Abstract
A sensitive and specific monoclonal antibody-based sandwich enzyme-linked immunosorbent assay (ELISA) was established to detect the Cronobacter sakazakii. A pair of monoclonal antibodies (mAbs) selected from mAbs produced by 11 murine hybridomas was selected for the sandwich ELISA procedure. Targets of two mAbs were 100 kDa and 42 kDa protein extracted from the bacteria, respectively, which were proved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting analysis. The limit of detection of this method was established as 1 × 104 cfu/mL, and the linear range from 1 × 105 to 1 × 108 cfu/mL. In the real sample test, 1 cfu/g C. sakazakii was detected in artificially contaminated powdered infant formula with 4 h enrichment.
Introduction
Cronobacter spp., formerly Enterobacter sakazakii (Nazarowec-White & Farber, Citation1997; Steigerwalt, Fanning, Fife-Asbury, & Brenner, Citation1976), were suggested to be created as new genus at 2007 (Iversen et al., Citation2007, Citation2008) which contain six species: Cronobacter sakazakii, Cronobacter malonaticus, Cronobacter turicensis, Cronobacter muytjensii, Cronobacter dublinensis (contain three subspecies), and Cronobacter genomospecies 1. In 2012, new suggestion was proposed to divide the genus into seven kinds and belong to enterobacteriaceae (Joseph, Cetinkaya, et al., Citation2012; Joseph, Sonbol, et al., Citation2012).
The Cronobacter spp. were motile, peritrichous, gram-negative bacillus. As an opportunistic food-borne pathogen, the Cronobacter spp. were frequently reported to cause severe neonatal meningitis (Arseni, Malamou-Ladas, Koutsia, Xanthou, & Trikka, Citation1987; Lai, Citation2001), enterocolitis (van Acker et al., Citation2001), and bacteremia (Noriega, Kotloff, Martin, & Schwalbe, Citation1990), leading to 40–80% mortality in infants and immunocompromised populations (Ruan, Li, Liu, Li, & Li, Citation2013). Because all the species of Cronobacter contain the clinical isolates and all the Cronobacter spp. could not been proved harmless to infant and immunocompromised populations, people considered all of the genus were pathogenic.
Conventional biochemical detection methods are based on the characteristic enzymatic properties of Cronobacter spp. (α-glycosidase-positive, oxidase-negative, and catalase-positive), and generation of yellow colonies (Iversen, Waddington, On, & Forsythe, Citation2004; Muytjens & Van Druten, Citation1984). In recent years, several molecular biological techniques have been applied for pathogen detection, including reverse transcription-PCR El-Sharoud, Darwish, & Batt, Citation2013), real-time PCR (Cai et al., Citation2013; Liu et al., Citation2006), PCR-enzyme-linked immunosorbent assay (PCR-ELISA; Li et al., Citation2013), loop-mediated isothermal amplification Liu, Zheng, Zhang, Hou, & Liu, Citation2009), and fluorescence in situ hybridization (Almeida et al., Citation2009). And biosensor techniques have additionally been employed to detect Cronobacter spp. (Dou, Tang, & Zhao, Citation2013; Zhang, Dou, Zhan, & Zhao, Citation2012).
Cronobacter spp. also could produce enterotoxin, which is a potential risk to neonates (Pagotto, Nazarowec-White, Bidawid, & Farber, Citation2003). Further studies on enterotoxin are proposed to facilitate effective detection of the bacterium (Raghav & Aggarwal, Citation2007).
Traditional biochemical detection methods are time- and labor-consuming and may provide inaccurate results. Detection methods based on DNA require extraction of the DNA strand and rely heavily on specific testing instruments and professional operators, rendering them unsuitable for field detection of large numbers of samples. Establishment of accurate and rapid detection methods for Cronobacter spp. is a considerable challenge.
ELISAs are simple, sensitive, and rapid methods commonly used for pathogen detection (Park et al., Citation2012; Xu et al., Citation2014). The sensitivity of ELISA method relied on the stability and sensitivity of quality antibody. In this study, we developed a rapid, sensitive, and cost-effective quantitation method for C. sakazakii based on a pair of monoclonal antibodies (mAbs) and successfully applied to detection of the pathogen in powdered infant formula.
Materials and methods
Bacteria strains and chemicals
Strains used for this study (C. sakazakii ATCC 29544, ATCC 29004, IQCC 10403.20, C. muytjensii ATCC 51329, three Escherichia coli, one Listeria, one Salmonella and one Staphylococcus aureus) were purchased from the China Center of Industrial Culture Collection (CICC), China Medical Culture Collection (CMCC), China Academy of Inspection and Quarantine of food safety of Microbial Culture Collection Management Center (IQCC), and American Type Culture Collection (ATCC, USA). Details of the strains are presented in Gelatin was purchased from Beijing BioDee Biotechnology Co., Ltd. (Beijing, China). Freund's Complete/Incomplete Adjuvant, 3,3′,5,5′-tetramethylbenzidine (TMB) and horseradish peroxidase (HRP) were acquired from Sigma (St. Louis, MO, USA). Enzyme immunoassay-grade HRP-labeled goat anti-mouse immunoglobulin was obtained from Hua Mei Co., Ltd (Shanghai, China). Bacterial protein extraction reagent (B-PER) was obtained from Thermo Scientific (Waltham, MA, USA). Sediment type TMB substrate solution was obtained from Tiangen Biotech Co., Ltd (Beijing, China). Powdered infant formula was purchased from a local supermarket. Modified lauryl sulfate tryptose broth-vancomycin medium (mLST-Vm, vancomycin 10 µg/mL) was obtained from Beijing Land Bridge Technology Co., Ltd (Beijing, China). TSA agar medium and nutrient agar medium were obtained from Hangzhou Microbial Reagent Co., Ltd. (Hangzhou, China). All other reagents and chemicals were from the National Pharmaceutical Group Chemical Reagent Co., Ltd. (Shanghai, China).
Table 1. The bacterial strains tested in this study.
Buffers and solutions
The following solutions were used for ELISA: (1) 0.05 M sodium carbonate buffer (pH 9.6) as coating buffer; (2) 0.2% (w/v) gelatin in coating buffer as blocking buffer; (3) 0.01 M phosphate-buffered saline containing 0.05% (v/v) Tween 20 (PBST, pH 7.4) as washing and standard dilution buffer; (4) 0.01 M phosphate-buffered saline containing 0.1% (w/v) gelatin as antibody dilution buffer; (5) 0.1 M citrate phosphate buffer (pH 5.0) containing 180 µL of 30% H2O2 (A solution) and ethylene glycol substrate solution containing 0.06% (w/v) 3,3′,5,5′ -TMB (B solution) mixed at a ratio of 5:1 as substrate solution, and (6) 2 M sulfuric acid as stop reagent
Preparation of monoclonal antibody against C. sakazakii
Preparation of strains standards
Cronobacter spp. strains were initially activated in modified lauryl sulfate tryptose broth-vancomycin medium (mLST-Vm, vancomycin 10 µg/mL) at 44°C for 24 h and inoculated on TSA agar at 37°C for 18–24 h. Inoculation was performed with one colony in nutrient broth at 37°C for 16 h. Salmonella, E. coli, Listeria species, and S. aureus were cultured at 37°C on nutrient agar, and a single colony of each species separately inoculated into buffered peptone water and 10% NaCl tryptic soy broth, followed by culture at 37°C. In all cases, the microorganisms obtained were counted, treated with boiling water for 30 min for inactivation, and adjusted the concentrations to 1010 cfu/mL by stroke-physiological saline solution as the standard.
Immunization procedure
As the type strain of C. sakazakii, we selected the C. sakazakii ATCC 29544 as the immunogen. At the first immunization, heat killed C. sakazakii ATCC 29544 was diluted by stroke-physiological and mixed with the same volume of Freund's Complete Adjuvant to a final concentration of 109 cfu/mL, and at the next immunization, mixed with the same volume of Freund's Incomplete Adjuvant to the same final concentration. Five female BALB/c mice (six weeks old) were subcutaneously injected with 150 µL of the 1:1 (v/v) mixture (109 cfu/mL), followed by administration of 100 µL antigen mixture every three weeks until high serum antibody titer was achieved, determined using indirect ELISA (Feng et al., Citation2013).
Preparation of mAbs
The mouse displaying the highest serum antibody titer was eyeball extirpation and spleen cells fused with SP2/0 myeloma cells. Positive hybridoma cell lines were obtained via indirect ELISA screening. After subcloning three times, 11 different cell lines of positive hybridomas were obtained. All mAbs were purified from ascites and conjugated with HRP using the sodium periodate method (Boorsma & Kalsbeek, Citation1975).
The ELISA procedure
Indirect ELISA
The indirect ELISA method was used to detect serum titers and screen positive hybridoma cell lines. Each well of the 96-well polystyrene ELISA plates was coated with 100 µL of inactivated C. sakazakii ATCC 29544 (107 cfu/mL) in coating buffer at 37°C for 2 h. Plates were rinsed thoroughly with 200 µL washing buffer three times and tapped dry, followed by treatment with 200 µL blocking buffer at 37°C for 2 h. The wash procedure was repeated and 100 µL of cell supernatant or mouse serum in antibody dilution buffer added to each well, followed by incubation at 37°C for 30 min. After washing, 100 µL of HRP-labeled goat anti-mouse immunoglobulin was added to each well, diluted 1:3000 with antibody dilution buffer, and incubated at 37°C for 30 min. Cells were washed, incubated in 100 µL of substrate solution, and reacted at 37°C for 15 min in the dark. Finally, 50 µL stop reagent was added to each well, and absorbance measured at 450 nm by microplate reader (BioTek, Winooski, VT, USA).
Monoclonal sandwich ELISA
The following monoclonal sandwich ELISA procedure was used to detect the pathogen and infant milk samples. Each well of the 96-well polystyrene ELISA plates was coated with 100 µL of capture mAb in coating buffer at 37°C for 2 h. After incubation, plates were rinsed thoroughly with 200 µL washing buffer and tapped dry three times, followed by blocking with 200 µL blocking buffer at 37°C for 2 h. The wash procedure was repeated, and inactivated bacterial standards or infant milk samples in 100 µL PBST added to each well, followed by incubation at 37°C for 1 h. The wash procedure was repeated and 100 µL detection mAb conjugated with HRP added to each well at 37°C for 1 h. After another wash step, 100 µL of substrate solution was added to individual wells and reacted at 37°C for 15 min in the dark. Finally, 50 µL stop reagent was added to the wells, and absorbance measured at 450 nm by microplate reader (Kuang et al., Citation2013; Peng et al., Citation2014).
Establishment of the sandwich ELISA method
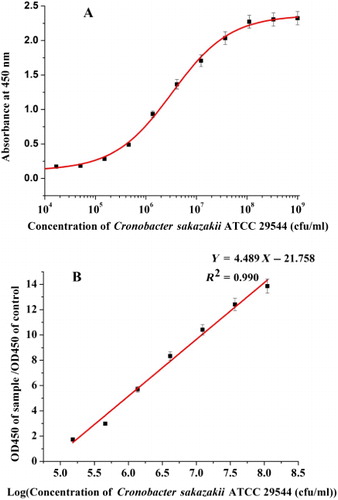
To establish the sandwich ELISA method, mAbs obtained from the previous experiment were conjugated with HRP and used as the capture and detection antibodies, with a view to selecting the optimal antibody combination for detection. After pairwise interaction analysis, combinations providing the highest positive/negative value (P/N) were selected (P/N is the ratio of the optical density values of the positive test sample to negative sample). Following the further optimization of coating buffer, blocking buffer, antibody dilution, and other conditions, the optimal combination of capture and detection antibodies was selected for development of the sandwich ELISA method. Based on optimization results, a standard curve was generated with OD450 value as the ordinate and concentrations of microorganism standards as the abscissa.
Cross-reactivity of the sandwich ELISA method
Four Cronobacter strains and six other pathogenic strains were tested by the method at a concentration of 108 cfu/mL.
Electrophoresis and immunoblotting
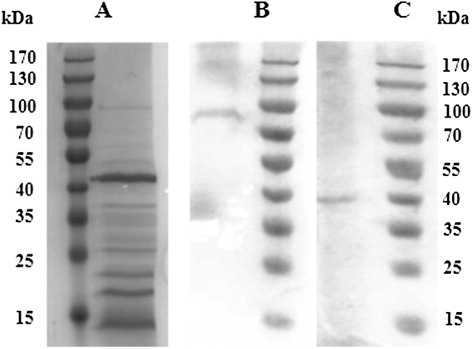
The properties of antibodies in sandwich ELISA method were analyzed using SDS-PAGE and immunoblotting methods. SDS-PAGE method was based on the performed description (Laemmli, Citation1970). Five percent stacking gel and 8% resolving gel were used in a mini-protean tetra system (Bio-Rad, Hercules, Cal, USA). The samples for SDS-PAGE were the proteins extracted from bacteria using B-PER reagent as follows. Pellet bacterial cells by centrifugation at 5000 rpm for 10 min. Add 4 mL of B-PER reagent per gram of cell pellet, pipette the suspension up and down until it is homogeneous. Incubate 10–15 min at room temperature. Centrifuge lysate at 15,000 rpm for 5 min to separate soluble proteins from the insoluble proteins. Both proteins were tested using the indirect ELISA based on the antibodies selected for the sandwich ELISA method. The target proteins were incubated with equal volume of loading buffer at 100°C for 10 min and loaded 10 ul into each well of the gel, then electrophoresis at 120 V for 90 min. Gels were stained using Coomassie blue and used for immunoblotting, respectively. The proteins transferred to polyvinylidene fluoride membranes by Trans-Blot semi-dry transfer cell (Bio-Rad, Hercules, Cal, USA; Lin et al., Citation2006). The membrane was blocked with 2.5% Bovine serum albumin (BSA) (w/v) in TBS (Tris buffer solution) containing 0.02% Tween-20 at room temperature for 2 h. Then the membrane incubated at 37°C with specific mAb in TBS containing 1% BSA (w/v) for 1 h and developed with goat anti-mouse-HRP in TBS containing 1% BSA (w/v) for 1 h. Sediment type TMB substrate solution was used for the coloring step.
Detection of powdered infant formula samples
The powdered infant formula tested using conventional method for Cronobacter to assess for environmental contamination (reference standard GB4789.40–2010). A measure of 10 g of negative powdered infant formula dissolved in 90 mL buffer peptone water and spiked with 1, 10, 103, or 104 cfu C. sakazakii ATCC 29544 and enrichment at 37°C for 4 h. A measure of 1 mL suspension of culture medium was centrifuged at 4°C and 10,000 rpm for 15 min. The sediment was re-dissolved in 100 µL PBST and tested using both optimized monoclonal sandwich ELISAs and conventional plate culture method (reference standard GB4789.40-2010).
Results and discussion
Pairwise interaction analysis
After obtaining 11 positive hybridoma cell lines, 9 combinations with higher P/N values (≥10.0) were selected for further optimization according to the pairwise interaction analysis (). With the same tested concentration of ATCC 29544 at 108 cfu/mL, the combinations taken higher P/N value may be more sensitive.
Table 2. The sandwich ELISA for pair-wise interaction analysis (P/N value).
Specificity of sandwich ELISA
All 10 strains samples were tested in the sandwich ELISA method based on 9 different combinations at the concentration of 108 cfu/mL. Just one combination was chosen with the great specificity. The mAb K13 was selected as the capture antibody and K15 as the detection antibody. Cross-reactivity with all the other no-C. sakazakii strains was not significant with this antibody combination, as shown in (P/N values < 2.1). These results demonstrated that the established sandwich ELISA could be used to detect C. sakazakii depended on the mAbs used in the method which were specific to the common antigens epitopes shared by C. sakazakii.
Table 3. The strains tested by optimized sandwich ELISA (n = 8).
Optimization of sandwich ELISA
With the establishment of the antibody combination, further optimization was taken. Sandwich ELISA with coating buffer (0.05 M sodium carbonate buffer, pH 9.6), blocking buffer (0.01 M phosphate-buffered saline containing 0.2% polyethylene glycol 20000), and standard dilution buffer (0.02 M phosphate-buffered saline containing 0.05% (v/v) Tween 20, pH 7.2) disclosed a linear dynamic range of 1 × 105 cfu/mL–1 × 108 cfu/mL ( and ). The limit of detection of C. sakazakii ATCC 29544 was 1 × 104 cfu/mL, based on the mean and three standard deviations of absorbance at 450 nm of negative samples (). The linear regression equation was y = 4.489x − 21.758 and linear correlation coefficient (R 2) was 0.990 ().
Note: The linear regression equation was y = 4.489x – 21.758. The linear correlation coefficient (R2) was 0.990.
Electrophoresis and immunoblotting analysis
The soluble proteins and insoluble proteins were tested by the indirect ELISA. The result shown that only soluble proteins could be identified by both mAb K13 and K15. By SDS-PAGE, the soluble proteins extracted from bacteria were ranged from 14k Da to 100 kDa, and the major components were 42 kDa protein, 22 kDa protein, 18 kDa protein, and 14 kDa protein (). In immunoblot analysis, the 100 kDa protein and 42 kDa protein were proved to be the target substances of mAb K13 and K15, respectively (, ). Other mAbs were tested by immunoblot but some exhibiting no reactivity in immunoblot. These phenomena may be dependent on the change of proteins steric configuration in electrophoresis under denaturing conditions or failed transfer process onto the membranes (Kathariou, Mizumoto, Allen, Fok, & Benedict, Citation1994).
The method developed by us was different from previous studies. Cruz-Córdova had studied the properties and pathogenesis of flagellum protein extracted from five different Cronobacter species (Cruz-Córdova et al., Citation2012). But the instability of flagellum on the surface of bacteria made the method based on flagellum test hard to achieve. Polyclonal antibody (PcAb) had been performed using sandwich ELISA method (Wang, Du, Lu, & Wang, Citation2013). Compared with PcAb, the properties of mAb were more stable and reliable on the mass production of marketization. In the current investigation, we successfully established a rapid sandwich ELISA method for the specific and sensitive detection of C. sakazakii. Exploited different epitopes led less cross-reactivity with other pathogenic in our method. And the abundant epitopes on the bacterial surface enhanced the sensitivity of our method relative to other available techniques.
Detection of C. sakazakii in artificially contaminated powdered infant formula samples
In our study, we detected the artificially contaminated powdered infant formula sample by the optimized monoclonal sandwich ELISA method and conventional culture method. The results are presented in While C. sakazakii ATCC 29544 (1 cfu/mL) was detected in powdered infant formula using both conventional and optimized sandwich ELISA methods after 4 h enrichment, our protocol required significantly less time than the 48–96 h required by the conventional plate culture method.
Table 4. The results of detecting C. sakazakii in powdered infant formula (n = 8).
Conclusions
A novel, rapid, and reliable method for detection of C. sakazakii in powdered infant formula was established in this study. The monoclonal antibody pair, K13 and K15, served as the capture and detection antibodies, respectively, and based on different epitopes of the pathogen. The limit of detection was 1 × 104 cfu/mL, and the linear range established as 1 × 105 cfu/mL–1 × 108 cfu/mL. In powdered infant formula samples, 1 cfu/g C. sakazakii could be detected after 4 h enrichment. The cross-reactivity experiment established the high specificity of the assay. We conclude that the newly developed sandwich ELISA procedure provides a rapid sensitive and reliable method for detecting C. sakazakii contamination in powdered infant milk formula.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Almeida, C., Azevedo, N. F., Iversen, C., Fanning, S., Keevil, C. W., & Vieira, M. J. (2009). Development and application of a novel peptide nucleic acid probe for the specific detection of Cronobacter genomospecies (Enterobacter sakazakii) in powdered infant formula. Applied and Environmental Microbiology, 75, 2925–2930. doi:10.1128/AEM.02470-08
- Arseni, A., Malamou-Ladas, E., Koutsia, C., Xanthou, M., & Trikka, E. (1987). Outbreak of colonization of neonates with Enterobacter sakazakii. Journal of Hospital Infection, 9(2), 143–150. doi:10.1016/0195-6701(87)90052-1
- Boorsma, D. M., & Kalsbeek, G. L. (1975). A comparative study of horseradish peroxidase conjugates prepared with a one-step and a two-step method. Journal of Histochemistry & Cytochemistry, 23, 200–207. doi:10.1177/23.3.47869
- Cai, X.-Q., Yu, H.-Q., Ruan, Z.-X., Yang, L.-L., Bai, J.-S., Qiu, D.-Y., … Zhu, X.-Q. (2013). Rapid detection and simultaneous genotyping of Cronobacter spp. (formerly Enterobacter sakazakii) in powdered infant formula using real-time PCR and high resolution melting (HRM) analysis. PloS One, 8(6), e67082. doi:10.1371/journal.pone.0067082.t001
- Cruz-Córdova, A., Rocha-Ramírez, L. M., Ochoa, S. A., Gónzalez-Pedrajo, B., Espinosa, N., Eslava, C., … Valencia-Mayoral, P. (2012). Flagella from five Cronobacter species induce pro-inflammatory cytokines in macrophage derivatives from human monocytes. PLoS ONE, 7(12), e52091.
- Dou, W., Tang, W., & Zhao, G. (2013). A disposable electrochemical immunosensor arrays using 4-channel screen-printed carbon electrode for simultaneous detection of Escherichia coli O157:H7 and Enterobacter sakazakii. Electrochimica Acta, 97, 79–85. doi:10.1016/j.electacta.2013.02.136
- El-Sharoud, W. M., Darwish, M. S., & Batt, C. A. (2013). A real-time PCR-based microfluidics platform for the detection of Cronobacter sakazakii in reconstituted milks. International Dairy Journal, 33(1), 67–74. doi:10.1016/j.idairyj.2013.06.010
- Feng, M., Yong, Q., Wang, W., Kuang, H., Wang, L., & Xu, C. (2013). Development of a monoclonal antibody-based ELISA to detect Escherichia coli O157:h77. Food and Agricultural Immunology, 24, 481–487. doi:10.1080/09540105.2012.716026
- Iversen, C., Lehner, A., Mullane, N., Bidlas, E., Cleenwerck, I., Marugg, J., … Joosten, H. (2007). The taxonomy of Enterobacter sakazakii: Proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evolutionary Biology, 7(1), 64. doi:10.1186/1471-2148-7-64
- Iversen, C., Mullane, N., McCardell, B., Tall, B. D., Lehner, A., Fanning, S., … Joosten, H. (2008). Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. International Journal of Systematic and Evolutionary Microbiology, 58, 1442–1447. doi:10.1099/ijs.0.65577-0
- Iversen, C., Waddington, M., On, S. L. W., & Forsythe, S. (2004). Identification and phylogeny of Enterobacter sakazakii relative to Enterobacter and Citrobacter species. Journal of Clinical Microbiology, 42, 5368–5370. doi:10.1128/JCM.42.11.5368-5370.2004
- Joseph, S., Cetinkaya, E., Drahovska, H., Levican, A., Figueras, M. J., & Forsythe, S. J. (2012). Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. International Journal of Systematic and Evolutionary Microbiology, 62, 1277–1283.
- Joseph, S., Sonbol, H., Hariri, S., Desai, P., McClelland, M., & Forsythe, S. J. (2012). Diversity of the Cronobacter genus as revealed by multilocus sequence typing. Journal of Clinical Microbiology, 50, 3031–3039. doi:10.1128/JCM.00905-12
- Kathariou, S., Mizumoto, C., Allen, R., Fok, A., & Benedict, A. (1994). Monoclonal antibodies with a high degree of specificity for Listeria monocytogenes serotype 4b. Applied and Environmental Microbiology, 60, 3548–3552.
- Kuang, H., Wang, W., Xu, L., Ma, W., Liu, L., Wang, L., & Xu, C. (2013). Monoclonal antibody-based sandwich ELISA for the detection of staphylococcal enterotoxin A. International Journal of Environmental Research and Public Health, 10, 1598–1608. doi:10.3390/ijerph10041598
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
- Lai, K. K. (2001). Enterobacter sakazakii infections among neonates, infants, children, and adults: Case reports and a review of the literature. Medicine, 80(2), 113–122. doi:10.1097/00005792-200103000-00004
- Li, Y., Cao, L., Zhang, C., Chen, Q., Lu, F., Bie, X., & Lu, Z. (2013). Development and evaluation of a PCR-ELISA assay for the detection and quantification of Cronobacter spp. International Dairy Journal, 33(1), 27–33. doi:10.1016/j.idairyj.2013.06.009
- Lin, M., Todoric, D., Mallory, M., Luo, B. S., Trottier, E., & Dan, H. (2006). Monoclonal antibodies binding to the cell surface of Listeria monocytogenes serotype 4b. Journal of Medical Microbiology, 55, 291–299.
- Liu, Y., Cai, X., Zhang, X., Gao, Q., Yang, X., Zheng, Z., … Huang, X. (2006). Real time PCR using TaqMan and SYBR Green for detection of Enterobacter sakazakii in infant formula. Journal of Microbiological Methods, 65(1), 21–31. doi:10.1016/j.mimet.2005.06.007
- Liu, C., Zheng, W., Zhang, H., Hou, Y., & Liu, Y. (2009). Sensitive and rapid detection of Enterobacter sakazakii in infant formula by loop-mediated isothermal amplification method. Journal of Food Safety, 29(1), 83–94. doi:10.1111/j.1745-4565.2008.00142.x
- Muytjens, H. L., & Van Druten, H. (1984). Enzymatic profiles of Enterobacter sakazakii and related species with special reference to the alpha-glucosidase reaction and reproducibility of the test system. Journal of Clinical Microbiology, 20, 684–686.
- Nazarowec-White, M., & Farber, J. (1997). Enterobacter sakazakii: A review. International Journal of Food Microbiology, 34(2), 103–113. doi:10.1016/S0168-1605(96)01172-5
- Noriega, F., Kotloff, K., Martin, M., & Schwalbe, R. (1990). Nosocomial bacteremia caused by Enterobacter sakazakii and Leuconostoc mesenteroides resulting from extrinsic contamination of infant formula. Pediatric Infectious Disease Journal, 9, 447–448. doi:10.1097/00006454-199006000-00018
- Pagotto, F. J., Nazarowec-White, M., Bidawid, S., & Farber, J. M. (2003). Enterobacter sakazakii: Infectivity and enterotoxin production in vitro and in vivo. Journal of Food Protection, 66, 370–375.
- Park, S., Shukla, S., Kim, Y., Oh, S., Hun Kim, S., & Kim, M. (2012). Development of sandwich enzyme-linked immunosorbent assay for the detection of Cronobacter muytjensii (formerly called Enterobacter sakazakii). Microbiology and Immunology, 56, 472–479. doi:10.1111/j.1348-0421.2012.00466.x
- Peng, J., Meng, X., Deng, X., Zhu, J., Kuang, H., & Xu, C. (2014). Development of a monoclonal antibody-based sandwich ELISA for the detection of ovalbumin in foods. Food and Agricultural Immunology, 25(1), 1–8. doi:10.1080/09540105.2012.716398
- Raghav, M., & Aggarwal, P. K. (2007). Purification and characterization of Enterobacter sakazakii enterotoxin. Canadian Journal of Microbiology, 53, 750–755. doi:10.1139/W07-037
- Ruan, J., Li, M., Liu, Y.-P., Li, Y.-Q., & Li, Y.-X. (2013). Rapid and sensitive detection of Cronobacter spp. (previously Enterobacter sakazakii) in food by duplex PCR combined with capillary electrophoresis–laser-induced fluorescence detector. Journal of Chromatography B, 921–922, 15–20.
- Steigerwalt, A. G., Fanning, G. R., Fife-Asbury, M. A., & Brenner, D. J. (1976). DNA relatedness among species of Enterobacter and Serratia. Canadian Journal of Microbiology, 22(2), 121–137. doi:10.1139/m76-018
- van Acker, J., de Smet, F., Muyldermans, G., Bougatef, A., Naessens, A., & Lauwers, S. (2001). Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. Journal of Clinical Microbiology, 39, 293–297. doi:10.1128/JCM.39.1.293-297.2001
- Wang, J., Du, X.-J., Lu, X.-N., & Wang, S. (2013). Immunoproteomic identification of immunogenic proteins in Cronobacter sakazakii strain BAA-894. Applied Microbiology and Biotechnology, 97, 2077–2091. doi:10.1007/s00253-013-4720-5
- Xu, X., Zhang, Y., Shi, M., Sheng, W., Du, X., Yuan, M., & Wang, S. (2014). Two novel analytical methods based on polyclonal and monoclonal antibodies for the rapid detection of Cronobacter spp.: Development and application in powdered infant formula. LWT – Food Science and Technology, 56, 335–340.
- Zhang, X., Dou, W., Zhan, X., & Zhao, G. (2012). A novel immunosensor for Enterobacter sakazakii based on multiwalled carbon nanotube/ionic liquid/thionine modified electrode. Electrochimica Acta, 61, 73–77. doi:10.1016/j.electacta.2011.11.092
