features (Ma & Gao, Citation2020a, Citation2020b), construct classifiers (Muni et al., Citation2006; Neshatian, Citation2010; Neshatian & Zhang, Citation2011) and solve practical industrial problems (Bi et al., Citation2021a, Citation2021b; Peng et al., Citation2020). The commonly used representation method of GP is based on tree structure. The terminal nodes (constants and variables) are randomly selected from a terminal set, and the function (mathematical and logical operators) is randomly selected from a function set to constitute internal nodes. GP randomly initialises the first generation of individuals and evaluates each individual using a fitness function. Genetic operators including selection, crossover and mutation are then performed to produce the next generation's population. This step is iterated until the termination criterion is reached. Then GP outputs the optimal individual. For feature selection, the terminal nodes contain the features selected by GP.
2.2. Correlation
The correlation-based feature evaluation criterion is proposed by Hall (Citation1999), which takes into account both the correlation between features and classes and the correlation between features. This evaluation criterion is proved to be effective to select low redundancy and high discrimination feature subsets (Hall & Smith, Citation1999; Ma & Gao, Citation2020b), and is adopted as the evaluation criterion in this paper. The following is the formula for calculating the correlation of a feature subset.
(1)
(1) where
is the correlation value of a set S containing k features,
denotes the average correlation between each feature and class, and
denotes the average correlation between each feature pair. They are calculated by formula (2).
and
are the information entropy of X and Y respectively,
is the conditional entropy.
(2)
(2)
2.3. Feature selection methods based on GP
Due to GP's flexible representation methods, GP can be used for feature selection. Recent studies have focused on feature selection methods using evolutionary algorithms (Canuto & Nascimento, Citation2012; Papa et al., Citation2017; Ribeiro et al., Citation2012; Tan et al., Citation2008) due to their global search ability. According to whether the classification algorithm is involved in the fitness function, feature selection is divided into filter-based (Davis et al., Citation2006; Lin et al., Citation2008; Neshatian & Zhang, Citation2009b, Citation2012; Purohit et al., Citation2010; Ribeiro et al., Citation2012) and wrapper-based (Hunt et al., Citation2012). The filter-based feature selection methods use information measures such as mutual information (Vergara & Estévez, Citation2014), information gain (Neshatian & Zhang, Citation2012), consistency and correlation (Hall, Citation1999; Neshatian & Zhang, Citation2009c) as the evaluation criterion, and need less running time than wrapper-based methods. Moreover, the models produced by filter-based methods are more general. So, filter-based feature selection is investigated further in this paper. The fundamental research goal of feature selection is to find the feature subset with the best classification performance and fewer features.
Lin et al. (Citation2008) constructed a classifier using layered genetic programming, which had the characteristic of feature selection and feature extraction. Neshatian and Zhang (Citation2012) combined information entropy and conditional entropy to evaluate the correlation between feature sets and classes for feature selection.
Purohit et al. (Citation2010) proposed a GP classifier construction method with feature selection and investigated a new crossover operator to discover the best crossover site. Davis et al. (Citation2006) proposed a two-stage feature selection method. In the first stage, a feature subset is selected using GP and GP classifiers are evolved using the selected features. Neshatian and Zhang (Citation2009b) proposed a filter-based multiple-objective feature selection method for binary classification task.
GP has flexible representation ability. In general, the GP-based feature selection method takes original features as the terminal set and terminal nodes as the selected features. However, some researchers have researched other representations. Hunt et al. (Citation2012) developed GP-based hyper-heuristics for adding and removing features. Each individual generated by GP is a series of operations on the feature set. Ribeiro et al. (Citation2012) used four feature selection criteria (information gain, chi-square, correlation and odds ration) as the terminal set of GP and three set operations (intersection, union and difference) as the function set. Viegas et al. (Citation2018) proposed a GP-based feature selection method for skewed dataset and validated the method on four high-dimensional datasets. Papa et al. (Citation2017) used binary strings as individual's terminal nodes, where 1 means selecting a certain feature and 0 means without selecting. AND, OR and XOR are regarded as non-terminal nodes. The output of the individual is also a binary string, which contains the selected features.
2.4. Feature ranking methods based on GP
The goal of feature ranking is to show the importance of the original features under certain evaluation criteria and is often used as a basis of feature selection.
Neshatian (Citation2010) proposed a feature ranking method based on GP, which used GP to evolve many weak classifiers and preserved the optimal weak classifiers. The score of each feature is proportional to the fitness value of the weak classifier that contains it. This method needs to run a large number of GPs, which is time-consuming.
Friedlander et al. (Citation2011) proposed a feature ranking method based on weight vectors. GP updates the weights for each feature at the end of each generation of evolution. After 10 generations of evolution, terminal nodes are no longer randomly selected during the generation of new subtrees in mutation operation, but are selected according to different probabilities based on feature weights.
Ahmed et al. (Citation2014) proposed a two-stage GP-based classification method which first ranked the original features then constructed classifiers. Neshatian and Zhang (Citation2009a) constructed GP programs to measure the goodness of feature subsets using a virtual program structure and an evaluation function.
At present, there have been many researches on GP-based feature selection methods, but few research focus on the large amount of evolutionary information generated during the evolution process. And some researchers have begun to use GP to rank the features (Friedlander et al., Citation2011; Neshatian, Citation2010), which indicates that the frequency of features appearing in terminal nodes can show the importance of features. Based on this motivation, some issues should be investigated on whether feature ranking can help quantify the importance of features and improve the classification performance, and whether a multi-criteria fitness function can help to further explore this area select more effective features.
3. Method
In this paper, our proposed methods are based on standard GP, and correlation is chosen as the fitness function. The fundamental purpose of this paper is to investigate whether the feature selection method based on correlation evaluation criteria using GP can effectively get rid of invalid features, and whether feature ranking and a multi-criteria fitness function can improve the classification performance. We have different variants of algorithms to verify the research objectives. FS denotes the feature selection method using GP. FRFS denotes the feature selection method combined with feature ranking. MFRFS denotes the algorithm that combines FRFS with a multi-criteria fitness function. The details of the algorithms are described below.
3.1. Feature selection using GP (FS)
This algorithm employs GP as the search algorithm and correlation as the evaluation criterion. Because the smaller the fitness of GP, the better. The fitness function of FS is shown in Formula (Equation3(3)
(3) ).
(3)
(3) where Correlation is the correlation value of the selected features and classes.
The output of FS is an individual that contains the optimal feature subset. The terminal nodes are the selected features. The parameter settings of GP are shown in Section 4.1.
3.2. FS combined with feature ranking (FRFS)
FS only considers the best individual generated by GP and does not make full use of the information during GP's evolutionary process. With the convergence of GP, there are a large number of individuals with the same best fitness. Our next step, feature ranking, is based on the assumption that the more frequent features occur in these individuals, the more valuable they are. All the individuals with the same best fitness during the evolution process after the jth generation are preserved. The occurrence of features in these individuals' terminal nodes are counted, and the features are ranked according to their occurrence. Our experiments found that GP starts to converge after 30 iterations, so the parameter j is set to 30.
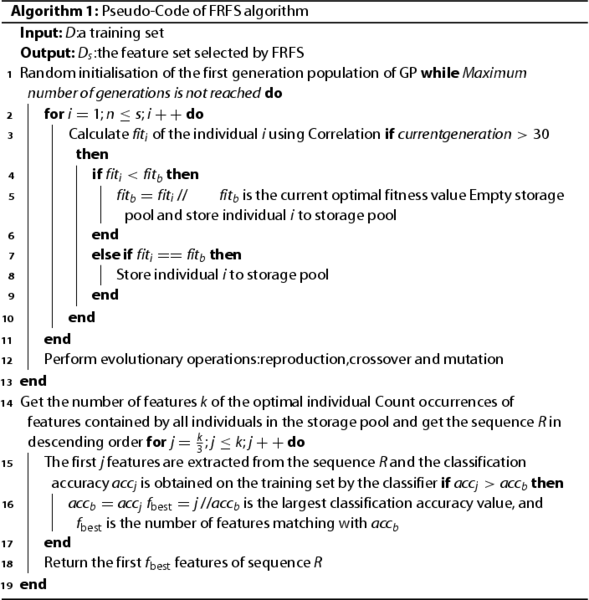
Suppose the number of features selected by FS is k. Our goal is to select the top features among top k features according to ranking. The Pseudo-Code of FRFS is shown in Algorithm 1. To determine how many features selected from k features are the best, first
and the
features are tested on the classification algorithms. Then the rest
features are added into
feature subset and tested one by one according to their ranking until the classification accuracy begins to decline. Then, the top
features are the selected features. In the paper, the parameter n is set to 3, that is, top one-third of k features are selected into
feature subset. We started with top
features instead of the top feature is to improve the search efficiency. We set the value of n to 3 based on our experiments.
3.3. FRFS combined with a multi-criteria fitness function (MFRFS)
In the process of GP evolution, we found that two individuals with the same fitness value are not necessarily the same individuals. To give priority to individuals with the same fitness value but less features, we use the multi-criteria fitness function as Formula 4. The method FRFS using the multi-criteria fitness function is named as MFRFS.
(4)
(4) where
is the number of features in an individual and α is the penalty coefficient between Correlation and
. The parameter α is discussed in Section 5.4.
4. Experimental design
4.1. Datasets and parameter settings
To verify the effectiveness of our proposed feature selection methods, 15 datasets are collected from UCI machine learning repository (Dheeru & Karra Taniskidou, Citation2017). The details of the datasets are shown in Table . In the table, #features indicates the number of original features, #instances indicates the number of instances and #classes indicates the number of class labels. The datasets have different number of classes and features, and are representative classification problems.
Table 1. Description of the fifteen datasets.
A random split of 7:3 is used and 70% of the dataset is used as the training set and 30% as the testing set (Ma & Gao, Citation2020b; Xue et al., Citation2013). The training set is used to select and rank the original features. The testing set is used to evaluate the classification performance of selected and ranked features by 10-fold cross-validation. To avoid stochastic characteristics of GP, 30 experiments are done independently (Ma & Teng, Citation2019). The average of classification accuracy and number of features of 30 different experiments are obtained.
K-nearest Neighbours (KNN, K = 5), C4.5 decision tree and Naive Bayes (NB) are used to evaluate the performance of the proposed methods. The ECJ platform (Luke, Citation2017) is used to run GP. The parameter settings of GP are shown in Table , which are the commonly used parameters of GP (Ma & Gao, Citation2020b; Ma & Teng, Citation2019). The function set is composed of four arithmetic operators, the terminal set is the original features. The population size is set to 500, initial GP experimental results indicate that this population size can evolve good solutions. The generations are set to 50 and the maximum tree depth is 17 to restrict irrelevant and redundant features can also reduce the depth of GP programs. Mutation probability and crossover probability are 10% and 90% respectively, which balance exploration and exploitation during the evolution. The parameter α in Formula 3 is set as 0.01, which is discussed in Section 5.4.
Table 2. Parameter setting of GP.
4.2. Benchmark techniques
To verify the effectiveness of the proposed MFRFS methods, we use five existing feature selection methods as benchmarks to compare, including ReliefF feature selection method (Kira & Rendell, Citation1992), linear forward feature selection method (LFS) (Gutlein et al., Citation2009) and three GSK-related methods (Agrawal, Ganesh, & Mohamed, Citation2021b; Agrawal, Ganesh, Oliva, et al., Citation2022).
4.2.1. ReliefF
ReliefF (Kira & Rendell, Citation1992) is an extension of Relief algorithm. Relief can only be used to solve binary classification problems. Both Relief and ReliefF are feature weighting algorithms, which assign different weights to features according to the correlation between each feature and class, and features with a weight less than a certain threshold will be deleted. The weight of a feature is proportional to its classification ability. ReliefF can be applied to solve multi-class classification problems. First, one sample is randomly selected from the dataset, and then k nearest neighbour samples are selected from all classes. Then, the weight of all features is calculated, and the operation is repeated n times. Finally, the final weight of each feature is obtained and the features are ranked according to their weights. To facilitate comparisons with the proposed MFRFS, the same number of features as MFRFS is selected on all datasets by ReliefF.
4.2.2. LFS
LFS (Gutlein et al., Citation2009) starts from an empty set and adds features one by one. It restricts the number of features in each step of the forward selection. When the evaluation criteria show that its performance remains unchanged or decreases after adding a feature, LFS will be terminated. LFS is proved to be faster than standard forward selection and can obtain good results (Gutlein et al., Citation2009). To compare with the proposed MFRFS method fairly, Correlation is also selected as the evaluation criteria.
4.2.3. GSK related methods
Gaining-sharing knowledge-based optimisation algorithm (GSK) (Agrawal, Ganesh, & Mohamed, Citation2021b) is a human-related metaheuristic algorithm. The basic principle of GSK is to simulate the process of human gaining and sharing knowledge in the life span. Recently, GSK has been used for feature selection and proved to be effective, so three GSK-related methods including BGSK (Agrawal, Ganesh, & Mohamed, Citation2021b), pBGSK (Agrawal, Ganesh, & Mohamed, Citation2021b) and PbGSK-V (Agrawal, Ganesh, Oliva, et al., Citation2022) were selected as benchmarks for comparison.
5. Experimental results and conclusion
To verify the effectiveness of our proposed methods, four experiments are done. (1) FRFS is compared with two baselines, i.e. original features (F0) and FS, to verify whether FRFS can get higher classification accuracy with fewer features. (2) MFRFS is compared with FRFS to verify whether the multi-criteria fitness function used by MFRFS can maintain the classification performance and reduce the number of features. (3) MFRFS is compared with five benchmarks, i.e.ReliefF, LFS, BGSK, pBGSK and PbGSK-V, to verify whether MFRFS can achieve better classification performance in terms of accuracy and number of features. (4) The parameter α used in MFRFS is discussed to determine an appropriate parameter setting.
To justify the significance of various feature selection methods, the performance differences between paired feature selection methods are shown on the boxplots in Figures and . The post-hoc analysis for Friedman test (Hollander & Wolfe, Citation1999) abbreviated as Post-Friedman test using different colours is also marked on the boxplots. If the Post-Friedman test achieves or
, the box plots are marked green or yellow respectively, which shows that the performance differences between paired feature processing methods are significantly different or borderline significantly different. Figures and show whether one of the paired methods is significantly better than the other.
Figure 1. The boxplots of performance differences between FRFS and two baselines marked by Post-Friedman-test: (a) KNN, (b) C4.5, (c) NB.
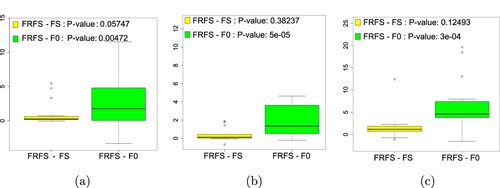
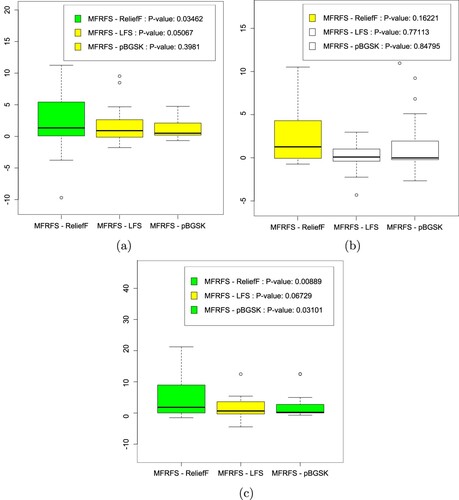
Figure 2. The box plots of performance differences between MFRFS and three benchmarks marked by Post-Friedman test: (a) KNN, (b) C4.5, (c) NB.
5.1. Comparison between FRFS and F0, FS
In this section, the experiments between FRFS and F0, FS were done and the experimental results are shown in Table . In the table, A-KNN, A-C4.5 and A-NB represent the average classification accuracy of 30 independent experiments using KNN, C4.5 and NB respectively. F-KNN, F-C4.5 and F-NB denote the average number of selected features of 30 independent experiments using KNN, C4.5 and NB respectively.
Table 3. The experimental results of FRFS, F0 and FS.
The box plots of performance differences between FRFS and two baselines marked by Post-Friedman test are shown in Figure . From the Friedman significance test, we can see that there are significant differences between FRFS and two baselines. FRFS is significantly better than F0 in all learning algorithms and is borderline significantly better than FS in all learning algorithms.
As shown in Table , FRFS can achieve smaller number of features than F0 and FS in all learning algorithms. FRFS can achieve better classification accuracy than F0 and FS in C4.5 on 12 out of 15 datasets, can achieve better classification accuracy than F0 and FS in NB on 11 out of 15 datasets, and can achieve better classification accuracy than F0 and FS in KNN on 10 out of 15 datasets.
FS can select several features which have stronger discrimination ability than original features on datasets such as audiology, planning-relax, seismic-bumps, statlog-image-segmentation and thoracic-surgery. However, comparing with FS, FRFS can further reduce the number of features and maintain the classification accuracy, which shows that the feature subset selected by FS is not optimal. For example, on a high-dimensional dataset Lymphoma, FS not only selects about 20 features from original 4026 features but also improves the classification accuracy. However, FRFS further selects about 10 from 20 features selected by FS and improves the classification accuracy. We can get the same results on robot-failure-lp4 dataset and the number of features selected by FRFS has been reduced to nearly 50% comparing with FS.
The experiments in this section confirm our assumption, that is, the more the original features appear in the GP individuals' terminal nodes, the more valuable these features are. Based on this assumption, feature ranking can help feature selection method further reduce the number of features while maintaining the classification accuracy.
5.2. Comparison between FRFS and MFRFS
In this section, MFRFS is compared with FRFS. The experimental results are shown in Table . As shown in Table , MFRFS can further reduce the number of features comparing with FRFS and can maintain or even improve the classification accuracy in C4.5, KNN and NB learning algorithms on 12, 9 and 8 datasets respectively.
Table 4. The experimental results of FRFS and MFRFS.
MFRFS further reduces the number of features by more than 50% on arrhythmia dataset. MFRFS can not only select one-third of the number of features but also improve the classification accuracy comparing with FRFS on Lymphoma, especially in NB learning algorithm. As shown in the experimental results, one feature is obtained by MFRFS on climate-simulation-craches, planning-relax and thoracic-surgery datasets, and is enough to obtain equal or higher classification accuracy than FRFS.
The experimental results show that the multi-criteria fitness function can restrict redundant features as GP's terminal nodes and can achieve the purpose of decreasing the number of selected features while maintaining the classification performance. However, appropriate parameter α in Formula 4 should be set and the effect of parameter α is discussed in Section 5.4.
5.3. Comparison between MFRFS and benchmarks
In this section, MFRFS is compared with five benchmark techniques, i.e. ReliefF, LFS, BGSK, pBGSK and PbGSK-V. To make a fair comparison with our proposed method, in GSK related methods, the parameter MAXNFE is set to 25,000, the number of generation is set to 500 when the dimension < = 20, and the number of generation is set to 250 when the dimension > 20. The experimental results are shown in Table . The box plots of performance differences between MFRFS and benchmarks are shown in Figure .
Table 5. Experimental results of different feature selection methods.
5.3.1. Comparison between MFRFS and ReliefF
Like MFRFS, ReliefF also needs to rank the features. In the experiment of ReliefF, we select the same number of features as MFRFS to verify whether MFRFS can achieve higher classification accuracy when the two methods take the same number of features.
On statlog-image-segmentation dataset, both MFRFS and ReliefF select three features, but the accuracy of MFRFS is 10% higher than that of ReliefF. MFRFS and ReliefF can achieve comparable classification accuracy with only one feature on climate-simulation-craches, seismic-bumps, thoracic-surgery and planning-relax datasets. On arrhythmia, hepatitis, robot-failure-lp4, robot-failure-lp5, spambase and statlog-heart datasets, MFRFS can achieve better classification accuracy with same number of features than ReliefF. From the experimental results in Table , we can see that MFRFS is more robust than ReliefF. The box plots marked by Post-Friedman test in Figure show that MFRFS is significantly better than ReliefF in KNN and NB learning algorithms, and is borderline significantly better than ReliefF in C4.5 learning algorithm. In general, the experimental results show that MFRFS can obtain better classification results than ReliefF.
5.3.2. Comparison between MFRFS and LFS
The comparison results between MFRFS and LFS are shown in Table . As shown in Table , in general, LFS can achieve comparable or better classification accuracy than MFRFS. The box plots marked by Post-Friedman test in Figure show that MFRFS is borderline significantly better than LFS in KNN and NB learning algorithms, however, LFS selects more features than MFRFS.
On arrhythmia and audiology datasets, the number of features selected by LFS is nearly four times that of MFRFS. However, on audiology dataset, LFS does not obtain a better classification accuracy with more features than MFRFS. On robot-failure-lp4 and robot-failure-lp5 datasets, features selected by LFS are nearly six times that of MFRFS. On the Lymphoma dataset, LFS even uses more than 20 times features that of MFRFS, and when NB is used as the learning algorithm, the accuracy of LFS is 10% less than that of MFRFS. On other datasets, MFRFS can select smaller number of features and achieve comparable or better classification accuracy than LFS. The experimental results show that LFS's dimensionality reduction performance is weak. The features selected by LFS still contains irrelevant and redundant features, which confirm that some invalid features are not helpful for classification. Our proposed MFRFS can select more effective features on the premise of maintaining the classification accuracy.
5.3.3. Comparison between MFRFS and GSK related methods (BGSK, pBGSK, PbGSK-V
 )
)
Compared with BGSK, MFRFS selects a smaller feature subset and achieves better classification performance on most cases in all learning algorithms. On the higher-dimensional dataset Lymphoma, BGSK cannot reduce the dimension of features better, and select more than 1300 features. We obtain similar experimental results on pBGSK, and in most cases, a larger feature subset is selected, especially on the Lymphoma dataset. The box plots marked by Post-Friedman test in Figure show that MFRFS is borderline significantly better than pBGSK in KNN learning algorithm and significantly better than pBGSK in NB learning algorithm.
As shown in Table , PbGSK-V selects minimum number of features among the six methods. However, this method cannot achieve good classification performance, especially on audiology, robot-failure-lp5, statlog-image-segmentation and spambase datasets. Only an average of less than 2 features is selected by PbGSK-V
in those datasets, but the classification accuracy is very low. Compared with MFRFS, PbGSK-V
reduces the number of features, but in most cases the overall classification accuracy of MFRFS is better than PbGSK-V
.
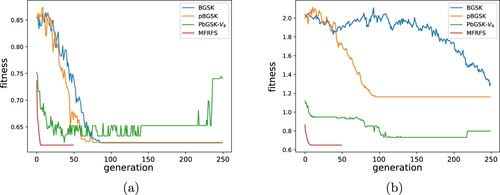
Figure shows the convergence behaviours of four EC-based methods on arrhythmia and spambase datasets. As shown in Figure , MFRFS starts to converge in about 10 generations and can obtain good solutions. The GSK related methods need more generations to converge.
5.4. The parameter setting of α in MFRFS
The parameter α in Formula 4 is used to restrict redundant and irrelevant features as GP's terminal nodes and further reduce the number of selected features. In Section 5.2, we have shown that MFRFS can further reduce the number of features comparing with FRFS. To show the impact of parameter α on experimental results and set an appropriate parameter, different values of parameter α are experimented and the experimental results are shown in Table . As shown in Table , the number of features selected by MFRFS is inversely proportional to the value of α, which means that the smaller the value of α, the more the number of features selected.
Table 6. Experimental results of different α values.
When , the classification performance of MFRFS is similar to FRFS, and the parameter α has little effect on classification performance. When
, it has a certain degree of dimensionality reduction effect. When
, it can be seen from Table that the overall performance is the best. When
, the number of features further decreases, but the average classification accuracy starts to decline to some extent. When
and 0.1, the number of features is obviously reduced a lot, but the accuracy at this time is also declined a lot.
Therefore, the parameter α has a great impact on the classification accuracy. If the parameter α is too small, it cannot achieve the purpose of dimensionality reduction comparing with FRFS. If the parameter α is too large, it will remove effective features and reduce the classification accuracy. Through the experiments, meets the purpose of decreasing the number of selected features while maintaining the classification performance.
5.5. Further discussion
In this paper, FRFS, a feature selection method combined with feature ranking using GP, is proposed to select smaller number of features comparing with feature selection method only (FS). Based on a multi-criteria fitness function, MFRFS is proposed to obtain smaller number of features comparing with FRFS. Here are some further discussions.
(1) Evolutionary computation algorithms will produce a large number of individuals with the best fitness. If only one of the best individuals is used for feature selection, some good individuals may be lost. In this paper, GP individuals after the 30th generation with the best fitness are preserved. The features are ranked according to their occurrence times in these individuals. Experiments show that feature ranking can help select more effective features.
To further demonstrate how feature ranking works, we take the arrhythmia dataset as an example. The results of feature ranking are shown below. A total of 12 best features were selected. The values before the “–” represent the index of the feature, and the values after the “–” represent the number of times the feature occurs in all the best individuals.
F14-60244 F250-59799 F109-58235 F182-42527 F212-38822 F195-38505
F241-37687 F7-31955 F208-26438 F38-24976 F97-23970 F10-21086
We can see that feature F14 is probably the best feature, and feature F250 is the second best, and so on. The purpose of feature ranking is to select the best feature set from the top 12 features.
According to the method described in Section 3.2, the classification accuracy on different number of top ranked features on training set and testing set in feature subset are shown in Table . For training set, we can see that the classification accuracy first increases and then decreases with the increase of the number of top features. When the number of top features is 9, 10 and 9 for KNN, C4.5 and NB, the classification accuracy is the highest respectively.
Table 7. Classification accuracy of different number of top features.
For the testing set, we also obtain the classification accuracies of the corresponding ranking features for KNN, C4.5 and NB learning algorithms respectively in Table , and feature ranking can achieve highest classification accuracy on corresponding ranking features in C4.5 and NB learning algorithms.
(2) Feature ranking is based on the assumption that the more the original features appear in the GP individuals' terminal nodes, the more valuable these features are. Experiments prove that the assumption is correct. GP can be used to quantify the importance of features and select more important feature sets.
(3) MFRFS can restrict the number of irrelevant and redundant features in the GP's terminal nodes by a multi-criteria fitness function. By adjusting the parameter α, MFRFS can achieve a smaller number of features while maintaining the classification accuracy compared with FRFS.
(4) In general, MFRFS can achieve better classification performance than the five benchmarks. However, MFRFS needs to store the individuals with the best fitness, rank the features according to the occurrence times of features appearing in the GP individuals' terminal nodes and search smaller number of features with higher classification performance from the ranked features. Therefore MFRFS has no advantages in time consumption and computational complexity, especially on datasets with more features. In practical applications, we need to find a balance between classification performance and computational complexity.
(5) Feature selection is a key data preprocessing process, which can solve “the curse of dimensionality” problems. Our proposed MFRFS can greatly reduce the dimension of features and obtain a better classification performance. The most important thing is that we can identify which features are more important through the feature ranking method in this paper. In industrial applications, our methods may greatly reduce the model's training time and improve the classification performance.
6. Conclusion and future work
This paper proposes a feature selection method with feature ranking (FRFS) using GP, which ranks the features according to the number of their occurrence in the individuals with the best fitness. Based on FRFS, to further reduce the number of selected features, another feature selection method using a multi-criteria fitness function (MFRFS) is proposed. Experiments on 15 datasets show that FRFS can achieve better classification performance and obtain smaller number of features than FS. Compared with FRFS, MFRFS can further remove redundant and irrelevant features, and select smaller number of features while maintaining the classification performance. Compared with other five existing feature selection methods, MFRFS can achieve better classification performance.
Feature selection is important to build an efficient classification model, and feature ranking has been proved to be effective for feature selection. The fitness function of our methods in this paper are based on Correlation. It is necessary to further investigate whether our methods are effective to other evaluation criteria. Moreover, the dimension of the selected datasets is relatively low. In the future, feature ranking methods on high dimensional datasets will be investigated.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Agrawal, P., Ganesh, T., & Mohamed, A. W. (2021a). Chaotic gaining sharing knowledge-based optimization algorithm: An improved metaheuristic algorithm for feature selection. Soft Computing, 25(14), 9505–9528. https://doi.org/10.1007/s00500-021-05874-3
- Agrawal, P., Ganesh, T., & Mohamed, A. W. (2021b). A novel binary gaining–sharing knowledge-based optimization algorithm for feature selection. Neural Computing and Applications, 33(11), 5989–6008. https://doi.org/10.1007/s00521-020-05375-8
- Agrawal, P., Ganesh, T., Oliva, D., & Mohamed, A. W. (2022). S-shaped and V-shaped gaining-sharing knowledge-based algorithm for feature selection. Applied Intelligence, 52 (1), 81–112. https://doi.org/10.1007/s10489-021-02233-5
- Ahmed, S., Zhang, M., & Peng, L. (2014). Improving feature ranking for biomarker discovery in proteomics mass spectrometry data using genetic programming. Connection Science, 26(3), 215–243. https://doi.org/10.1080/09540091.2014.906388
- Al-Sahaf, H., Bi, Y., Chen, Q., Lensen, A., Mei, Y., Sun, Y., & Zhang, M. (2019). A survey on evolutionary machine learning. Journal of the Royal Society of New Zealand, 49(2), 205–228. https://doi.org/10.1080/03036758.2019.1609052
- Bi, Y., Xue, B., & Zhang, M. (2021a). Genetic programming for image classification: An automated approach to feature learning. Springer Nature.
- Bi, Y., Xue, B., & Zhang, M. (2021b). Multi-objective genetic programming for feature learning in face recognition. Applied Soft Computing, 103(4), 107152. https://doi.org/10.1016/j.asoc.2021.107152
- Canuto, A. M. P., & Nascimento, D. S. C. (2012). A genetic-based approach to features selection for ensembles using a hybrid and adaptive fitness function. The 2012 International Joint Conference on Neural Networks (IJCNN), (pp.1-8), Brisbane, QLD, Australia, June 2012.
- Davis, R. A., Charlton, A. J., Oehlschlager, S., & Wilson, J. C. (2006). Novel feature selection method for genetic programming using metabolomic 1H NMR data. Chemometrics and Intelligent Laboratory Systems, 81(1), 50–59. https://doi.org/10.1016/j.chemolab.2005.09.006
- Dheeru, D., & Karra Taniskidou, E. (2017). UCI machine learning repository. University of California, Irvine, School of Information and Computer Sciences
- Espejo, P. G., Ventura, S., & Herrera, F. (2010). A survey on the application of genetic programming to classification. IEEE Transactions on Systems, Man, and Cybernetics, Part C (Applications and Reviews), 40(2), 121–144. https://doi.org/10.1109/TSMCC.2009.2033566
- Friedlander, A., Neshatian, K., & Zhang, M. (2011). Meta-learning and feature ranking using genetic programming for classification: Variable terminal weighting. IEEE Congress on Evolutionary Computation, (pp. 5–8), New Orleans, LA, USA, June 2011.
- Gutlein, M., Frank, E., Hall, M., & Karwath, A. (2009). Large-scale attribute selection using wrappers. IEEE symposium on computational intelligence and data mining, (pp. 332–339), Nashville, TN, USA, May 2009.
- Hall, M. (1999). Correlation-based feature selection for machine learning (Unpublished doctoral dissertation). The University of Waikato.
- Hall, M., & Smith, L. (1999). Feature selection for machine learning: Comparing a correlation-based filter approach to the wrapper. Twelfth International Florida Artificial Intelligence Research Society Conference, (pp. 235–239), Orlando, Florida, USA, May 1999.
- Hancer, E. (2019). Fuzzy kernel feature selection with multi-objective differential evolution algorithm. Connection Science, 31(4), 323–341. https://doi.org/10.1080/09540091.2019.1639624
- Hollander, M., & Wolfe, D. A. (1999). Nonparametric statistical methods (2nd ed.). Wiley-Interscience.
- Hunt, R., Neshatian, K., & Zhang, M. (2012). A genetic programming approach to hyper-heuristic feature selection. The 9th international conference on Simulated Evolution and Learning, (pp. 320–330), Berlin, Heidelberg, December 2012.
- Kira, K., & Rendell, L. A. (1992). The feature selection problem: Traditional methods and a new algorithm. The tenth national conference on artificial intelligence, (pp. 129–134), San Jose California, July 1992.
- Koza, J. R. (1992). Genetic programming: On the programming of computers by means of natural selection. MIT Press.
- Koza, J. R., Andre, D., Bennett, F. H., & Keane, M. A. (1999). Genetic programming III: Darwinian invention and problem solving. IEEE Transactions on Evolutionary Computation, 7(4), 451–453. https://doi.org/10.1162/evco.1999.7.4.451
- Liang, Y., Zhang, M., & Browne, W. N. (2017). Image feature selection using genetic programming for figure-ground segmentation. Engineering Applications of Artificial Intelligence, 62(12), 96–108. https://doi.org/10.1016/j.engappai.2017.03.009
- Lin, J. Y., Ke, H. R., Chien, B. C., & Yang, W. P. (2008). Classifier design with feature selection and feature extraction using layered genetic programming. Expert Systems with Applications, 34(2), 1384–1393. https://doi.org/10.1016/j.eswa.2007.01.006
- Loughran, R., Agapitos, A., Kattan, A., Brabazon, A., & O'Neill, M. (2017). Feature selection for speaker verification using genetic programming. Evolutionary Intelligence, 10(1–2), 1–21. https://doi.org/10.1007/s12065-016-0150-5
- Luke, S.. (2017). ECJ then and now. Genetic and Evolutionary Computation Conference Companion Pages, (pp.1223–1230), Berlin Germany, July 2017.
- Ma, J., & Gao, X. (2020a). Designing genetic programming classifiers with feature selection and feature construction. Applied Soft Computing, 97(4), 106826. https://doi.org/10.1016/j.asoc.2020.106826
- Ma, J., & Gao, X. (2020b). A filter-based feature construction and feature selection approach for classification using genetic programming. Knowledge-Based Systems, 196(6), 105806. https://doi.org/10.1016/j.knosys.2020.105806
- Ma, J., & Teng, G. (2019). A hybrid multiple feature construction approach using genetic programming. Applied Soft Computing, 80(2), 687–699. https://doi.org/10.1016/j.asoc.2019.04.039
- Mei, Y., Nguyen, S., Xue, B., & Zhang, M. (2017). An efficient feature selection algorithm for evolving job shop scheduling rules with genetic programming. IEEE Transactions on Emerging Topics in Computational Intelligence, 1(5), 339–353. https://doi.org/10.1109/TETCI.2017.2743758
- Mohamed, A. W., Hadi, A. A., & Mohamed, A. K. (2020). Gaining-sharing knowledge based algorithm for solving optimization problems: A novel nature-inspired algorithm. International Journal of Machine Learning and Cybernetics, 11(7), 1501–1529. https://doi.org/10.1007/s13042-019-01053-x
- Muni, D. P., Pal, N. R., & Das, J. (2006). Genetic programming for simultaneous feature selection and classifier design. IEEE Transactions on Systems, Man and Cybernetics, Part B (Cybernetics), 36(1), 106–117. https://doi.org/10.1109/TSMCB.2005.854499
- Nagra, A. A., Han, F., Ling, Q. H., Abubaker, M., Ahmad, F., Mehta, S., & Apasiba, A. T. (2020). Hybrid self-inertia weight adaptive particle swarm optimisation with local search using C4.5 decision tree classifier for feature selection problems. Connection Science, 32(1), 16–36. https://doi.org/10.1080/09540091.2019.1609419
- Neshatian, K. (2010). Feature manipulation with genetic programming (Unpublished doctoral dissertation). Victoria University of Wellington.
- Neshatian, K., & Zhang, M. (2009a). Genetic programming for feature subset ranking in binary classification problems. The 12th European Conference on Genetic Programming, (pp. 121–132), Berlin, Heidelberg, April 2009.
- Neshatian, K., & Zhang, M. (2009b). Pareto front feature selection: Using genetic programming to explore feature space. The 11th annual genetic and evolutionary computation conference, (pp. 1027–1034), Montreal, Québec, Canada, July 2009.
- Neshatian, K., & Zhang, M. (2009c). Unsupervised elimination of redundant features using genetic programming. Australasian joint conference on artificial intelligence, (pp. 432–442), Berlin, Heidelberg, November 2009.
- Neshatian, K., & Zhang, M. (2011). Using genetic programming for context-sensitive feature scoring in classification problems. Connection Science, 23(3), 183–207. https://doi.org/10.1080/09540091.2011.630065
- Neshatian, K., & Zhang, M. (2012). Improving relevance measures using genetic programming. European conference on genetic programming, (pp. 97–108), Berlin, Heidelberg, April 2012.
- Papa, J. P., Rosa, G. H., & Papa, L. P. (2017). A binary-constrained geometric semantic genetic programming for feature selection purposes. Pattern Recognition Letters, 100(24), 59–66. https://doi.org/10.1016/j.patrec.2017.10.002
- Patel, S. P., & Upadhyay, S. H. (2020). Euclidean distance based feature ranking and subset selection for bearing fault diagnosis. Expert Systems with Applications, 154(8), 113400. https://doi.org/10.1016/j.eswa.2020.113400
- Peng, B., Wan, S., Bi, Y., Xue, B., & Zhang, M. (2020). Automatic feature extraction and construction using genetic programming for rotating machinery fault diagnosis. IEEE Transactions on Cybernetics, (99), 1–15. https://doi.org/10.1109/TCYB.2020.3032945
- Purohit, A., Chaudhari, N., & Tiwari, A. (2010). Construction of classifier with feature selection based on genetic programming. In Proceedings of the IEEE congress on evolutionary computation.
- Ribeiro, I., Andrade, G., Viegas, F., Madeira, D., Rocha, L., Salles, T., & Gonçalves, M. (2012). Aggressive and effective feature selection using genetic programming. In Proceedings of IEEE congress on evolutionary computation.
- Sreeja, N. K. (2019). A weighted pattern matching approach for classification of imbalanced data with a fireworks-based algorithm for feature selection. Connection Science, 31(2), 143–168. https://doi.org/10.1080/09540091.2018.1512558
- Tan, F., Fu, X., Zhang, Y., & Bourgeois, A. G. (2008). A genetic algorithm-based method for feature subset selection. Soft Computing, 12(2), 111–120. https://doi.org/10.1007/s00500-007-0193-8
- Too, J., & Abdullah, A. R. (2020). Binary atom search optimisation approaches for feature selection. Connection Science, 32(4), 406–430. https://doi.org/10.1080/09540091.2020.1741515
- Too, J., Sadiq, A. S., & Mirjalili, S. M. (2021). A conditional opposition-based particle swarm optimisation for feature selection. Connection Science, 77(3), 1–23. https://doi.org/10.1080/09540091.2021.2002266
- Vergara, J. R., & Estévez, P. A. (2014). A review of feature selection methods based on mutual information. Neural Computing and Applications, 24(1), 175–186. https://doi.org/10.1007/s00521-013-1368-0
- Viegas, F., Rocha, L., Gonçalves, M., Mourão, F., Sá, G., Salles, T., & Sandin, I. (2018). A genetic programming approach for feature selection in highly dimensional skewed data. Neurocomputing, 273(3), 554–569. https://doi.org/10.1016/j.neucom.2017.08.050
- Xue, B., Zhang, M., & Browne, W. (2013). Particle swarm optimization for feature selection in classification: A multi-objective approach. IEEE Transactions on Cybernetics, 43(6), 1656–1671. https://doi.org/10.1109/TSMCB.2012.2227469
- Xue, B., Zhang, M., & Browne, W. N. (2014). Particle swarm optimisation for feature selection in classification: Novel initialisation and updating mechanisms. Applied Soft Computing, 18(C), 261–276. https://doi.org/10.1016/j.asoc.2013.09.018