Abstract
The prevalence of fish allergies has become a serious health problem and has increased alarmingly over the past few years. To contain this problem, we would need to identify all commonly consumed fish allergens. To date, however, no report has identified largemouth bass allergens. This study attempted to identify and purify the major allergen implicated in the allergic response to largemouth bass (Micropterus salmoides), a freshwater fish widely consumed in China. Fifteen patients with a positive history of type I allergy to fish were recruited from skin-prick test and the allergy screen. Total protein extracts and purified allergenic protein from bass were tested for their immunoglobulin E–binding properties. Immunoblot assay resulted in strong reactivity with the 17-kDa protein in all patients. In summary, nucleoside diphosphate kinase B was identified as a novel fish allergen in largemouth bass. This finding is important for allergy diagnoses and the treatment of freshwater fish–allergic disorders.
1. Introduction
Fish are an important component of the world's food supply and are rich in nutrients, such as proteins, lipid-soluble vitamins and polyunsaturated fatty acids. However, fish are one of the most frequently reported sources of food allergens (Hajeb & Selamat, Citation2012; Lehrer, Ayuso, & Reese, Citation2003; Waserman & Watson, Citation2011). Sensitised individuals may present various symptoms, such as gastrointestinal problems (oral allergy syndrome, vomiting, cramps, diarrhoea) and skin problems (rash, urticaria). A close cross-reactivity exists within different species of fish. It is estimated that 50% of individuals allergic to some type of fish are at risk for reacting to a second species.
Since 1971, a number of food allergens have been identified in various saltwater fish. All of these allergens belong to the family of parvalbumins, which are the most common fish allergens.
Freshwater fish are a major source of dietary protein in China, which has the highest fish consumption in the world (Lin, Citation2010). Freshwater fish accounted for 41.5% of the total fishery industry in China (Wang et al., Citation2006). Therefore, the prevalence of freshwater fish allergies represents a serious health risk that has increased significantly over the past few years. Largemouth bass (Micropterus salmoides), which belongs to the family Centrarchidae, was selected for our research due to its nutrition, high allergic potential and widely distributed consumption throughout China. Largemouth bass in China is usually processed into various products, such as canned, oil-fried and fermented fish, but their biological and immunological properties have not been well characterised.
For studies on major fish allergens, many researchers have focused on saltwater fish. Therefore, the present study identified and characterised major allergens in edible freshwater fish to elucidate their roles in food allergies. As an important species among freshwater fish, the largemouth bass has both high economic value and high nutritional content.
The principal aim of this study was to use two-dimensional gel electrophoresis (2-DE) in combination with matrix-assisted laser desorption/ionisation-time of flight mass spectrometry (MALDI-TOF MS) after tryptic digestion to determine the suitability of this method for qualitatively characterising and identifying the major allergen of largemouth bass (Micropterus salmoides).
2. Materials and methods
2.1. Preparation of fish protein extraction
Largemouth bass specimens were purchased at a local supermarket. The specimens were euthanised and stored at −80°C until use. Fish protein extracts were made by mixing homogenised fish muscle (50 g) with 100mL of 20 mM Tris-HCl buffer (pH 7.5), stirring at 4°C for 2 hours. The aqueous fraction was clarified by centrifugation at 12,000 rpm for 20 minutes. The resulting supernatant was filtered through a 0.45 µm filter (Millipore, Bedford, MA). The extracts were stored at −20°C until needed. The protein content of the crude fish muscle tissue was determined by the Kjeldahl method, and the soluble protein of the fish extracts was determined using the commercial Bio-Rad Protein Assay Dye Reagent (BioRad, Italy) (Pico de Coana et al., Citation2010).
2.2. Patients and sera/patient characteristics
The study included 15 patients who had been previously diagnosed as allergic to fish and were chosen from the Daqing Oilfield General Hospital, Daqing, Heilongjiang, PR China. For this study, all patients had a convincing clinical history and hypersensitivity. Patient sera data were obtained based on an appropriate concentration of specific immunoglobulin E (IgE) to fish allergy (). The control serum was collected from non-atopic laboratory volunteers. The negative control serum was made by 12 non-atopic volunteers’ serum pool. Informed consent was obtained from each volunteer, and sera from these patients were stored at −80°C until use.
Table 1. Description of fish-allergic subjects.
2.3. Sodium dodecyl sulphate polyacrylamide gel electrophoresis
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to Laemmli with the use of an AE6400 Dual Mini Slab electrophoresis apparatus (Atto Corporation, Tokyo, Japan). Fish protein samples were mixed in a 1:1 ratio with 50 mM Tris buffer (pH 6.8) containing 1% SDS, 1% β-mercaptoethanol, 20% (v/v) glycerin and 0.1% bromophenol blue. To ensure proper protein separation and visualisation, aliquots of fish protein sample (5 µL, 15 mg/mL) were boiled for 5 minutes before being resolved on acrylamide gels (90×73×1 mm) consisting of 12.5% separating gel and 4% stacking gel. The gels were electrophoresed at a constant current of 30 mA for 2–3 hours and analysed by staining with Coomassie Blue R-250 or used for immunoblotting. Blue Plus II Protein Marker molecular weight markers (TianGen Biotech CO.Ltd., Beijing, China) with molecular weights of 14.4, 20.0, 26.0, 33.0, 45.0, 66.2 and 94.0 kDa were used for reference.
2.4. Two-dimensional electrophoresis
The fish extracts were mixed and analysed by 2-DE, as described previously (Beyer, Bardina, Grishina, & Sampson, Citation2002). Briefly, approximately 500 µg of the protein sample was applied to a 17 cm gel strip containing an immobilised pH gradient (3.5–10) (GE Healthcare Sciences, Uppsala, Sweden) for isoelectricpoint comparison. Isoelectric focusing was performed in the following conditions: 250 V ramp for 30 minutes, held at 1000 V for 1 hour, ramped to 10,000 V in 5 hours and held at 10,000 V for 60 kV for 3 hours. After isoelectric focusing, the strip was equilibrated (Tris 0.05 mol/L, urea 6 mol/L, glycerol 30%, SDS 1%, bromophenol blue 0.005% and 1% DTT). For convenience, the strip was kept frozen at −80°C between the two equilibration steps (Kjaersgard & Jessen, Citation2004). SDS-PAGE with 2-DE was performed using a laboratory-made 12.5% (w/v) SDS-PAGE gel at a constant current of 30 mA for 2–3 hours and followed by Coomassie Brilliant Blue (CBB) staining.
2.5. Western blot analysis
For the immunoblot analysis, proteins quantified using a Bio-Rad Protein Assay (Bio-Rad, Hercules, CA, USA) were separated by SDS-PAGE and transferred onto a nitrocellulose membrane (Pall Corporation, Mexico), as described previously (Spier et al., Citation2001), using a transfer unit (Atto Corporation, Japan) at a constant voltage of 100V for 1.5 hours. To reduce nonspecific binding, the blotted membranes were incubated in 2% bovine serum albumin for 2 hours before being probed at 4°C overnight with patient sera (1:10 dilution) (Liu, Luo, & Li, Citation2011).
The strips incubated with non-atopic volunteer sera served as the control. Bound IgE antibodies were detected with an HRP-labeled goat anti-human IgE antibody (1:4000 dilution, Sigma). The blots were developed by adding 4-chloro-1-naphthol buffer (Sigma).
2.6. Protein in-gel digestion
After staining with CBB, protein spots of interest were excised and destained with 50 mM ammonium bicarbonate and 50% acetonitrile (ACN). Gel pieces were then dried completely by speed vacuum concentration. In-gel digestion was performed with 25 ng/µL trypsin (Promega Corp., Madison, WI, USA) in 50 mM ammonium bicarbonate at 37°C for 16 hours. The supernatants were collected into a new Eppendorf tube (EP) tube, and the tryptic peptides were extracted from the gel with 5% trifluoroacetic acid (TFA) at 37°C for 1 hour. The extracts were pooled and dried completely by speed vacuum. The dried extracts were desalted using the Ziptip C18 method, and the extracts were then dissolved with 0.1% methane acid in preparation for subsequent MALDI-TOF analysis.
2.7. Peptide analysis by matrix-assisted laser desorption ionisation-time of flight analysis
The in-gel tryptic digests resulting from the 2DE separation were analysed using an ABI 4700 MALDI TOF mass spectrometer (Applied Biosystems, Foster City, CA, USA). The instrument's parameters were set as follows: spectra were acquired in the positive ion mode, and the detector was set to reflector mode. Accelerating voltage was set to 20 kV, and the laser intensity was set to 2000 laser shots. Acquisition was performed in the m/z range 700–4000. A total of 100 shots were performed per spectrum, and to increase the signal-to-noise ratio, 10–15 spectra were accumulated per sample. The samples were prepared using a saturated solution of 4-hydroxycinnamic acid in 50% (v/v) ACN and 0.1% (v/v) TFA. A mass database search was performed using Mascot (Matrix Science, London, UK) (http://www.matrixscience.com), which also obtained results for the MALDI. Mascot was also used to search for peptide mass fingerprinting (PMF) spectra in the National Center for Biotechnology Information non-redundant (NCBInr) database. The parameters for the database search are shown in .
Table 2. Database search parameters.
2.8. Purification of the 17-kDa allergen
Muscle tissue from largemouth bass was homogenised in 20 mM Tris-HCl (pH 7.5) buffer using a homogeniser (Kinematica, PT-2100, Switzerland). After centrifugation at 8000 rpm for 20 minutes, the supernatant was collected and fractionated with 30–90% ammonium sulphate. After centrifugation at12000 rpm for 20 minutes, the resulting precipitate was dissolved in 20 mM Tris-HCl (pH 7.5) and dialysed in the same buffer.
The dialysed solution was subsequently applied to DEAE-Sepharose CL-6B (1.6 cm×20 cm), which was previously equilibrated with 20 mM Tris-HCl (pH 7.5). After the column was washed extensively with equilibrated buffer, the binding proteins were eluted with a 0–0.3 M NaCl linear gradient at a flow rate of 1 mL/min. Fractions of interest were respectively pooled and then applied to a Sephacryl S-200 gel filtration column, which was equilibrated with 20 mM Tris-HCl containing 0.15 M NaCl (pH 7.5). The column was eluted at a flow rate of 0.5 mL/min, and fractions containing purified 17-kDa allergen were pooled. All procedures were performed at 4°C.
3. Results and discussion
3.1. Identification of the major allergen from largemouth bass
The 12 mg/mL protein component of the crude extract from largemouth bass was analysed by SDS-PAGE. In , lane 1 shows the Coomassie-stained SDS-PAGE protein profiles from the raw largemouth bass extracts. There are 15 protein bands with molecular weights ranging from 8 to 120 kDa, and many dominant bands were observed at 12, 17, 27, 34, 44, 54 and 96 kDa. A protein of approximately 12kDa, corresponding to the well-characterised fish allergen parvalbumin, was also present.
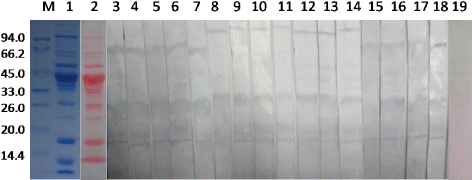
The visualisation of proteins transferred to nitrocellulose with Ponceau is shown in lane 2. Lane 3 shows the immunoblot results incubated with pooled fish-allergic serum. The specific IgE showed high binding capacity with proteins with molecular weights of 17, 27, 34, 68 and 96 kDa. No IgE binding was observed with the negative control serum (, lane19).
IgE-binding profiles differed among the fish-allergic patient sera when evaluated individually (, lanes 4–18). Fish-allergic subjects were selected based on a convincing history of allergic symptoms experienced after eating fish and on their specific IgE levels to fish species (). Symptoms included oral allergy syndrome (throat, mouth and lip itching and swelling), wheezing, urticaria and conjunctivitis. Sera 5–11 had immunoreactions with 96-kDa proteins; 6 of the 15 patients had serum that showed positive IgE-binding to the 68-kDa proteins. The 27-kDa profiles showed positive IgE-binding to almost all sera except P12 and P14. Sera 3, 4 and 10 showed immunoreactions with proteins at 34, 43 and 54 kDa, respectively. Interestingly, the 17-kDa proteins showed a positive reaction with sera from all 15 fish-allergic patients.
Most fish allergy studies performed in western countries have shown that parvalbumin is the major allergen in many seawater fish. However, in this study, the 12-kDa protein corresponding to parvalbumin was not considered to be the main IgE-binding protein in largemouth bass (). This observation implies that racial differences in the response to fish allergens should be taken into account. In China, parvalbumin may not play an important role in fish allergies.
3.2. 2D electrophoresis and immunoblot analysis
Largemouth bass extract was analysed by 2DE, an efficient method for separating several thousand different protein spots simultaneously. Proteins were first separated according to their isoelectricpoint. The first dimensional electrophoresis showed that most of these protein spots had pI values ranging from 3.5–8. The pI of the 17-kDa protein was approximately 6.5. Proteins were subsequently separated according to their molecular weight. The second dimensional gel showed a large number of protein spots ranging from 8 to 120 kDa ().
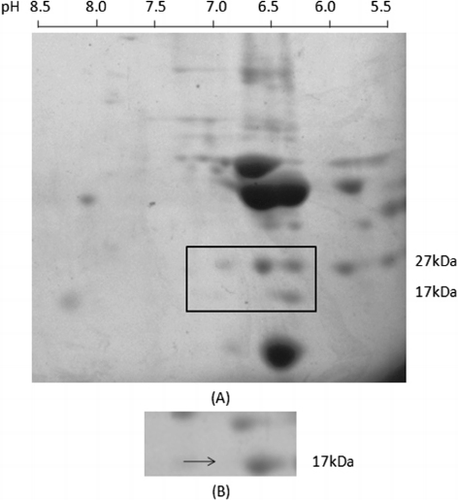
Proteins on 2D gels were transferred to a nitrocellulose membrane for further characterisation of their reaction with allergic patient sera as a source of specific IgE. Allergen-IgE complexes were visualised using horseradish peroxidase conjugated anti-human IgE antibody. There was only one spot comprising the 17-kDa protein that showed a strong reaction with patients’ sera (). A reaction with the 27-kDa protein spots was also detected but not further characterised.
3.3. MALDI-TOF MS analysis of the 17-kDa major allergen of largemouth bass
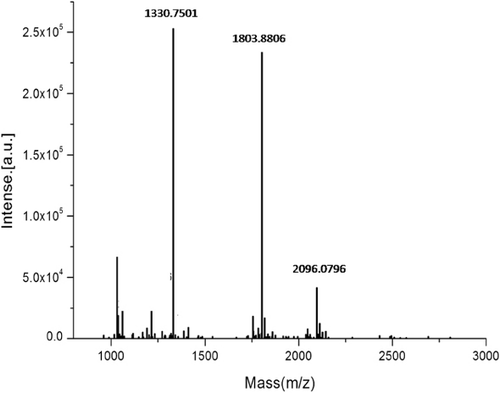
The 17-kDa protein spot recognised by the immune-blot was excised from the 2D gel. After in-gel digestion with trypsin, the resulting peptide mixture was analysed by MALDI-TOF MS to obtain its PMF and structural information related to the peptide sequence. The PMF of the 17-kDa protein showed multiple peaks ranging from 960 to 2800 Da ().
The Mascot search tool was selected to provide the number of hits for peptides. By comparing the number of hits for peptide with the theoretically expected tryptic peptide masses in the NCBInr databases, protein data that scored greater than 56 were identified as highly significant (P<0.05). According to the peptide mass analysis, there were five proteins with scores greater than 56. This search was matched with NDPK B from Malabar grouper (Epinephelus coioides), NDPK 3 from zebrafish (Danio rerio), NDPK 3 from channel catfish (Ictalurus punctatus) and NDPK 3 from zebra finches (Taeniopygia Guttata). The proteins’ description and additional relevant information are summarised in . Interestingly, Mascot research results showed that the top five matched proteins were all NDPK.
Table 3. Summary report of the top five proteins matched by the Mascot search.
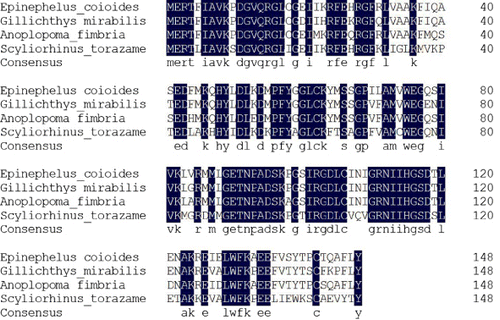
The masses showed the highest correlation with the NDPK B from Epinephelus coioides, with a score of 324. The amino acid sequence of the largemouth bass 17-kDa allergen that matched, completely or partially, with the Malabar grouper NDPK B is summarised in . As shown in , peptides with 48 amino acid residues in total had significant sequence homology to NDPK B from Epinephelus coioides. The NDPK B had a 32% sequence coverage rate with the 17-kDa largemouth allergen.
Table 4. Effective peptides from MS compared with data in NCBInr.
As shown in , the alignment score for the identity of the four NDPK-B sequenced proteins was 86.91%.Compared with the NDPK among fish species, Epinephelus coioides had approximately 87.92% identity with Gillichthys mirabilis (AAG13336.1) and 90.6% identity with Anoplopoma fimbria (ACQ58455.1). The alignment score revealed the homology of each two species. Epinephelus coioides had higher homology with Gillichthys mirabilis than that with Anoplopoma fimbria. A homology search in the GenBank databases revealed that NDPK-B shares a relatively high homology in the fish family. NDPK is a housekeeping enzyme that maintains the intracellular levels of all deoxynucleotide triphospates used in biosynthesis except adenosine triphosphate (Mehta & Orchard, Citation2009). NDPK-B has a role in the regulation of a wide variety of cellular processes (Hippe et al., Citation2009; Wieland, Citation2007), including activation of T-cell receptor–stimulated Ca2 + influx, interaction with the K+ channel and maintenance of a negative membrane potential by T lymphocyte Ca2 +signalling (Di et al., Citation2010). However, NDPK-B has not been reported as a food allergen. Lactate dehydrogenase, creatine kinase, enolase and NDPK B were the most vulnerable sarcoplasmic proteins. Enolase has been recognised as a class of highly conserved fungal allergens (Simon-Nobbe et al., Citation2000), such as house dust mite allergen Der f (Crack, Chan, McPherson, & Ogg, Citation2011), Hevea brasiliensis allergen Hev b 9 (Wagner et al., Citation2000) and blunt snout bream fish allergen (Liu, Krishnan, Xue, & Liu, Citation2011). Birch pollen allergen Bet v 1 (Gronlund & Gafvelin, Citation2010) and kiwifruit allergen (Oberhuber et al., Citation2008) are both creatine kinases. In our research, the isolated 17-kDa allergen was characterised by its molecular weight, isoelectric point and MALDI-TOF MS. According to the World Health Organization nomenclature, this allergen is called Mics1 (Meyer zum Gottesberge, Meyer zum Gottesberge, & Maass, Citation1987). The largemouth bass NDPK-B also functions as a novel fish allergen.
3.4. Purification of the NDPK B
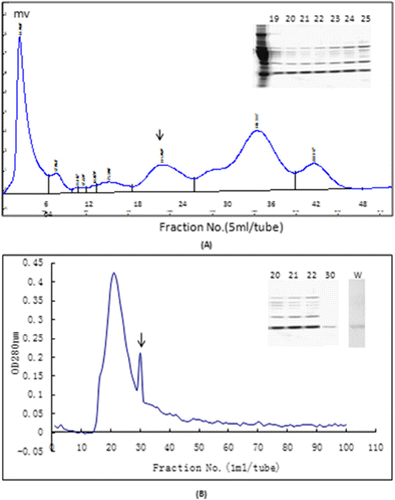
As shown in , NDPK B was highly purified for the first time from largemouth bass muscle tissue by column chromatography on DEAE-Sepharose CL-6B (A) and Sephacryl S-200 (B). After ammonium sulphate fractionation, NDPK B fractions were eluted at an NaCl concentration of 0.1−0.4 mol/L. Using SDS-PAGE detection, we obtained fractions 21, 22 and 23 due to their NDPK B content. These three fractions were pooled and further separated by Sephacryl S-200.Fraction pool was divided into two major peaks, and fraction 30 displayed a highly purified NDPK B. Purified NDPK-B was separated in 12% polyacrylamide gels with 4% stacking gels, resulting in the appearance of a single protein band with a molecular mass of approximately17 kDa (B).
4. Conclusions
In summary, we have identified a novel 17-kDa largemouth bass allergen (designated Mics1) from Micropterus salmoides by 2D immunoblotting using sera from fish allergic patients. This allergen is a major IgE-binding protein belonging to nucleoside diphosphate kinase B. This allergen, however, has not been reported as an allergenic protein in food. Therefore, we inferred that NDPK B from largemouth bass maybe a new allergen in fish. In further studies, we intend to confirm this putative new allergen in fish and characterise its allergenic potency.
Acknowledgements
This work was supported by grant from the National Natural Scientific Foundation of China (31072248).
References
- Beyer, K., Bardina, L., Grishina, G., & Sampson, H. A. (2002). Identification of sesame seed allergens by 2-dimensional proteomics and Edman sequencing: Seed storage proteins as common food allergens. Journal of Allergy and Clinical Immunology, 110(1), 154–159. 10.1067/mai.2002.125487
- Crack, L. R., Chan, H. W., McPherson, T., & Ogg, G. S. (2011). Identification of an immunodominant region of the major house dust mite allergen Der p 2 presented by common human leucocyte antigen alleles. Clinical and Experimental Dermatology, 37(3), 266–276.10.1111/j.1365-2230.2011.04227.x
- Di, L., Srivastava, S., Zhdanova, O., Sun, Y., Li, Z., & Skolnik, E. Y. (2010). Nucleoside diphosphate kinase B knock-out mice have impaired activation of the K+ channel KCa3.1, resulting in defective T cell activation. Journal of Biological Chemistry, 285(50), 38765–38771. 10.1074/jbc.M110.168070
- Gronlund, H., & Gafvelin, G. (2010). Recombinant Bet v 1 vaccine for treatment of allergy to birch pollen. Human Vaccines, 6(12), 970–977. 10.4161/hv.6.12.13348
- Hajeb, P., & Selamat, J. (2012). A contemporary review of seafood allergy. Clinical Reviews in Allergy and Immunology, 42(3), 365–385. 10.1007/s12016-011-8284-9
- Hippe, H. J., Wolf, N. M., Abu-Taha, I., Mehringer, R., Just, S., Lutz, S., … Wieland, T. (2009). The interaction of nucleoside diphosphate kinase B with Gbetagamma dimers controls heterotrimeric G protein function. Proceedings of the National Academy of Sciences of the United States of America, 106(38), 16269–16274. 10.1073/pnas.0901679106
- Kjaersgard, I. V., & Jessen, F. (2004). Two-dimensional gel electrophoresis detection of protein oxidation in fresh and tainted rainbow trout muscle. Journal of Agriculture and Food Chemistry, 52(23), 7101–7107. 10.1021/jf049573b
- Lehrer, S. B., Ayuso, R., & Reese, G. (2003). Seafood allergy and allergens: A review. Marine Biotechnology (New York), 5(4), 339–348.
- Lin, H. (2010). Fish research in China: Basic and applied. Foreword. Fish Physiology and Biochemistry, 36(2), 123–124. 10.1007/s10695-008-9242-1
- Liu, R., Krishnan, H. B., Xue, W., & Liu, C. (2011). Characterization of allergens isolated from the freshwater fish blunt snout bream (Megalobrama amblycephala). Journal of Agricultural and Food Chemistry, 59(1), 458–463. 10.1021/jf103942p
- Liu, X., Luo, Y., & Li, Z. (2011). Effects of pH, temperature, enzyme-to-substrate ratio and reaction time on the antigenicity of casein hydrolysates prepared by papain. Food and Agricultural Immunology, 23(1), 69–82. 10.1080/09540105.2011.604770
- Mehta, A., & Orchard, S. (2009). Nucleoside diphosphate kinase (NDPK, NM23, AWD): Recent regulatory advances in endocytosis, metastasis, psoriasis, insulin release, fetal erythroid lineage and heart failure; translational medicine exemplified. Molecular and Cellular Biochemistry, 329(1–2), 3–15. 10.1007/s11010-009-0114-5
- Meyer zum Gottesberge, A. M., Meyer zum Gottesberge, A., & Maass, B. (1987). Autoradiography study of the uptake of 3H-2-deoxyglucose in the inner ear in normal and unilaterally sympathectomized albino rats. Laryngologie, Rhinologie, Otologie (Stuttg), 66(8), 398–403. 10.1055/s-2007-998688
- Oberhuber, C., Bulley, S. M., Ballmer-Weber, B. K., Bublin, M., Gaier, S., & DeWitt, A. M. (2008). Characterization of Bet v 1-related allergens from kiwifruit relevant for patients with combined kiwifruit and birch pollen allergy. Molecular Nutrition and Food Research, 52(Suppl. 2), S230–S240.
- Pico de Coana, Y., Parody, N., Fuertes, M. A., Carnes, J., Roncarolo, D., Ariano, R., Alonso, C. (2010). Molecular cloning and characterization of cup a 4, a new allergen from Cupressus arizonica. Biochemical and Biophysical Research Communications, 401(3), 451–457. 10.1016/j.bbrc.2010.09.079
- Simon-Nobbe, B., Probst, G., Kajava, A. V., Oberkofler, H., Susani, M., & Crameri, R. (2000). IgE-binding epitopes of enolases, a class of highly conserved fungal allergens. Journal of Allergy and Clinical Immunology, 106(5), 887–895. 10.1067/mai.2000.110799
- Spier, A. W., Meurs, K. M., Coovert, D. D., Lehmkuhl, L. B., O'Grady, M. R., & Freeman, L. M. (2001). Use of western immunoblot for evaluation of myocardial dystrophin, alpha-sarcoglycan, and beta-dystroglycan in dogs with idiopathic dilated cardiomyopathy. American Journal of Veterinary Research, 62(1), 67–71. 10.2460/ajvr.2001.62.67
- Wagner, S., Breiteneder, H., Simon-Nobbe, B., Susani, M., Krebitz, M., Niggemann, B., Hoffmann-Sommergruber, K. (2000). Hev b 9, an enolase and a new cross-reactive allergen from hevea latex and molds. Purification, characterization, cloning and expression. European Journal of Biochemistry, 267(24), 7006–7014. 10.1046/j.1432-1327.2000.01801.x
- Wang, Q., Yang, G., Wu, X., Xu, X., Lai, C., & Ning, Y. (2006). Fish resources and their conservation strategies in Hepu Dugong State Nature Reserve and its adjacent waters. Ying Yong Sheng Tai Xue Bao, 17(9), 1715–1720.
- Waserman, S., & Watson, W. (2011). Food allergy. Allergy, Asthma and Clinical Immunology, 7(Suppl. 1), S7.
- Wieland, T. (2007). Interaction of nucleoside diphosphate kinase B with heterotrimeric G protein betagamma dimers: Consequences on G protein activation and stability. Naunyn Schmiedeberg's Archives of Pharmacology, 374(5–6), 373–383. 10.1007/s00210-006-0126-6