Abstract
Deoxynivalenol (DON), or vomitoxin, is a mycotoxin of the trichothecene group produced by Fusarium species. The present work described construction of a phage display library based on variable domain of heavy chain of HCAbs (VHHs) and panning for binders of DON. Two pair PCR primers designed based on the VHHs conserved domains were used for cloning coding sequences of complementary DNA (cDNA). After total RNA extraction, semi-nested PCR, subcloning and electro transformation, the primary library was generated containing 1.9×107 independent clones. Phage display library was rescued by infection of the primary library with helper phage KM13, and the titre of the library was 5.6×1012 CFU/ml. Three clones with specific DON-binding affinity were obtained after three rounds of panning. Since the library was derived from non-immune animal, it has potential to be used for panning binders of other mycotoxins or low molecular weight haptens. The binders with specific affinity provided new recourses for developing detection methods or tools of mycotoxins in foodstuffs.
Introduction
Deoxynivalenol (DON), or vomitoxin, is a mycotoxin of the trichothecene group produced by Fusarium species. DON commonly contaminates cereal-based foods (Pestka & Smolinski, Citation2005) and is the most common of the trichothecenes detected in cereal crops (Lombaert, Citation2002).
Immunoassay of DON in food stuffs has been developed with different kinds of antibodies, such as polyclonal antibodies (Abouzied, Azcona-Olivera, Yoshizawa, & Pestka, Citation1993), monoclonal antibodies (Yoshizawa et al., Citation2004), egg yolk antibodies (Schneider, Pichler, & Krska, Citation2000), single-chain antibodies (Wang et al., Citation2007) and single-domain antibody fragments (Doyle et al., Citation2008). Furthermore, antibodies with specific DON-binding affinity can be applied in immunoaffinity chromatography (Klinglmayr, Nobauer, Razzazi-Fazeli, & Cichna-Markl, Citation2010) or other sample clean-up methods (Bohm, Cichna-Markl, Brenn-Struckhofova, & Razzazi-Fazeli, Citation2008) based on the specific binding affinity between antibodies and DON.
Using recombinant DNA technology, collections of billions of gene fragment- or cDNA-encoded (poly)peptides can be expressed on the surface of filamentous phages. This phage display technology, coupled with in vitro selection, enables rapid identification and optimisation of proteins based on their structural or functional properties (Bratkovic, Citation2010). One of the major advantages of phage display technology of antibody fragments compared with standard hybridoma technology is that the generation of specific binders to a particular antigen can be performed within weeks (Carmen & Jermutus, Citation2002). The sequences of the binders can be easily obtained, providing convenience for affinity maturation or expression. There are many successful examples of isolating binders for proteins or haptens by phage display technology (e.g., Daly et al., Citation2002; Du, Wu, Zhang, Dong, & Wang, Citation2010).
Hamers-Casterman et al. demonstrated the existence in camelids of functional heavy chain IgGs (heavy-chain antibodies, HCAbs) that are devoid of light chains. The antigen-binding site of HCAbs is comprised in one single domain, referred to as variable domain of heavy chain of HCAbs (VHHs). These findings open new perspectives in the engineering of antibodies (Hamers-Casterman et al., Citation1993).
In the present study, a phage display library was constructed using peripheral blood lymphocytes (PBL) from non-immunised alpacas as a source of variable domain coding sequences. DON affinity binders were isolated and characterised, providing new reagents for developing detection methods for DON in foodstuffs. Additionally, the library has potential to be used for isolating binders of other mycotoxins or low molecular weight haptens.
Materials and methods
Preparation of alpaca lymphocytes
Blood was obtained from the jugular vein of two male non-immunised alpacas (Lama pacos). White blood cells were isolated from about 15 ml of citrated blood of each animal. PBL was partially purified by Ficoll-1.077 (Sangon, Shanghai, China) gradient centrifugation and stored in RNAiso (TAKARA, Dalian, China) at −80°C.
Library construction
Total RNA extraction from PBL was performed using an RNAiso total RNA isolation reagent (TAKARA). The quality and quantity of total RNA were tested by agarose gel electrophoresis and UV spectrophotometer at 260 nm and 280 nm according to the manufacturer's protocol. The library was constructed according to Maass, Sepulveda, Pernthaner, and Shoemaker (2007) with following modifications. First-strand complementary DNA (cDNA) was synthesised with oligo dT primers or random hexamer primers using PrimeScript™ RT-PCR Kit (TAKARA). The cDNA served as a template for the first PCR (PCR1) using PrimeSTAR HS DNA polymerase (TAKARA) with two pair primers (AlpVh-LD and AlpVHH-R1, AlpVh-LD and AlpVHH-R2). The primers used were: AlpVh-LD (5′-TTGGTCCTGGCTGC-3′) which anneals to the leader sequence of the heavy chain immunoglobulin, AlpVHH-R1 (5′-CGAGTGCGGCCGCGGGG TCTTCGC TGTGGTGCG-3′) which anneals to the short hinge coding region and AlpVHH-R2 (5′-CGAGTGCGGCCGC TTGTGGTTTTGGTGTCTTGGG-3′) which anneals to the long hinge coding region. The first PCR (PCR1) protocol consisted of an initial denaturation step at 98°C for 15 s followed by 35 cycles of 98°C for 10 s, 50°C for 20 s and 72°C for 40 s, and a final step extension at 72°C for 10 min. The products of PCR1 were purified via agarose electrophoresis and used as templates for the second PCR (PCR2). PCR2 was performed using a semi-nested degenerate primer AlpVh-F1 (5′-TCGCGGCCCAGCCGGCCATGGCCCAGKTGCAGCTCGTGGAGTCNGG NGG-3′), which anneals to the framework 1, and AlpVHH-R1 or AlpVHH-R2, respectively. The PCR2 reaction was firstly cycled 5 times (98°C 10 s, 50°C 20 s and 72°C 40 s) and then 30 times (98°C 10 s, 68°C 40 s) followed by a final step extension at 72°C for 10 min. The PCR2 products were pooled and purified by phenol-chloroform extraction and ethanol precipitated.
After digestion with Sfi I and Not I (TAKARA), the purified second PCR products were then loaded on a 1.2% agarose gel. The band between 250 bp and 600 bp were purified using a gel extraction kit (TAKARA) and ligated into similarly digested phagemid pHEN1 (Kong, Liu, Yang, & Ma, Citation2006). The ligation product were incubated at 65°C for 10 min to inactivate T4 DNA ligase and purified by phenol-chloroform extraction following ethanol precipitation. The ligated DNA was electroporated into freshly prepared Escherichia coli TG1 electrocompetent cells. Transformants were scraped off the plates with Laurie Bertani (LB) medium supplemented with 30% glycerol, then pooled and stored as 0.5 ml aliquots at –80°C. The library size was estimated by serial dilutions of transformation products in LB plates supplemented with 0.1 mg/ml ampicillin. Diversity of the library was analysed by colony PCR. Randomly picked clones were amplified using the universal primers pHEN-R and M13-R (5′-GCCCCATTCAGATCCTCTTC-3′ and 5′-AGCGGATAACAATTTCACACAGGA-3′). The helper phage KM13 was used for library rescue. Titration, purification and infection of KM13 were performed using standard methods (Sambrook & Russell, Citation2001).
Library panning
The phage library was subjected to three rounds of panning on 96-well strip plates (Corning, Lowell, MA). DON was conjugated to modified bovine serum albumin (MBSA) according to Deng, You and Xu (Citation2007). DON-MBSA was coated (100 µg/ml in PBS, 100 µl/well) overnight at 4°C. Blank control wells were filled with phosphate buffered saline (PBS, pH 7.4) in each round of panning. Blocking buffer (3%, w/v BSA in PBS) was used for panning rounds 1 and 3. In panning rounds 2, the blocking buffer was switched to ovalbumin (OVA, 2%, w/v in PBS) for limiting non-specific binding. The phage library (100 µl, 1×1012 CFU/ml) was pre-incubated with the respective blocking buffer for 1 h at 37°C before it was added into antigen- or PBS-coated wells. After incubating at 37°C for 1.5 h, the wells were then washed with PBS containing 0.1% Tween 20 (PBST) 4 times followed by 10 times with PBS.
Bound phages were eluted by adding 100 µl glycine-HCl (0.1 mol/l, pH 2.2) for 8 min and then neutralised with 50 µl Tris–HCl (1 mol/l, pH 8.5). A 75 µl aliquot of the eluted phage was used to infect 2 ml culture of log-phase E. coli TG1 cells for subsequent amplification. The remaining eluted phage was mixed with an equal volume glycerol and stored at −80°C.
Indirect phage-ELISA
Single independent colonies from each round of panning were rescued using helper phage KM13. The rescued phage particles were tested against DON-MBSA by indirect phage-ELISA. Ninety-six-well ELISA microplates (Greiner Bio-One, Frichenhausen, Germany) were coated with DON-MBSA (5 µg/ml in PBS, pH 7.4, 100 µl/well) overnight at 4°C, while the blank wells were coated with PBS (pH 7.4, 100 µl/well). Plates were filled with 3% BSA for 2 h at 37°C, washed three times with PBST and incubated with purified phage particles. After one-hour incubation at 37°C, plates were washed with PBST six times, followed by addition of anti-M13 horseradish peroxidase (HRP) conjugate (1:5000; GE Healthcare, London, UK) which had been pre-incubated with 3% BSA for 15 min at 37°C. The phages bound to DON were detected by the addition of 100 µl/well of substrate (3,3′,5,5′-tetramethyl benzidine, TMB). After 30 min incubation at 37°C, 50 µl/well H2SO4 (2 M) was added to stop the reaction, and the absorbance at 450 nm was measured by a plate reader (Thermo, Waltham, MA). The original phage library and the amplified phages of the second round panning served as negative control (NC) and positive control (PC), respectively.
Indirect-competitive phage-ELISA
The positive colonies of indirect phage-ELISA were subjected to competitive phage-ELISA. Preparation, incubation and washing of plates were conducted as described earlier. Purified phage particles were mixed with equal volume of serial diluted DON (200 ng/ml, 100 ng/ml, 50 ng/ml and 25 ng/ml in PBS, pH 7.4; Sigma, St. Louis, MO, USA) before added into blocked plates. After incubation and washing, the phages bound to DON were detected as described previously. The clones which were inhibited from binding when DON was present in solution were sent to Genscript (Nanjing, China) for sequencing.
Results
Library construction
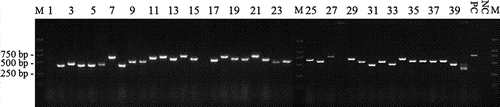
Total RNA was isolated from about 3×107 lymphocytes collected from blood of non-immunised alpacas and used to synthesise cDNA employing oligo dT or random hexamer primers. The semi-nested PCR products were digested and subcloned into the phagemid vector pHEN1. Dilution plating of electro-transformed E. coli indicated a total dimension of 2×107 colonies. Forty independent colonies were analysed by colony PCR. About 95% of analysed clones had insertions of different lengths (). These data also indicated that the library size was about 1.9×107, including redundancy and non-functional clones. The library was rescued by the helper phage KM13 and the titre of the phage display library was 5.6×1012 CFU/ml.
Panning and phage-ELISA
The phage display library was panned against DON-MBSA through three rounds of panning. About 1×1011 phage particles were input for each round of panning, and the titre of eluted phage was increased in later rounds of panning.
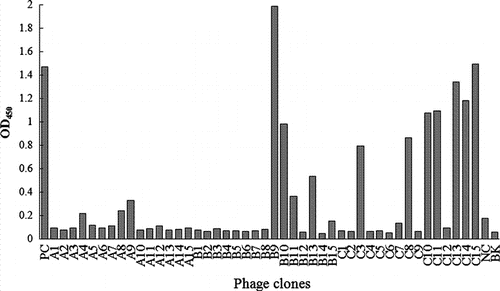
Phage-ELISA was conducted on 15 randomly selected clones from each round of panning against DON-MBSA (). Fourteen clones out of 45 tested phage clones showed absorbance higher than the absorbance of NC. Most of them were presented in the elution of 2 and 3 rounds of panning, indicating the accumulation of binding phage particles in panning process.
Figure 2. Phage-ELISA of phage clones selected randomly from each round of panning. A1–A15: phage clones picked from the first round of panning; B1–B15: phage clones picked from the second round of panning; C1–C15: phage clones picked from the third round of panning; PC: positive control; NC: negative control; BK: blank.
Indirect competitive phage-ELISA and sequencing
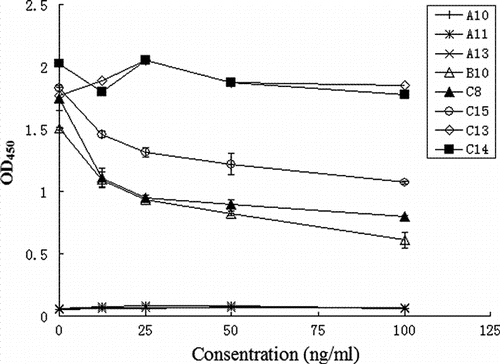
Seventeen phage particles including 14 phage-ELISA positive clones and 3 phage-ELISA negative clones were employed for indirect competitive phage-ELISA. The absorbance of three clones decreased as the concentration of DON increased (). Thus, DON inhibited the binding of the clones with DON-MBSA, indicating that the phage particles can specifically bind with DON. The absorbance decreased by half when the concentration increased to 100 ng/ml of DON. Meanwhile, the presence of DON did not affect the binding of clone C13 and C14 (). The three phage-ELISA negative clones (A10, A11, A13) that served as NCs produced absorbance below 0.1, similar to the blank.
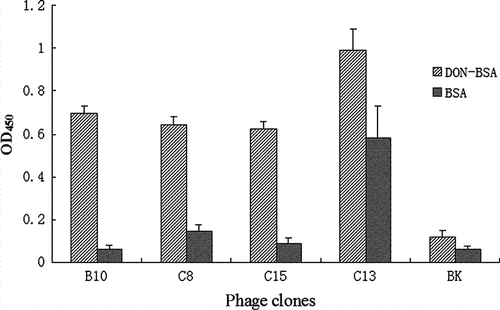
Further phage-ELISA analysis showed that the DON restrained clones could not bind to BSA (the carrier protein of artificial antigen). Thus, C13 and C14 could bind to both BSA and DON-MBSA ().
The positive clones were sequenced and analysed with program ClustalW for multiple sequence alignment. The deduced amino acid sequences are highly conserved (). The consensus sequence was analysed using a BLASTp programme with standard parameters on the GenBank (http://blast.ncbi.nlm.nih.gov), resulting in no hits. The amino sequences of the positive clones (B10, C8, C15) were submitted to GenBank (accession numbers HM622287, HM622288 and HM622289).
Discussion
DON toxin and its derivatives (i.e., nivalenol [NIV], 15-acetyl-deoxynivalenol [15-AcDON] and 3-acetyl-deoxynivalenol [3-AcDON]) are low molecular weight, highly stable trichothecene mycotoxins (Widestrand & Pettersson, Citation2001; Wolf & Bullerman, Citation1998). These mycotoxins are known to cause severe toxicosis in crops, humans and farm animals, for example, feed refusal and vomiting, growth retardation, inhibition of seedling and green plant regeneration (Rocha, Ansari, & Doohan, Citation2005). A stable and commercialised artificial antigen of DON, designated DON-MBSA, was synthesised for library panning. Considering the conjugation ratio might differ, same batch of DON-MBSA was used.
Immunoassay technologies, based on the interaction between antibody and antigen, can provide rapid, cost-effective and high throughput methods for detection of toxins in foodstuffs (Garet, Gonzalez-Fernandez, Lago, Vieites, & Cabado, Citation2010; Zheng, Richard, & Binder, Citation2006). The binding specificity and affinity of antibody directly affect the accuracy and limit of detection. Moreover, generating toxin-specific antibody is time consuming and laborious since it typically involves the repeated immunisation of animals and the used of cell fusion and culture techniques. Alternatively, phage display, based on expressing of proteins or peptides on the surface of filamentous phages, offers a powerful approach to isolation binders or the most interesting lead candidates by combining the generation of billions of components with a fast in vitro panning procedure. Construction of a non-immunised phage display library avoids the time-consuming immunisation procedure. Potentially, the same library could be used for panning against different antigens, including toxins, which are harmful to experimental animals.
The peripheral leukocytes from two non-immunised animals were used as sources of RNA for RT-PCR (Muyldermans et al., Citation2009). As the diversity of library was limited to the variety of leukocytes, larger sampling from several different animals would be better than that from single animal. The primers used for the semi-nested PCR were designed according to Maass (Maass et al., Citation2007) with minor modification. In order to amplify the repertoire coding sequence, a relatively low annealing temperature was employed for amplification. PCR1 product got a smear on the agarose gel ranging from 300 bp to 1000 bp. In theory, the VHH coding sequence should be around 400 bp; however, the PCR2 products showed a bright band between 250 bp and 600 bp. And all of these PCR products were subcloned into phagemid. Colony PCR results showed that the inserts with difference length were ligated into phagemid (). Interestingly, after three rounds of panning, none of phage particles encoding VHH with DON-binding affinity was isolated but clones encoding polypeptides. This may be due to the low annealing temperature (50°C) in PCR processes, which lead to error priming, and the number of phage particles displaying VHHs was discounted.
However, (poly)peptides as ligand of probes were widely used in the field of molecular imaging (Lee, Xie, & Chen, Citation2010). The indirect competitive phage-ELISA and indirect phage-ELISA confirmed that the binders can specifically interact with DON ( and ). The high conservation of the amino sequences of DON binders also implied the DON-specific binding affinity (). Results suggested that these binders have potential for developing detection methods or tools of DON in foods. In addition, the binder B10 was expressed in E. coli and it was confirmed that the recombinant protein can bind to DON (data not shown). Since the library was derived from non-immune animal, it could potentially be used for panning binders of other mycotoxins or low molecular weight haptens.
Acknowledgements
This work was supported by a grant of Innovation Fund for Technology Based Firms (09C26213604449) and by a grant of the Education Department of Jiangxi Province (GJJ09442). In addition, this work was supported by Natural Science Foundation of Jiangxi Province. Thanks to Dr. Cao Huabing of Jiangxi Agricultural University for the help with collecting blood samples and Dr. Ma Weijun for providing phagemid and helper phage.
References
- Abouzied , M.M. , Azcona-Olivera , J.I. , Yoshizawa , T. and Pestka , J.J. 1993 . Production of polyclonal antibodies to the trichothecene mycotoxin 4,15-diacetylnivalenol with the carrier-adjuvant cholera toxin . Applied and Environmental Microbiology , 59 : 1264 – 1268 .
- Bohm , C. , Cichna-Markl , M. , Brenn-Struckhofova , Z. and Razzazi-Fazeli , E. 2008 . Development of a selective sample clean-up method based on immuno-ultrafiltration for the determination of deoxynivalenol in maize . Journal of Chromatography. A , 1202 : 111 – 117 .
- Bratkovic , T. 2010 . Progress in phage display: Evolution of the technique and its application . Cellular and Molecular Life Sciences , 67 : 749 – 767 .
- Carmen , S. and Jermutus , L. 2002 . Concepts in antibody phage display . Briefs in Functional Genomics and Proteomics , 1 : 189 – 203 .
- Daly , S.J. , Dillon , P.P. , Manning , B.M. , Dunne , L. , Killard , A. and O'Kennedy , R. 2002 . Production and characterization of murine single chain Fv antibodies to aflatoxin B-1 derived from a pre-immunized antibody phage display library system . Food and Agricultural Immunology , 14 : 255 – 274 .
- Deng , S. , You , S. , & Xu , Y. 2007 . Preparation of artificial antigen of mycotoxin deoxynivalenol . Food Science , 28 , 149 – 152 . (In Chinese)
- Doyle , P.J. , Arbabi-Ghahroudi , M. , Gaudette , N. , Furzer , G. , Savard , M.E. Gleddie , S. 2008 . Cloning, expression, and characterization of a single-domain antibody fragment with affinity for 15-acetyl-deoxynivalenol . Molecular Immunology , 45 : 3703 – 3713 .
- Du , X.J. , Wu , Y.N. , Zhang , W.W. , Dong , F. and Wang , S. 2010 . Construction and quality examination of murine naive T7 phage display antibody library . Food and Agricultural Immunology , 21 : 81 – 90 .
- Garet , E. , Gonzalez-Fernandez , A. , Lago , J. , Vieites , J.M. and Cabado , A.G. 2010 . Comparative evaluation of enzyme-linked immunoassay and reference methods for the detection of shellfish hydrophilic toxins in several presentations of seafood . Journal of Agricultural and Food Chemistry , 58 : 1410 – 1415 .
- Hamers-Casterman , C. , Atarhouch , T. , Muyldermans , S. , Robinson , G. , Hamers , C. Songa , E.B. 1993 . Naturally-occurring antibodies devoid of light-chains . Nature , 363 : 446 – 448 .
- Klinglmayr , C. , Nobauer , K. , Razzazi-Fazeli , E. and Cichna-Markl , M. 2010 . Determination of deoxynivalenol in organic and conventional food and feed by sol-gel immunoaffinity chromatography and HPLC-UV detection . Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences , 878 : 187 – 193 .
- Kong , B. , Liu , H. , Yang , S.L. , & Ma , W.J. 2006 . Large recombinant protein displayed on filamentous phage surface and its interaction with small molecule . Chinese Journal of Biotechnology , 22 , 19 – 24 . (In Chinese)
- Lee , S. , Xie , J. and Chen , X. 2010 . Peptide-based probes for targeted molecular imaging . Biochemistry , 49 : 1364 – 1376 .
- Lombaert , G.A. 2002 . Methods for the determination of deoxynivalenol and other trichothecenes in foods . Advances in Experimental Medicine and Biology , 504 : 141 – 153 .
- Maass , D.R. , Sepulveda , J. , Pernthaner , A. and Shoemaker , C.B. 2007 . Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs) . Journal of Immunological Methods , 324 : 13 – 25 .
- Muyldermans , S. , Baral , T.N. , Retamozzo , V.C. , De Baetselier , P. , De Genst , E. Kinne , J. 2009 . Camelid immunoglobulins and nanobody technology . Veterinary Immunology and Immunopathology , 128 : 178 – 183 .
- Pestka , J.J. and Smolinski , A.T. 2005 . Deoxynivalenol: toxicology and potential effects on humans . Journal of Toxicology and Environmental Health, Part B: Critical Reviews , 8 : 39 – 69 .
- Rocha , O. , Ansari , K. and Doohan , F.M. 2005 . Effects of trichothecene mycotoxins on eukaryotic cells: a review . Food Additives and Contaminants , 22 : 369 – 378 .
- Sambrook , J. and Russell , D.W. 2001 . Molecular cloning: A laboratory manual , 3rd ed. , New York : Cold Spring Harbor Laboratory Press .
- Schneider , L. , Pichler , H. and Krska , R. 2000 . An enzyme linked immunoassay for the determination of deoxynivalenol in wheat based on chicken egg yolk antibodies . Fresenius Journal of Analytical Chemistry , 367 : 98 – 100 .
- Wang , S.H. , Du , X.Y. , Huang , Y.M. , Lin , D.S. , Hart , P.L. and Wang , Z.H. 2007 . Detection of deoxynivalenol based on a single-chain fragment variable of the antideoxynivalenol antibody . FEMS Microbiology Letters , 272 : 214 – 219 .
- Widestrand , J. and Pettersson , H. 2001 . Effect of time, temperature and solvent on the stability of T-2 toxin, HT-2 toxin, deoxynivalenol and nivalenol calibrants . Food Addititives and Contaminants , 18 : 987 – 992 .
- Wolf , C.E. and Bullerman , L.B. 1998 . Heat and pH alter the concentration of deoxynivalenol in an aqueous environment . Journal of Food Protection , 61 : 365 – 367 .
- Yoshizawa , T. , Kohno , H. , Ikeda , K. , Shinoda , T. , Yokohama , H. Morita , K. 2004 . A practical method for measuring deoxynivalenol, nivalenol, and T-2 + HT-2 toxin in foods by an enzyme-linked immunosorbent assay using monoclonal antibodies. Bioscience, Biotechnology . and Biochemistry , 68 : 2076 – 2085 .
- Zheng , M.Z. , Richard , J.L. and Binder , J. 2006 . A review of rapid methods for the analysis of mycotoxins . Mycopathologia , 161 : 261 – 273 .