Abstract
An indirect enzyme-linked immunosorbent assay (ELISA) has been developed for the identification of albacore (Thunnus alalunga) and its differentiation from other less-valued scombrid species such as yellowfin tuna (Thunnus albacares), bullet tuna (Auxis rochei), atlantic bonito (Sarda sarda), bigeye tuna (Thunnus obesus), little tunny (Euthynnus alleteratus) and skipjack tuna (Katsuwonus pelamis). The assay uses polyclonal antibodies raised in rabbits against soluble muscle protein extract from fresh albacore. These polyclonal antibodies were rendered species-specific by blocking them with the heterologous soluble muscle proteins, allowing discrimination between fresh albacore and the rest of the scombrid species, except for yellowfin tuna. A total of 40 commercial albacore fresh and frozen fillets were analysed, revealing an incorrect labelling in 32.5% of the albacore samples. However, positive samples (67.5%) could be albacore or yellowfin tuna and should require a DNA assay as discriminatory technique.
Introduction
Increases in fish international trade, rising worldwide seafood consumption and varying levels of supply and demand of certain fish species have led to cases of economic fraud, in which one seafood species is illegally substituted for another. In this context, traceability of food products has become a government priority for comprehensive and integrated food safety and quality policies, as reflected in the key European Union food legislation (European Parliament and Council [EC] (Citation2002), No. 178/2002). In the scope of fish traceability, Regulation 178/2002 is complemented by another ones (European Council [EC] (Citation2000), No. 104/2000 and European Commission [EC] (Citation2001), No. 2065/2001), which establishes labelling laws regarding mandatory information such as species identification, production method and geographic origin of fish at every step of the production and retailing chain. However, in spite of these legal requisites, renaming and mislabelling of seafood occur globally to a significant extent, undermining fisheries management and conservation efforts and, consequently, deceiving consumers (Jacquet & Pauly, Citation2008).
Fish of Scombridae family, which include species of tunas, mackerel and bonito, is one of the most popular groups of food fishes forming the basis of important commercial and recreational fisheries. The geographic distribution of the individual species differs, as do their commercial value, and many of these fish are present as main or secondary ingredient in various foods which are prone to frauds. In this context, albacore fish (Thunnus alalunga) is a premium-priced white meat tuna distributed worldwide and greatly appreciated among consumers. Comparing scombrid species prices in MercaMadrid (Madrid, Spain) wholesale food market, albacore price is 1.5 times higher than yellowfin tuna (Thunnus albacares) and bigeye tuna (Thunnus obesus) price, and five times higher than bullet tuna (Auxis rochei), atlantic bonito (Sarda sarda), little tunny (Euthynnus alleteratus) and skipjack tuna (Katsuwonus pelamis). Although according to European law, only Thunnus alalunga can be commercially labelled as “white meat tuna” or “albacore” (European Council [EC] (Citation2000), No. 104/2000), albacore fish is often a target candidate for substitution by other less valuable Scombridae species but similar in sensory attributes.
One of the prerequisites for complying with European traceability and labelling regulations is the existence of techniques that can be used to incorporate fish species identification fully and on a routine basis into a traceability scheme (Martinsohn, Citation2011). Different protein-based methods have been applied for the identification of fish species, such as electrophoretic and chromatographic techniques (Berrini, Tepedino, Borromeo, & Secchi, Citation2006; Dreyfuss, Cutrufelli, Mageau, & McNamara, Citation1997; Gill, Citation1997; Le Fresne, Popova, Le Vacon, & Carton, Citation2011; Martinez, Šližytė, & Daukšas, Citation2007). These methods have shown considerable value in certain instances, but they are not convenient for routine sample analyses because they are relatively costly, time-consuming and complex to perform. Consequently, in the last years the identification of fish and seafood, as well as meat, milk and other food products has been performed preferentially by genetic (Armani, Castigliego, Tinacci, Gianfaldoni, & Guidi, Citation2012; Carrera et al., Citation1998; Chuang, Chen, & Shiao, Citation2012; Dalmasso et al., Citation2007; Espiñeira, Gonzalez-Lavín, Vieites, & Santaclara, Citation2009; Pardo & Pérez-Villareal, Citation2004) and immunological techniques (Carrera et al., Citation1996).
DNA methods, in particular those based on the polymerase chain reaction (PCR) are highly specific and reliable tools that are being increasingly used for fish authentication and traceability purposes (Asensio, Citation2007; Bottero & Dalmasso, Citation2011). However, these techniques are somewhat expensive and the feasibility of their routine use in the food industry is still dependent on the lowering of instrument and running costs. As an alternative, immunological assays can be used to reduce difficulty and assay cost. Particularly, enzyme-linked immunosorbent assay (ELISA) is the most widely used technique for regulatory purposes in species authentication because of its specificity, simplicity, sensitivity and high throughput screening (Asensio, Gonzalez, Pavon, Garcia, & Martin, Citation2008; Fæste & Plassen, Citation2008; Taylor, Patel, & Jones, Citation1994). Moreover, advances in ELISA technologies accomplished in the last years have led to rapid development of different easy-to-use stable commercial kits containing all the necessary components for rapid species verification at any point of the supply chain (Bonwick & Smith, Citation2004).
Based upon these considerations, this study reports the application of a polyclonal antibody-based indirect ELISA for the identification of albacore and its differentiation from other less-valued scombrid species such as yellowfin tuna, bigeye tuna, bullet tuna, atlantic bonito, little tunny and skipjack tuna. Because fish species substitution is a widespread problem worldwide, such an assay would provide a powerful tool to discourage illegal practices as well as to protect consumers from economic loss and reduce the potential health risks associated with fish fraud.
Materials and methods
Preparation of antigenic extracts
Authentic fresh muscle samples of albacore, yellowfin tuna, bullet tuna, atlantic bonito, bigeye tuna, little tunny and skipjack tuna individuals were obtained from MercaMadrid (Madrid, Spain) wholesale food market and were identified attending to morphological characteristics (Bauchot & Pras, Citation1993).
Antigenic sarcoplasmic protein extracts from five authentic samples of albacore were obtained as follows: representative 200 g fresh muscle samples were thoroughly homogenised in 1 L of a saline solution (8.5 g of NaCl per L) at 20 °C using a mechanical blender (Heidolph Elektro GmbH & CoKG, Kelheim, Germany). Soluble sarcoplasmic proteins (SP) were extracted from the muscle by gentle agitation of the homogenate for 2 h at 20 °C. The sarcoplasmic extract was centrifuged at 2500 g for 30 min at 10 °C and the supernatant was filtered through a Whatman no. 1 filter paper and lyophilised.
The protein content of the lyophilised extracts was calculated with the BCA Protein Assay Kit (Pierce, Rockford, IL, USA) using bovine serum albumin as protein standard.
Heterologous sarcoplasmic protein extracts from five reference fresh samples of each yellowfin tuna, bullet tuna, atlantic bonito, bigeye tuna, little tunny and skipjack tuna were obtained using the same procedure.
Production and blocking of polyclonal antiserum
Polyclonal antibodies against muscle SP from authentic fresh albacore (referred as anti-ASP antibodies) were raised in New Zealand white male rabbits. Immunisation commenced by subcutaneous injection of the albacore lyophilised protein extract (5 mg) in 0.5 mL of saline solution (9 g of NaCl/L) (Braun Medical, S.A., Barcelona, Spain), emulsified in 0.5 mL of Freund's Complete Adjuvant (Sigma-Aldrich, Saint Louis, MO, USA). Six booster doses made in Freund's Incomplete Adjuvant were applied by subcutaneous injections every 16 days. After 110 days, the rabbits were bled and the blood was allowed to clot for 1 h at room temperature and then overnight at 4 °C. Serum was recovered by centrifugation at 3000 g for 10 min at 4 °C and stored frozen at −20 °C.
Polyclonal anti-ASP antibodies were diluted 1/5000 in phosphate-buffered saline (PBS: 0.14 M ClNa, 0.0015 M KH2PO4, 0.081 M NaHPO4·12H2O, 0.0027 M KCl, pH 7.2) containing 2% Tween 20 (PBST). Antibodies were then made species-specific by mixing them with 0.5 mg/mL of the lyophilised antigenic extracts from the heterologous species atlantic bonito, bullet tuna and bigeye tuna, followed by incubation for 24 h at 37 °C and centrifugation for 10 min at 2500 g and 20 °C. After centrifugation, the supernatant containing the blocked anti-ASP antibodies was used in ELISA indirect assay.
Preparation of fish samples
Liquid extracts containing SP were obtained from authentic fresh samples of each albacore, yellowfin tuna, bullet tuna, atlantic bonito, bigeye tuna, little tunny and skipjack tuna following the previously described procedure for preparation of antigenic extracts, eliminating the lyophilisation step. Filtered liquid extracts were diluted 1/1, 1/10, 1/50 and 1/100 in PBS before they were analysed by the ELISA technique.
In addition, to check the effect of thermal treatments on the technique's ability to discriminate the target species, all authentic fish samples were homogenised in a blender with 1:2 parts of 0.05 M PBS, pH 7.2 (PBS). The homogenates were heated for 30 min in an autoclave at 121 °C, centrifuged at 1000 g and the supernatant was filtered through a Whatman no. 1 filter paper and kept at −20 °C until use in the ELISA procedure (Rencová, Svoboda, & Necidová, Citation2000).
Forty commercial samples consisting in 20 fresh and 20 frozen fillets from different individuals labelled as “albacore” were obtained from a wide representation of local markets and supermarkets in Madrid (Spain). All the samples were transported to the laboratory under controlled conditions. At arrival, they were immediately processed or stored frozen at −20 °C until use.
Indirect ELISA
Flat-bottomed micro-ELISA plates (Costar, Corning, NY, USA) were coated with 0.1 mL of the filtered muscle sample extracts, diluted in PBS, pH 7.2. Plates were incubated for 1 h at 20 °C and wells were then washed five times with PBST (PBS containing 1% Tween 20) before coating them with 0.3 mL of 2% skimmed milk powder in PBS. After 30 min incubation at 20 °C, plates were washed five times with PBST. Then, 0.1 mL aliquots of the blocked anti-ASP antibodies diluted in PBST (1/5000) were added to the wells. The plates were incubated on a plate shaker for 1 h at 20 °C. After washing five more times with PBST to remove heterologous antigens and unattached antibodies, 0.1 mL aliquots of peroxidase-conjugated goat anti-rabbit immunoglobulins (DAKO, DK 2600 Glostrup, Denmark) diluted in PBST (1/2000) were added to the wells. The plates were incubated with shaking for 1 h at 20 °C and wells were washed five more times with distilled water before addition of 0.15 mL of a ready to use substrate solution of 3,3’,5,5’-tetramethylbenzidine (Sigma, Saint Louis, Missouri, USA). After 10 min incubation, the reaction was stopped by the addition of 0.05 mL of 1 M H2SO4 to each well. The yellow colour developed by conversion, and the substrate was measured at 450 nm with a spectrophotometer (DigiScan Reader, Asys Hitech, Austria) using a Digiwin V 3.2 software.
Results and discussion
Fish authenticity and traceability are subjects of great concern to the food industry, as incorrect labelling can represent a commercial fraud with both economic and public health repercussions (Martinsohn, Citation2011). To enforce compliance with legislation, manufacturers and food control agencies are challenged to develop and implement suitable analytical methods for fish identification which can prevent dishonest misdescriptions and enable rapid product withdrawal. Among these methods, ELISA is a simple, effective and flexible means of detecting fish species substitution at any point of the supply chain, providing the trials with speed and automation and reducing the cost of sophisticated equipment (Asensio, Gonzalez, Garcia, & Martin, Citation2008; Fæste & Plassen, Citation2008).
Both polyclonal and monoclonal antibodies have been used in different ELISA-based approaches for fish species identification, each having their own useful advantages and applications. Monoclonal antibodies are a homogeneous population of hybridoma-based antibodies showing defined biological activity and a consistently high specificity (Harlow & Lane, Citation1999). However, polyclonal antibodies may offer improved signal strength by recognition of a mixture of different antigen epitopes, and more tolerance to small changes in the nature of the antigens. Moreover, large quantities are relatively quick and inexpensive to produce compared with monoclonal antibodies. Polyclonal antibodies have been successfully produced against a number of fish targets to be used in different ELISA formats for species identification. These include, for example, antibodies against parvalbumin protein from atlantic cod (Fæste & Plassen, Citation2008), tropomyosin protein from crustacean (Fuller, Goodwin, & Morris, Citation2006) or muscle soluble proteins from different fish species like grouper, wreck fish and Nile perch (Asensio, Citation2007), clams (Fernández et al., Citation2002) and different flat fish species (Cespedes et al., Citation1999). In the present work, we report the development of an indirect ELISA using polyclonal antibodies obtained against albacore native SP, and its application to differentiate this specie from other less commercially valued Scombridae species susceptible to be fraudulently labelled as albacore at the market level.
As can be seen in , the polyclonal anti-ASP antibodies exhibited strong ELISA reactivity against the liquid antigenic extracts from the heterologous fish yellowfin tuna, bullet tuna, atlantic bonito, bigeye tuna, little tunny and skipjack tuna. This result was expected, since the antigenic extract used for immunisation contained all the soluble muscle proteins from albacore and there might be many shared epitopes in the same proteins from other related Scombridae species. Therefore, to be used in the indirect ELISA, the anti-ASP polyclonal antibodies were rendered species-specific by mixing them with lyophilised antigenic extracts from the heterologous species in a process known as “blocking” (Asensio et al., Citation2003; Song, Xue, & Han, Citation2011). After performing several experiments using all the heterologous fish species included in the study, optimal blocking results were obtained by mixing the anti ASP-antibodies (diluted 1/5000) with 0.5 mg/mL of the antigenic extracts from atlantic bonito, bullet tuna and bigeye tuna. Clearly, these three heterologous fish species (atlantic bonito, bullet tuna and bigeye tuna) contain proteins which show the most shared epitopes from the rest of the studied Scombridae species.
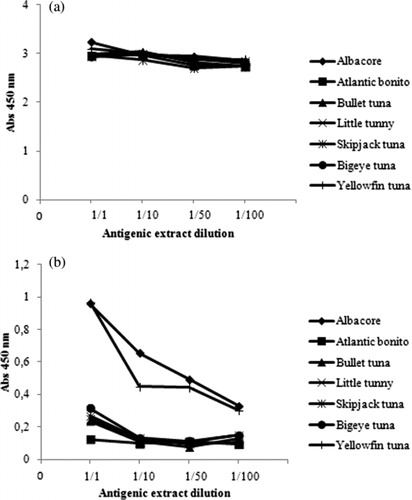
The effectiveness of the blocking procedure for the anti-ASP antibodies is shown in . As can be seen, once the cross-reacting antibodies were blocked by the heterologous proteins, the anti-ASP antiserum recognised the antigenic extract against which it was produced (albacore), although it showed strong cross-reactivity with yellowfin tuna antigenic extract. These results indicated that the blocking of the anti-ASP antibodies with the lyophilised heterologous species antigenic protein extracts eliminated most of the cross-reactions found with heterologous species but it was no sufficient to discriminate albacore from yellowfin tuna. Although this is a disadvantage in devising authenticity tests for albacore, the simplicity, low cost and the short time required during this ELISA test make it suitable for field screening purposes to differentiate albacore and yellowfin tuna from the rest of scombrid species.
On the other hand, the polyclonal antibodies obtained against native muscle SP from authentic albacore did not show ELISA reactivity against heat-treated antigenic extracts from albacore, yellowfin tuna, bullet tuna, atlantic bonito, bigeye tuna, little tunny and skipjack tuna (results not shown), and does not enable its use for the authentication of albacore heat-treated products.
Once the immunoassay conditions were set-up, the applicability of the ELISA technique was further evaluated through the analysis of 40 commercial fish samples (fillets) labelled as albacore (20 fresh and 20 frozen samples), obtained from a wide representation of local marketplaces in Madrid (Spain). Species misdescription at this level should be prevented, since fresh and frozen albacore are considered by fish experts as one of the best choices for many high-quality recipes (Chuang, Chen, & Shiao, Citation2012; Michelini et al., Citation2007). Results obtained using the indirect ELISA described showed 13 samples (32.5%), out of the 40 fish samples analysed, incorrectly assigned (negative values) in the fish market, all of them frozen samples. The absorbance values of negative samples were similar to those of the background readings (0.1–0.2), while positive samples reached values higher than 1.0. However, positive samples (67.5%) could be albacore or yellowfin tuna as the anti-ASP antibodies cannot discriminate between these two species. Therefore, a DNA assay will be necessary to differentiate albacore from yellowfin tuna.
In fact, DNA methods based on PCR technology have been successfully developed for the rapid identification of Scombridae species in raw and processed samples (Chuang, Chen, & Shiao, Citation2012; Michelini et al., Citation2007; Pardo & Pérez-Villareal, Citation2004; Quinteiro et al., Citation1998; Ram, Ram, & Baidoun, Citation1996; Rehbein et al., Citation1999). These include, for example, PCR followed by phylogenetic analysis for the authentication of scombrid species using the mitochondrial cytochrome b and cytochrome c oxidase I as molecular marker (Cawthorn, Steinman, & Witthuhn, Citation2011; Espiñeira, Gonzalez-Lavín, Vieites, & Santaclara, Citation2009); the PCR-restriction fragment length polymorphism (PCR-RFLP) (Aguilar, Alonso, & Barrero, Citation2012; Lin & Hwang, Citation2007); or the real-time PCR (Chuang, Chen, & Shiao, Citation2012).
Despite the fact that the developed ELISA method was unable to distinguish albacore from yellowfin tuna, its application allows fast screening for a high number of samples, reducing considerably the amount of post-DNA analysis for the authentication of albacore specimens. Both procedures, ELISA and PCR-based techniques, are complementary and reliable tools for the specific detection of inappropriately labelled albacore fillets.
References
- Aguilar, A., Alonso, G., & Barrero, M. (2012). Identification of commercial species of tuna (Thunnus spp.) in Venezuela using PCR technique. Revista Cientifica-Facultad De Ciencias Veterinarias, 22, 368–375.
- Armani, A., Castigliego, L., Tinacci, L., Gianfaldoni, D., & Guidi, A. (2012). Multiplex conventional and real-time PCR for fish species identification of bianchetto (juvenile form of Sardina pilchardus), rossetto (Aphia minuta), and icefish in fresh, marinated and cooked products. Food Chemistry, 133, 184–192. doi:10.1016/j.foodchem.2011.12.076
- Asensio, L. (2007). PCR-based methods for fish and fishery products authentication. Trends in Food Science & Technology, 18, 558–566. doi:10.1016/j.tifs.2007.04.016
- Asensio, L., Gonzalez, I., Garcia, T., & Martin, R. (2008). Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA). Food Control, 19(1), 1–8. doi:10.1016/j.foodcont.2007.02.010
- Asensio, L., Gonzalez, I., Pavon, M. A., Garcia, T., & Martin, R. (2008). An indirect ELISA and a PCR technique for the detection of Grouper (Epinephelus marginatus) mislabeling. Food Additives and Contaminants, 25, 677–683. doi:10.1080/02652030701765731
- Asensio, L., Gonzalez, I., Rodriguez, M. A., Hernandez, P. E., Garcia, T., & Martin, R. (2003). Development of a monoclonal antibody for grouper (Epinephelus marginatus) and wreck fish (Polyprion americanus) authentication using an indirect ELISA. Journal of Food Science, 68, 1900–1903. doi:10.1111/j.1365-2621.2003.tb06990.x
- Bauchot, M. L., & Pras, A. (1993). Guía de los peces de mar de España y Europa [Guide sea fish in Spain and Europe]. Barcelona: Ediciones Omega, S.A.
- Berrini, A., Tepedino, V., Borromeo, V., & Secchi, C. (2006). Identification of freshwater fish commercially labelled “perch” by isoelectric focusing and two-dimensional electrophoresis. Food Chemistry, 96, 163–168. doi:10.1016/j.foodchem.2005.04.007
- Bonwick, G. A., & Smith, C. J. (2004). Immunoassays: Their history, development and current place in food science and technology. International Journal of Food Science & Technology, 39, 817–827. doi:10.1111/j.1365-2621.2004.00855.x
- Bottero, M. T., & Dalmasso, A. (2011). Animal species identification in food products: Evolution of biomolecular methods. The Veterinary Journal, 190(1), 34–38. doi:10.1016/j.tvjl.2010.09.024
- Carrera, E., García, T., Céspedes, A., González, I., Sanz, B., Hernández, P. E., & Martín, R. (1998). Identification of Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) by using polymerase chain reaction amplification and restriction analysis of the mitochondrial cytochrome b gene. Journal of Food Protection, 61, 482–486.
- Carrera, E., Martín, R., García, T., González, I., Sanz, B., & Hernández, P. E. (1996). Development of an enzyme-linked immunosorbent assay for the identification of smoked salmon (Salmo salar), trout (Oncorhynchus mykiss) and bream (Brama raii). Journal of Food Protection, 59, 521–524.
- Cawthorn, D. M., Steinman, H. A., & Witthuhn, R. (2011). Establishment of a mitochondrial DNA sequence database for the identification of fish species commercially available in South Africa. Molecular Ecology Resources, 11, 979–991. doi:10.1111/j.1755-0998.2011.03039.x
- Cespedes, A., Garcia, T., Carrera, E., Gonzalez, I., Fernandez, A., Asensio, L., … Martin, R. (1999). Indirect enzyme-linked immunosorbent assay for the identification of sole (Solea solea), European plaice (Pleuronectes platessa), flounder (Platichthys flesus), and Greenland halibut (Reinhardtius hippoglossoides). Journal of Food Protection, 62, 1178–1182.
- Chuang, P. S., Chen, M. I., & Shiao, J. C. (2012). Identification of tuna species by a real-time polymerase chain reaction technique. Food Chemistry, 133, 1055–1061. doi:10.1016/j.foodchem.2012.01.076
- Dalmasso, A., Fontanella, E., Piatti, P., Civera, T., Secchi, C., & Bottero, M. T. (2007). Identification of four tuna species by means of real-time PCR and melting curve analysis. Veterinary Research Communications, 31, 355–357. doi:10.1007/s11259-007-0036-1
- Dreyfuss, M. S., Cutrufelli, M. E., Mageau, R. P., & McNamara, A. M. (1997). Agar-gel immunodiffusion test for rapid identification of pollock surimi in raw meat products. Journal of Food Science, 62, 972–975. doi:10.1111/j.1365-2621.1997.tb15018.x
- Espiñeira, M., Gonzalez-Lavín, N., Vieites, J. M., & Santaclara, F. J. (2009). Development of a method for the identification of scombroid and common substitute species in seafood products by FINS. Food Chemistry, 117, 698–704. doi:10.1016/j.foodchem.2009.04.087
- European Commission. (2001). Regulation (EC), No. 2065/2001. Laying down detailed rules for the application of Council Regulation (EC) No. 104/2000 as regards informing consumers about fishery and aquaculture products. Official Journal of the European Communities, L278, 6–8.
- European Council. (2000). Regulation (EC) No. 104/2000. On the common organisation of the markets in fishery and aquaculture products. Official Journal of the European Communities, L17, 22–52.
- European Parlamient and Council. (2002). Regulation (EC) No. 178/2002. Laying down the general principles and requirements of food law, establishing the European Food Safety Authority and laying down procedures in matters of food safety. Official Journal of the European Communities, L31, 1–24.
- Fæste, C. K., & Plassen, C. (2008). Quantitative sandwich ELISA for the determination of fish in foods. Journal of Immunological Methods, 329, 45–55. doi:10.1016/j.jim.2007.09.007
- Fernández, A., García, T., Asensio, L., Rodríguez, M. A., González, I., Lobo, E., … Martín, R. (2002). Identification of the clam species Ruditapes decussatus (grooved carpet shell), Venerupis romboides (yellow carpet shell) and Venerupis pullastra (pullet carpet shell) by ELISA. Food and Agricultural immunology, 14, 65–71. doi:10.1080/09540100220137673
- Fuller, H. R., Goodwin, P. R., & Morris, G. E. (2006). An enzyme-linked immunosorbent assay (ELISA) for the major crustacean allergen, tropomyosin, in food. Food and Agricultural Immunology, 17, 43–52. doi:10.1080/09540100600572651
- Gill, T. A. (1997). Advanced analytical tools in seafood science. In J. B. Luten, T. Borresen, & J. Oehlenschlager (Eds.), Seafood from producer to consumer, integrated approach to quality (vol. 38, pp. 479–490). Amsterdam: Elsevier Science Bv.
- Harlow, E., & Lane, D. (1999). Using antibodies. A laboratory manual. New York, NY: Cold Spring Harbor Laboratory Press.
- Jacquet, J. L., & Pauly, D. (2008). Trade secrets: Renaming and mislabeling of seafood. Marine Policy, 32, 309–318. doi:10.1016/j.marpol.2007.06.007
- Le Fresne, S., Popova, M., Le Vacon, F., & Carton, T. (2011). Application of denaturing high-performance liquid chromatography (DHPLC) for the identification of fish: A new way to determine the composition of processed food containing multiple species. Journal of Agricultural and Food Chemistry, 59, 12302–12308. doi:10.1021/jf2030242
- Lin, W. F., & Hwang, D. F. (2007). Application of PCR-RFLP analysis on species identification of canned tuna. Food Control, 18, 1050–1057. doi:10.1016/j.foodcont.2006.07.001
- Martinez, I., Šližytė, R., & Daukšas, E. (2007). High resolution two-dimensional electrophoresis as a tool to differentiate wild from farmed cod (Gadus morhua) and to assess the protein composition of klipfish. Food Chemistry, 102, 504–510. doi:10.1016/j.foodchem.2006.03.037
- Martinsohn, J. T. (2011). Fish species identification. In JRC European Commission (Ed.), Deterring illegal activities in the fisheries sector. Genetics, genomics, chemistry and forensics to fight IUU fishing and in support of fish product traceability (pp. 32–33). Luxemburg: Publications Office of the European Union.
- Michelini, E., Cevenini, L., Mezzanotte, L., Simoni, P., Baraldini, M., De Laude, L., & Roda, A. (2007). One-step triplex-polymerase chain reaction assay for the authentication of yellowfin (Thunnus albacares), bigeye (Thunnus obesus), and skipjack (Katsuwonus pelamis) tuna DNA from fresh, frozen, and canned tuna samples. Journal of Agricultural and Food Chemistry, 55, 7638–7647. doi:10.1021/jf070902k
- Pardo, M. A., & Pérez-Villareal, B. (2004). Identification of commercial canned tuna species by restriction site analysis of mitochondrial DNA products obtained by nested primer PCR. Food Chemistry, 86, 143–150. doi:10.1016/j.foodchem.2003.09.024
- Quinteiro, J., Sotelo, C. G., Rehbein, H., Pryde, S. E., Medina, I., Pérez-Martín, R. I., … Mackie, I. M. (1998). Use of mtDNA direct polymerase chain reaction (PCR) sequencing and PCR-restriction fragment length polymorphism methodologies in species identification of canned tuna. Journal of Agricultural and Food Chemistry, 46, 1662–1669. doi:10.1021/jf970552+
- Ram, J. L., Ram, M. L., & Baidoun, F. F. (1996). Authentication of canned tuna and bonito by sequence and restriction site analysis of polymerase chain reaction products of mitochondrial DNA. Journal of Agricultural and Food Chemistry, 44, 2460–2467. doi:10.1021/jf950822t
- Rehbein, H., Mackie, I. M., Pryde, S., Gonzales-Sotelo, C., Medina, I., Perez-Martin, R., … Rey-Mendez, M. (1999). Fish species identification in canned tuna by PCR-SSCP: Validation by a collaborative study and investigation of intra-species variability of the DNA-patterns. Food Chemistry, 64, 263–268. doi:10.1016/S0308-8146(98)00125-3
- Rencová, E., Svoboda, I., & Necidová, L. (2000). Identification by ELISA of poultry, horse, kangaroo, and rat muscle specific proteins in heat-processed products. Veterinary Medicine, 45, 353–356.
- Song, H., Xue, H., & Han, Y. (2011). Detection of cow's milk in Shaanxi goat's milk with an ELISA assay. Food Control, 22, 883–887. doi:10.1016/j.foodcont.2010.11.019
- Taylor, W. J., Patel, N. P., & Jones, J. L. (1994). Antibody based methods for assessing seafood authenticity. Food and Agricultural Immunology, 6, 305–314. doi:10.1080/09540109409354842
