Abstract
An immunochromatographic lateral flow strip test using the competitive format was developed for the determination of chlorothalonil (CTN) residues in cucumber. The limit of detection was less than 100 ng/mL by visual assessment and was found to be 91.78 ± 0.17 ng/mL for quantitative detection using a test strip reader. The recoveries of test samples were from 89.26% to 102.02% and the coefficient of variation (%) was less than 4.75%. Parallel analysis of cucumber samples with CTN showed comparable results from the strip test and high performance liquid chromatography. The strip test requires 5–8 min to complete and provides a very useful screening method for the quantitative and qualitative detection of CTN residues in cucumber.
1. Introduction
Chlorothalonil (2,4,5,6-tetrachloroisophthalonitrile, CTN) is one of the most popular fungicides that belongs to the group of halogenated benzonitriles (Wu, Cheng, Cao, & Yu, Citation2012). It works by inhibiting the activities of glutathione and glutathione-dependent enzymes (Bessi, Cossu-Leguille, Zaid, & Vasseur, Citation1999; DeLorenzo & Serrano, Citation2003; Long & Siegel, Citation1975). Over the past two decades, CTN has been widely applied in agriculture to control fungal diseases, but residual CTN was detected in vegetables, crops, soils, and environmental water and aroused health concerns due to its high toxicity (Bruynzeel & van Ketel, Citation1986; Garron, Knopper, Ernst, & Mineau, Citation2012; Sherrard, Murray-Gulde, Rodgers, & Shah, Citation2003). In addition, environmental CTN is also highly toxic to fish, birds, and aquatic invertebrates (Hrenn, Jahn, & Schwack, Citation2002; Liang et al., Citation2010). Consequently, it is listed as a probable human carcinogen by the National Environmental Protection Agency (USA).
Classical analytical methods for detecting CTN are high performance liquid chromatography (HPLC; Galera, Vidal, & Frenich, Citation1997), liquid chromatography–mass spectrometry (LC–MS; Chaves, Shea, & Danehower, Citation2008; Yamamoto et al., Citation2009), and gas chromatography (Kurz et al., Citation2008; Vargyas, Walls, Bramstedt, & Eilrich, Citation2000). These methods require extensive sample preparation and well-trained personnel to operate sophisticated instruments and interpret the results. Consequently, these traditional methods, although highly accurate, are time-consuming, costly, and generally not suitable for screening large numbers of samples or real-time detection. Therefore, an efficient, rapid, and sensitive method for the screening of CTN residues is highly desirable.
Recently, the immunochromatographic lateral flow strip test (ILFST) has become a popular diagnostic tool for detecting various analytes including viruses, bacteria, hormones, parasite antigens, and haptens (Guo et al., Citation2015; Zhang, Guo, & Wang, Citation2009; Zhi et al., Citation2010). It allows a one-step (only addition of specimen), rapid (less than 5 min), and cost-effective analysis and is suitable for screening large numbers of samples on-site. Hence, the aim of the present study is to develop an ILFST for the rapid detection of CTN residues in cucumber. We produced a monoclonal antibody (mAb) 1E8 specific for CTN and utilized a competitive format to establish an ILFST in which free CTN in the sample compete with CTN-bovine serum albumin (BSA) on the test line for binding to colloidal gold-labeled anti-CTN mAb. The strip test is rapid, simple, and cost-effective and is shown to be suitable for the detection of CTN residues on-sites.
2. Materials and methods
2.1. Chemicals and buffers
CTN, Quintozene, Chlorthal-dimethyl, Carbendazim, Kasugamycin, Isocarbophos, Ractopamine, Maduramicin, Doxycycline HCl, Salbutamol, Triadimefon, Cefalexin, and N,N′-carbonyldiimidazole (CDI) were purchased from Sigma (St Louis, MO, USA); BSA and ovalbumin (OVA) were bought from BDH (VWR International Ltd., UK); goat anti-mouse IgG antibody (whole molecule) was obtained from Sino-American Biotechnology Co. (Luoyang, China).
2.2. Apparatus
Microplate Readers 450/550 were from Bio-Rad (Richmond, CA, USA); Milli-Q water was obtained from Millipore (Bedford, MA, USA); an XYZ Biostrip Dispenser, CM 4000 Cutter, and TSR3000 membrane strip reader were purchased from Bio-Dot (USA); and Ultrasonic Cell Disrupter System was from Ningbo Scientz Biotechnology Co., Ltd.
2.3. Preparation of CTN-BSA and CTN-OVA
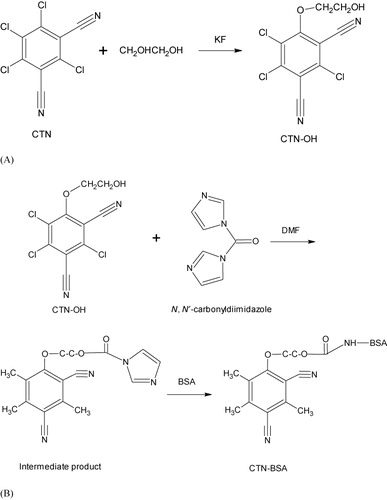
A derivative of CTN, CTN-hydroxyl-trichloroisophthalonitrile (OH), was synthesized by ethylene glycol method. Briefly, 10.00 mg CTN and 3.00 mg potassium fluoride (KF) were dissolved in ethylene glycol and stirred at 90°C for 24 h. After cooling to room temperature (RT), the solution was poured on ice to induce precipitation. The precipitate was collected, washed with double distilled water, and air-dried.
CTN-BSA and CTN-OVA conjugates were made from the CTN-OH using CDI. Briefly, 8.50 mg CTN-OH was dissolved in 500 μL N,N-dimethylformamide and 4.00 mg CDI was added. The solution was stirred for 3 h at RT. A 15.00 mg BSA dissolved in 1.00 mL phosphate buffered saline (PBS) (0.01 mol/L, pH 7.4) was added to the above solution and stirred overnight at 4°C. The conjugates were purified by dialysis against PBS to remove the uncoupled free hapten.
2.4. Production of mAb against CTN and conjugation with colloidal gold
Anti-CTN mAb 1E8 was produced by immunizing BALB/c mice with CTN-BSA and screened by CTN-OVA in indirect and competitive enzyme-linked immunosorbent assays (ELISAs) according to the previous report (Liu, Yan, Zhang, Kuang, & Xu, Citation2014). 1E8 was labeled with colloidal gold with a mean diameter of 15 nm produced by reduction of gold chloride with 1% sodium citrate. When preparing the colloidal gold-labeled mAb, colloidal gold solution was adjusted to pH 9.0 with 0.2 mol/L sodium carbonate. Two milliliter of mAb solution (2 μg/mL) was incubated with 10 mL of colloidal gold solution (pH 9.0) for 30 min at RT. After the addition of 1 mL of 10% BSA solution in 20 mmol/L sodium borate (pH 9.0), the mixture was incubated at RT for another 10 min, and the labeled mAb was then washed using repeated centrifugation (25,000 g) at 10°C for 30 min with 20 mmol/L sodium borate containing 1% BSA and 0.1% sodium azide. The precipitate was finally resuspended in PBST (PBS containing 0.05% Tween-20) and stored at 4°C for use.
2.5. Preparation of conjugate pad and immobilization of capture reagents
The 1E8-colloidal gold conjugate solution was prepared by diluting the colloidal gold-labeled anti-CTN mAb 1E8 with 20 mmol/L sodium borate buffer containing 8.75% (w/v) sucrose, 8.75% (w/v) BSA, 0.6 mol/L NaCl, 10 mmol/L ethylene diamine tetraacetic acid, and 0.1% (w/v) NaN3 to a final concentration of 2 μg/mL. A conjugate pad was made by dispensing a 7 × 300 mm glass fiber (Millipore) with the conjugate solution and then drying for 1 h at 56°C. CTN-BSA (1 mg/mL) and goat anti-mouse IgG (1 mg/mL) were dispensed onto the nitrocellulose membrane as the test and control lines, respectively. The test and control lines were situated 0.5 cm apart at the center of the membrane. These reagents were applied in the form of dots at 50 dots/mL/cm on the membrane. After drying for 1 h at 40°C, the membrane was blocked with 2% (w/v) BSA and then dried, sealed, and stored under dry conditions.
2.6. Assembly of the test strip
Sample and absorbent pads of C048 (Millipore) were made from nonwoven, 100% pure cellulose fiber. The sample pad was cut to 15 × 300 mm and saturated with a buffer (pH 8.0) containing 20 mmol/L sodium borate, 2.0% (w/v) sucrose, 2.0% (w/v) BSA, and 0.1% (w/v) NaN3 and then dried and stored as described above. The absorbent pad was cut to 40 × 300 mm. The sample pad, conjugate pad, blotted membrane, and absorption pad were assembled on the plastic backing support board sequentially with a 1–2 mm overlap and covered by color film at both ends. The master card was cut to 2.8 mm width strips using a CM 4000 Cutter (Bio-Dot). Strips were then sealed in a plastic bag in the form of desiccant gel and stored at 4°C.
2.7. Performance of the strip test
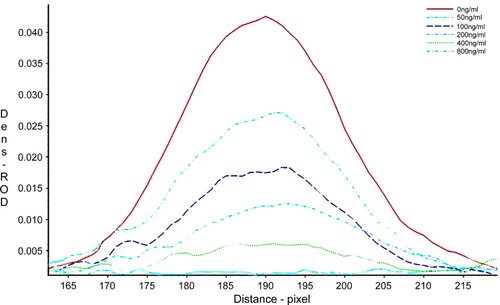
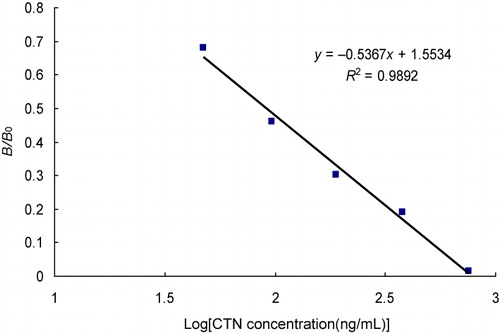
The sensitivity of the strip test was determined by testing 0, 50, 100, 200, 400, and 800 ng/mL of CTN samples. The assays were carried out in triplicate, and the relative optical density (ROD) of the test line was measured with a TSR3000 Membrane Strip Reader (Bio-Dot). A calibration curve was constructed by plotting the G/D-area-ROD or G/peak-ROD values obtained against the log10 of the CTN sample concentrations. G/D-area-ROD means density value of the sampled line points multiplied by the area of the sampling window on the image. G/peak-ROD is the maximum density value of sampled line points. The linearity of the analytes was assessed by the coefficient of determination (R 2). The limit of detection (LOD) was quantitatively defined as the amount of CTN in the standard sample solution that caused a 20% decrease of the G/peak-ROD or G/D × area-ROD compared with that produced by the blank sample. In qualitative testing with the naked eye, the LOD was determined by the minimal amount of CTN which produced a clearly visible difference in intensity of the test strip in comparison with the negative control line where no CTN was added in the sample.
To examine the specificity of the test strip CTN and putative competitors were added to cucumber samples. The competitors Quintozene, Chlorthal-dimethyl, Carbendazim, Kasugamycin, Isocarbophos, Ractopamine, Maduramicin, Doxycycline HCl, Salbutamol, Triadimefon, and Cefalexin were added at the concentration of 5000 ng/mL, respectively. Intra-assay precision was estimated by using one batch of the test strips for replication analysis (n = 3) of the doped CTN samples at 100, 200, and 400 μg/kg. For inter-assay precision, three batches of the test strips were used to detect the given samples. Precision was expressed as coefficient of variation (CV, %).
2.8. Comparison between the strip test and HPLC
Cucumber samples with CTN of 60.3, 355.7, and 724.9 ng/mL were authenticated by LC–MS and provided by the Supervision and Verification Center of the Ministry of Agriculture, Zhengzhou, China. The concentrations of CTN in the cucumber samples represent low, medium, and high levels, respectively. These samples were analyzed in triplicate using the strip test and HPLC for strip test validation. A Student's t-test statistical analysis was used to test the differences.
3. Results and discussion
3.1. Principle of the strip test
The ILFST for detecting CTN in cucumber is based on a competitive format. Colloidal gold was labeled with anti-CTN mAb 1E8 and used as the probe. CTN-BSA conjugate and goat anti-mouse IgG were dispensed onto the nitrocellulose membrane as test and control lines, respectively. When applied to the strip, free CTN in the sample would compete with CTN-BSA on the test line for binding to mAb-gold conjugate. The color of the test line is in a reverse relationship with the concentration of CTN within the sample. For semi-quantitative and qualitative detection, the color of the test line can be evaluated directly by naked eyes. For quantitative assay, the optical density of a test line can be measured with a test strip reader, and the level of the CTN residue can be calculated according to the regression equation from the quantitative calibration curve.
3.2. Hapten conjugation and characterization of anti-CTN mAb 1E8
A derivative of CTN, CTN-OH, was synthesized by ethylene glycol method (). CTN-OH was coupled via its hydroxyl group to the free amino groups of the protein carriers by CDI method resulting in the immunogen (CTN-BSA) and the coating antigen (CTN-OVA; ). The subclass of the mAb 1E8 was identified as IgG1 and the affinity constant (Ka) was 3.82 × 1010 L/mol. ELISA demonstrated that the 1E8 had 1.73% cross-reactivity with Quintozene and negligible cross-reactivity with other tested compounds (<0.11%) including Chlorthal-dimethyl, Carbendazim, Kasugamycin, Isocarbophos, Ractopamine, Maduramicin, Doxycycline HCl, Salbutamol, Triadimefon, and Cefalexin.
3.3. Sensitivity of strip test
Most of immunoassays use polyclonal antibodies in their detection and were found to be cross-reactive to other homologous molecules. The strip test in this study is based on an anti-CTN mAb 1E8 with high-affinity. The sensitivity of the strip test was determined CTN standard solutions at different concentrations. The intensity of the control line was near constant for all strips, validating strip performance. The intensity of the test line was reduced as the concentrations of CTN in the samples increased. The visual LOD of the strip test was less than 100 ng/mL. The intensity of the test line was also recorded by a test strip reader and plotted as a function of various concentrations of CTN (). The peak area decreased with the increase of CTN concentration, and no obvious signal for the sample of 800 ng/mL could be detected. A quantitative calibration curve of the detection was constructed by drawing the logarithmic values of the scanned peak area values against the concentration of CTN (). The LOD for quantitative detection was calculated as 91.78 ± 0.17 ng/mL. The maximum residue limit for total CTN in cucumber and tomatoes in USA, Japan, and China is 5 mg/kg and is 1 mg/kg in the European Union. Hence, the strip test fulfills the standard for practical use.
3.4. Specificity of the strip test
When competitors were added at 5000 ng/mL, the color of test line was the same as that of the negative control sample. Quantitative results from the strip reader demonstrated that the strip test has 3.43% cross-reactivity with Quintozene and minimal cross-reactivity with the other tested compounds (<0.92%; ). Therefore, the strip test for CTN has high specificity, showing negligible cross-reactivity to Quintozene, Chlorthal-dimethyl, Carbendazim, Kasugamycin, Isocarbophos, Ractopamine, Maduramicin, Doxycycline HCl, Salbutamol, Triadimefon, and Cefalexin.
Table 1. Cross-reactivity of the strip test with competitors.
3.5. Recoveries of CTN in cucumber samples
To determine the accuracy, cucumber extracts containing 100, 200, and 400 μg/kg of CTN were tested. Each test was performed in triplicate with a single batch of test strips and the optical density of the test line measured using the test strip reader and sample values calculated from the standard curve. As shown in , for intra-assay and inter-assay reproducibility, recoveries were from 89.95% to 101.73% and 89.26% to 102.02%, respectively. The CVs (%) were less than 4.75%.
Table 2. Recovery and precision of the strip test for CTN spiked in cucumber samples.
3.6. Comparison between the strip test and HPLC
A comparison between the test strip and the HPLC was performed with three levels of the actual samples. The differences of the test strip to actual samples were from 2.54% to 5.17%, and the HPLC was from 1.49% to 4.21%. The difference of the test strip and HPLC was from 1.38% to 5.24% (). For both methods the results were almost identical, and statistical analysis using a t test did not show a significant difference between the two methods.
Table 3. Comparison between the strip test and HPLC.
4. Conclusions
An ILFST for the determination of CTN residues in cucumber was developed using the competitive format for the rapid, sensitive, and one-step detection of a trace amount of CTN in cucumber samples. The ILFST is colloidal gold based and can be used for visual (qualitative) or strip reader (semi-quantitative or quantitative) assessment simultaneously. The LOD for qualitative detection by visual inspection was less than 100 ng/mL, while the LOD of the quantitative procedure was 91.78 ± 0.17 ng/mL. In summary, the strip test has the potential to be used as an on-site screening tool for detecting CTN.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bessi, H., Cossu-Leguille, C., Zaid, A., & Vasseur, P. (1999). Effects of chlorothalonil on glutathione and glutathione-dependent enzyme activities in Syrian hamster embryo cells. Bulletin of Environmental Contamination and Toxicology, 63, 582–589. doi:10.1007/s001289901020
- Bruynzeel, D. P., & van Ketel, W. G. (1986). Contact dermatitis due to chlorothalonil in floriculture. Contact Dermatitis, 14(1), 67–68. doi:10.1111/j.1600-0536.1986.tb01160.x
- Chaves, A., Shea, D., & Danehower, D. (2008). Analysis of chlorothalonil and degradation products in soil and water by GC/MS and LC/MS. Chemosphere, 71, 629–638. doi:10.1016/j.chemosphere.2007.11.015
- DeLorenzo, M. E., & Serrano, L. (2003). Individual and mixture toxicity of three pesticides; atrazine, chlorpyrifos, and chlorothalonil to the marine phytoplankton species Dunaliella tertiolecta. Journal of Environmental Science and Health. Part B, 38, 529–538. doi:10.1081/PFC-120023511
- Galera, M. M., Vidal, J., & Frenich, A. G. (1997). Evaluation of multiwavelength chromatograms for the quantification of mixtures of pesticides by high-performance liquid chromatography-diode array detection with multivariate calibration. Journal of Chromatography A, 778(1–2), 139–149. doi:10.1016/S0021-9673(97)00371-3
- Garron, C., Knopper, L. D., Ernst, W. R., & Mineau, P. (2012). Assessing the genotoxic potential of chlorothalonil drift from potato fields in Prince Edward Island, Canada. Archives of Environmental Contamination and Toxicology, 62, 222–232. doi:10.1007/s00244-011-9699-2
- Guo, J. G., Liu, L. Q., Xue, F., Xing, C. R., Song, S. S., Kuang, H., & Xu, C. L. (2015). Development of a monoclonal antibody-based immunochromatographic strip for cephalexin. Food and Agricultural Immunology, 26, 282–292. doi:10.1080/09540105.2014.907242
- Hrenn, H., Jahn, C., & Schwack, W. (2002). Formation of protein-bound residues of the fungicide chlorothalonil in tomatoes (Lycopersicon esculentum Mill.). European Food Research and Technology, 214(2), 138–142. doi:10.1007/s00217-001-0431-8
- Kurz, M. H. S., Gonçalves, F. F., Adaime, M. B., da Costa, I. F. D., Primel, E. G., & Zanella, R. (2008). A gas chromatographic method for the determination of the fungicide chlorothalonil in tomatoes and cucumbers and its application to dissipation studies in experimental greenhouses. Journal of the Brazilian Chemical Society, 19, 1129–1135. doi:10.1590/S0103-50532008000600012
- Liang, B., Li, R., Jiang, D., Sun, J., Qiu, J., Zhao, Y., … Jiang, J. (2010). Hydrolytic dechlorination of chlorothalonil by Ochrobactrum sp. CTN-11 isolated from a chlorothalonil-contaminated soil. Current Microbiology, 61, 226–233. doi:10.1007/s00284-010-9603-8
- Liu, L. Q., Yan, H. J., Zhang, X., Kuang, H., & Xu, C. L. (2014). Development of an anti-chlorothalonil monoclonal antibody based on a novel designed hapten. Food and Agricultural Immunology, 26, 410–419.
- Long, J. W., & Siegel, M. R. (1975). Mechanism of action and fate of the fungicide chlorothalonil (2,4,5,6-tetrachloroisophthalonitrile) in biological systems. 2. In vitro reactions. Chemico-Biological Interactions, 10, 383–394. doi:10.1016/0009-2797(75)90069-1
- Sherrard, R. M., Murray-Gulde, C. L., Rodgers, J. H., Jr., & Shah, Y. T. (2003). Comparative toxicity of chlorothalonil: Ceriodaphnia dubia and Pimephales promelas. Ecotoxicology and Environmental Safety, 56, 327–333. doi:10.1016/S0147-6513(02)00073-8
- Vargyas, L. D., Walls, G. E., Bramstedt, W. R., & Eilrich, G. L. (2000). Simultaneous determination of chlorothalonil and hexachlorobenzene in technical and formulated materials by capillary gas chromatography: Collaborative study. Journal of AOAC International, 83, 1047–1052.
- Wu, X., Cheng, L., Cao, Z., & Yu, Y. (2012). Accumulation of chlorothalonil successively applied to soil and its effect on microbial activity in soil. Ecotoxicology and Environmental Safety, 81(1), 65–69. doi:10.1016/j.ecoenv.2012.04.017
- Yamamoto, A., Miyamoto, I., Kitagawa, M., Moriwaki, H., Miyakoda, H., Kawasaki, H., & Arakawa, R. (2009). Analysis of chlorothalonil by liquid chromatography/mass spectrometry using negative-ion atmospheric pressure photoionization. Analytical Sciences: The International Journal of the Japan Society for Analytical Chemistry, 25, 693–697. doi:10.2116/analsci.25.693
- Zhang, G. P., Guo, J. Q., & Wang, X. N. (2009). Immunochromatographic lateral flow strip tests. Methods in Molecular Biology, 504, 169–183. doi:10.1007/978-1-60327-569-9_12
- Zhi, A. M., Li, B. B., Liu, Q. T., Hu, X. F., Zhao, D., Hou, Y. Z., … Zhang, G. P. (2010). Development of a lateral-flow immunochromatographic test device for the rapid detection of difloxacin residues. Food and Agricultural Immunology, 21, 335–345. doi:10.1080/09540105.2010.504766