Abstract
A pair of monoclonal antibodies (mAb) from 10 murine hybridomas secreting Escherichia coli O157:H7 (E. coli O157:H7)-specific mAbs were selected for the development of the sandwich ELISA to detect E. coli O157:H7. On the basis of pairwise interaction analysis, mAb-1 was selected as a capture antibody while mAb-6 was used as a detection antibody. The buffer system which provided the greatest difference between the specific E. coli O157:H7-positive antigen and the negative control was chosen. This sandwich ELISA showed good linearity when the concentration of E. coli O157:H7 was in the range of 105–108 cfu/mL, and the sensitivity was 1×104 cfu/mL. With 8-h enrichment of bacteria, this ELISA was found to detect 0.4 cfu/g E. coli O157:H7 in artificially contaminated green tea samples.
Introduction
Escherichia coli O157:H7 (E. coli O157:H7) is one of the most frequently reported food-borne pathogens, causing gastroenteritis, haemolytic-uraemic syndrome and thrombotic thrombocytopenic purpura throughout the world (O'Loughin, Citation1997). It is a big challenge to establish an accurate and rapid detection method for this pathogen.
During the past few years, several methods have been used to detect E. coli O157:H7. Some assay methods based on DNA have been established, including polymerase chain reaction (PCR) (Fode-Vaughan, Maki, Benson, & Collins, Citation2003; Gordillo, Cordoba, Andrade, Luque, & Rodriguez, Citation2011), reverse transcription PCR (Di & Tumer, Citation2010; Liu, Wang, Fung, & Li, Citation2010), quantitative real-time PCR (Elizaquível, Gabaldón, & Aznar, Citation2011; Fedio et al., Citation2011), and DNA microarrays (Douglas, Fred, & Darrell, Citation2001).
The nucleic acid-based assay requires good technical expertise and a nucleic acid extraction step. In contrast, the antibody-based measures, such as immuno-chromatographic strip, immuno-biosensor or ELISA can be applied in field detection with little technical knowledge or instruments and high-volume testing. An immuno-strip based on polyclonal antibody was reported to detect 1.8×103 cfu/mL E. coli O157:H7 in water (Park, Kim, Paek, Hong, & Kim, Citation2008). Numerous immuno-sensors have been developed for detection and enumeration of E. coli O157:H7 by immobilisation of the reaction to the surface of a device known as a transducer with sensitivity ranging 10–107 cfu/mL (Tokarskyy & Marshall, Citation2008).
A sandwich ELISA with a polyclonal antibody as a capture antibody and a monoclonal antibody (mAb) as a testing antibody was developed by Johnson et al. (Citation1995) to detect E. coli O157:H7 in meat samples. Incorporating immunocapture step and 6-h enrichment in trypticase soy broth, Johnson et al. reduced the non-specificity of this immunoassay and obtained a low detection limit of 0.14 cfu/g. Kerr developed an ELISA for E. coli O157:H7; however, it showed broad cross-reactivity with E. coli O15, Citrobacter freundii, Salmonella urbana and Vibrio cholerae O1 Inaba (Kerr et al., Citation2001). Owing to poor specificity of antibody for the Lipopolysaccharide of E. coli O157, the sensitivity was 105 cfu/mL for E. coli O157:H7.
China is one of the main providers of tea in the world. With the growing concern over pollutes in tea, more strict safety requirements in tea trade have been imposed by many importers (Yue, Kuang, Sun, Wu, & Xu, Citation2010). It is reported that some countries including America, Canada have tried to detect pathogenic bacteria in imported tea (Wang, Citation2008). To develop a low-cost and simple method to quantify E. coli O157:H7 in green tea samples, we attempted to develop a sandwich ELISA based on monoclonal antibodies.
Materials and methods
Bacterial strains
Two E. coli O157:H7 were purchased from the China Medical Culture Collection (CMCC) and China Center of Industrial Culture Collection (CICC). Four other E. coli strains, one Salmonella species and one Staphylococcus aureus were purchased from the American Type Culture Collection (ATCC, USA), CMCC or CICC. Seven clinical E. coli strains isolated from patients came from Chinese Academy of Military Medical Sciences. All the details are shown in . The bacteria grew on SMAC agar (containing cefixime 0.05 ng/mL, potassium tellurite 2.5 µg/mL) or nutrient agar plates at 37°C overnight.
Table 1. Bacterial strains tested in this study.
Immunisation procedure
EC broth medium inoculated with E. coli O157:H7 was treated in boiling water for 1 h, and bacterial cells were centrifuged at 1500 g for 10 min at 4°C. The pellets were washed three times in normal saline and subsequently suspended in the same buffer at a concentration of 109 cfu/mL. The resulting antigen was mixed at a 1:1 ratio with Freund complete adjuvant at the first immunisation and with Freund incomplete adjuvant at the next immunisation. Six female BALB/c mice (6 weeks old) were injected subcutaneously with 200 µL of 1:1 (v/v) mixture every 2 weeks. After three injections, booster immunisation with antigen in normal saline was carried out, and the mice were sacrificed 3 days later.
Monoclonal antibody production
The spleen cells taken from one immunised mouse were fused with SP2/0 myeloma cells, and positive hybridoma cells were screened by indirect ELISA (Deng et al., Citation2012). Positive hybridoma was cloned by three cycles of sub-cloning by limited dilution.
All hybridoma cell lines secreting mAb against BSA were injected into 8-week-old BALB/c mice to generate ascites fluid. The mAb was purified from ascitic fluid by sequential precipitation using caprylic acid and ammonium sulphate (Deng et al., Citation2012).
Establishment of the sandwich ELISA
To compare the binding of mAbs to different binding epitopes, a pairwise interaction analysis was carried out. Each mAb was labelled with horseradish peroxidase (HRP) and then utilised as either the capture or detection antibody to determine the optimum combination. The optimal combination that provided the greatest difference between the specific E. coli O157:H7-positive antigen and the negative control was chosen. On the basis of the chosen capture antibody and the detection antibody, a sandwich ELISA was developed. The ELISA was performed in 96-well polystyrene ELISA plates (Thermo Electron Corporation, Shanghai, China). The assay procedures were the same as used by Deng et al. (Citation2012). Various buffer systems including coating buffer, blocking buffer, antibody dilution buffer and assay solution were optimised to obtain the lowest non-specific binding.
Green tea sample detection
Two and a half grams of green tea was soaked in 22.5-mL hot sterile water (80–90°C) for 30 min, and the resulting solution was filtered; 0.1 mL of tea solution was diluted to 1 mL with assay buffer, which was used for ELISA analysis, or 1-mL tea solution was mixed with 9-mL EC broth for culture at 37°C for 8 h. Then, 1-mL culture medium was centrifuged at 1500 g for 10 min, and the pellet was re-dissolved in 0.1-mL assay buffer for determination. A test sample was considered positive if the ratio (P/N) of the optical density value in the test well to that of the negative control well was ≥2.1.
Results and discussion
Pair-wise interaction analysis
On the basis of the screening results, 10 positive hybridoma cell lines secreting mAb against E. coli O157:H7 were obtained. All of them had a high titre with E. coli O157:H7 (1:106). The 10 mAbs (No.1–No.10) were labelled with HRP and then utilised as either the capture or detection antibody to determine the optimum combination. According to the P/N values, mAb-1 and mAb-6 were selected to use as capture antibody and detection antibody, respectively (). Significantly lower absorbance was observed when any of the other combinations of capture and detection antibody were employed. There was no cross-reactivity with the other non-E.coli bacteria listed in . Weak recognition of E.coli O146 was found from mAb-1, but no cross-reactions occurred in the sandwich ELISA on the basis of mAb-1 and mAb-6, indicating a good specificity for E.coli O157:H7.
Table 2. Sandwich ELISA for pair-wise interaction analysis (P/N value).
Optimisation of sandwich ELISA
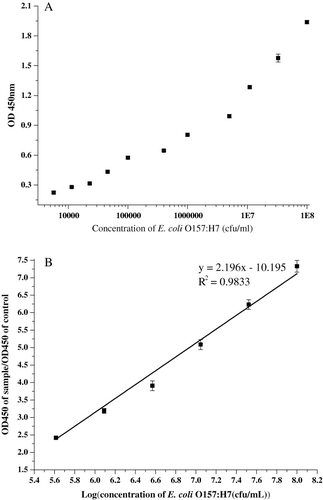
The highest ratio of positive antigen to negative control was achieved by the combination of the coating buffer (carbonated-bicarbonate buffer, 50 mmol, and pH 9.6), blocking solution (carbonated-bicarbonate buffer containing 1% casein) and assay buffer (0.02 M phosphate-buffered saline containing 1.8% NaCl, pH 7.2). The linear dynamic range for this method was between 105 and 108 cfu/mL ( and ). With the limit between negative and positive results set at the mean and two standard deviations for the concentrations of the negative control wells, a maximum sensitivity of this sandwich ELISA could be achieved 1×104 cfu/ml of E. coli O157:H7 (). The sandwich ELISA successfully distinguishes serotype O157:H7 from all E. coli stains in .
Detection of green tea samples
As shown in , the results from sandwich ELISA coincided well with conventional culture method. With 8 h enrichment, 1 cfu E. coli O157:H7 from 2.5-g tea samples could be detected. Compared with 24–96 h enrichment process of culture method, the proposed immunoassay here was fast and high throughout.
Table 3. Detection of E. coli O157:H7 in artificially contaminated green tea samples.
Conclusion
A sensitive sandwich ELISA was developed based on two mAbs that recognised different epitopes on E. coli O157:H7. The immunoassay showed good specificity with a limit of detection of 104 cfu/mL and linear range of 105–108 cfu/mL. In a test of contaminated green tea samples, the results from this ELISA system was in good accordance with GOLD standard culture methods. Thus, the mAb sandwich ELISA developed herein may provide a stable, precise and sensitive method for quantifying E. coli O157:H7 in food.
Acknowledgements
This work is financially supported by the National Natural Science Foundation of China (21071066, 20835006, 91027038, 21101079, 21175034), the Key Programs from MOST (2012BAC01B07, 2012BAD29B05, 2012AA06A303, 2012BAD29B04, 2011BAK10B07, 2011BAK10B05, 2011BAK10B01, 2010AA06Z302, 2010DFB3047, 2009BAK61B04, 2011ZX08012-001, 2010GB2C100167, 2012BAK17B10, 2012BAK08B01), and grants from Natural Science Foundation of Jiangsu Province, MOF and MOE (BE2011626, BK2010001, BK2010141, 201110060, 201110016, 201110061, 201210036, 201310135, 311002).
References
- Deng, X.F., Liu, L.Q., Ma, W.W., Xu, C.L., Wang, L.B., & Kuang, H. (2012). Development and validation of a sandwich ELISA for quantification of peanut agglutinin (PNA) in foods. Food and Agricultural Immunology, 23, 265–272.
- Di, R., & Tumer, N.E. (2010). Real-time reverse transcription PCR detection of viable shiga toxin-producing Escherichia coli o157:H7 in food. Journal of Food Safety, 30, 51–66.
- Douglas, R.C., Fred, J.B., & Darrell, P.C. (2001). Detecting and genotyping Escherichia coli O157:H7 using multiplexed PCR and nucleic acid microarrays. International Journal of Food Microbiology, 67, 71–80.
- Elizaquível, P., Gabaldón, J.A., & Aznar, R. (2011). Quantification of Salmonella spp., Listeria monocytogenes and Escherichia coli O157:H7 in non-spiked food products and evaluation of real-time PCR as a diagnostic tool in routine food analysis. Food Control, 22, 158–164.
- Fedio, W.M., Jinneman, K.C., Yoshitomi, K.J., Zapata, R., Wendakoon, C.N., Browning, P., et al. (2011). Detection of E. coli O157:H7 in raw ground beef by Pathatrix immunomagnetic-separation, real-time PCR and cultural methods. International Journal of Food Microbiology, 148, 87–92.
- Fode-Vaughan, K.A., Maki, J.S., Benson, J.A., & Collins, M.L.P. (2003). Direct PCR detection of Escherichia coli O157:H7. Letters in Applied Microbiology, 37, 239–243.
- Gordillo, R., Cordoba, J.J., Andrade, M.J., Luque, M.I., & Rodriguez, M. (2011). Development of PCR assays for detection of Escherichia coli O157:H7 in meat products. Meat Science, 88, 767–773.
- Johnson, R.P., Durham, R.J., Johnson, S.T., MacDonald, L.A., Jeffrey, S.R., & Butman, B.T. (1995). Detection of Escherichia coli O157:H7 in meat by an enzyme-linked immunosorbent assay, EHEC-Tek. Applied and Environmental Microbiology, 61, 386–388.
- Kerr, P., Chart, H., Finlay, D., Pollock, D.A., Mackie, D.P., & Ball, H.J. (2001). Development of a monoclonal sandwich ELISA for the detection of animal and human Escherichia coli O157 strains. Journal of Applied Microbiology, 90, 543–549.
- Liu, Y., Wang, C., Fung, C., & Li, X.Y. (2010). Quantification of viable but nonculturable Escherichia coli O157:H7 by targeting the rpoS mRNA. Analytical Chemistry, 82, 2612–2615.
- O'Loughin, E. (1997). Escherichia coli O157:H7. The Lancet, 349, 1553.
- Park, S., Kim, H., Paek, S.H., Hong, J.W., & Kim, Y.K. (2008). Enzyme-linked immuno-strip biosensor to detect Escherichia coli O157:H7. Ultramicroscopy, 108, 1348–1351.
- Tokarskyy, O., & Marshall, D.L. (2008). Immunosensors for rapid detection of Escherichia coli O157:H7 – Perspectives for use in the meat processing industry. Food Microbiology, 25, 1–12.
- Wang, L. (2008). E.coli O157:H7 detection in tea in food science. Chongqing: Southwest University, p. 51.
- Yue, N., Kuang, H., Sun, L., Wu, L., & Xu, C. (2010). An empirical analysis of the impact of EU's new food safety standards on China's tea export. International Journal of Food Science & Technology, 45, 745–750.