ABSTRACT
The accessibility of selenium from naturally enriched sources such as cereals crops can potentially be used as selenium supplements to support nutritional requirements. Dietary selenium supplementation, as Se-rich wheat extracts, on RAW264.7 macrophage cells enhanced the antioxidant capacity via augmentation of cellular selenoprotein glutathione peroxidase 1 (GPx-1) expression in the absence or presence of lipopolysaccharide (LPS) treatment. Cells were supplemented with Se in the form of sodium selenite (SS), seleniferous wheat extract (SeW) and seleniferous wheat extract with rMETase treatment (SeW + rMET) at three different concentrations. Cells supplemented with SS and SeW + rMET showed increase in GPx-1 expression as compared to SeW-treated cells. SeW + rMET, further, downregulated the LPS-induced expression of cyclooxygenase-2, microsomal PGE synthase-1 and inducible nitric oxide synthase w.r.t. Se-deficient cells, while the expression of haematopoietic PGD synthase was upregulated. This demonstrates SeSup effectively modulates the expression of inflammatory responses, indicating the potential benefits of dietary selenium supplementation.
Introduction
Macrophages are the prime responders to diverse stimuli for cell-to-cell communication. Activation of these innate immune cells is usually accompanied by oxidative burst, as seen by increased reactive oxygen and nitrogen species (RONS), which affects the pathogen. In addition, macrophage activation is also associated with the release of various mediators, including arachidonic acid (AA)-derived prostaglandins such as PGE2, TXA2 and PGD2, and its metabolite 15d-PGJ2 (Ricciotti & FitzGerald, Citation2011). However, any disruption in RONS encountering enzymes will lead to massive oxidation by cell populations in inadequate mitigation of pathogens. This represents a potential toxic insult, resulting in DNA damage and inactivation of enzymes, leading to recurrent infections and persistent inflammation. The basic reactions involved in oxidative burst are through chemically reactive secondary messenger molecules such as NO•, O2• and H2O2. These superoxide radicals may be rapidly reduced to H2O2 by SOD (Conrado, D’Angelantonio, Torreggiani, Pecci, & Fontana, Citation2014), and further to H2O by antioxidant/anti-inflammatory enzymes like selenoproteins such as glutathione peroxidase and selenium-independent enzymes such as H-PGDS, which belongs to the sigma-class of glutathione S-transferase with non-selenium-dependent peroxidase activity (Higgins & Hayes, Citation2011).
The biological effects of dietary Se is through its incorporation into selenoproteins that influence various cells and their functions such as activation, migration, proliferation capacity and signalling strength like cytokine production and phagocytosis (Huang, Rose, & Hoffmann, Citation2012). The ex-vivo supplementation of Se boosts up the innate/adaptive immune responses, which further directly/indirectly regulate the cellular redox tone by inhibiting the expression of pro-inflammatory genes and, hence, reducing the inflammatory response (Rayman, Citation2012).
Dietary Se supplementation is proved to elevate the transcription of anti-inflammatory enzymes (GPx and H-PGDS) and downregulate the transcription of pro-inflammatory inducible enzymes (COX-2 and iNOS), pro-inflammatory cytokines (IL-1, IL-2, IL-6 and TNF-α) and pro-inflammatory chemokines (IL-8, MIP-1α, MCP1, RANTES and eotaxin) (Gandhi, Nagaraja, & Prabhu, Citation2013). Se can potentially help in switching the AA pathway from pro-inflammatory expression to anti-inflammatory responses and reduce the risk of cancer (Lu et al., Citation2016), arthritis (Onal, Naziroglu, Colak, Bulut, & Flores-Arce, Citation2011) and other cardiovascular diseases (Bukkens et al., Citation1990). In the recent past, our group has reported the significant hyperaccumulation of Se in staple crops ranging from 20 to 123 μg/g in wheat, maize and rice matrices (Dhanjal, Sharma, & Nagaraja, Citation2016). In Se-rich wheat (Cubadda et al., Citation2010) and rice (Premarathna et al., Citation2012), 90% of Se content is in the form of seleniomethionine (SeMet). The in vitro gastrointestinal digestion studies show a better accessibility of Se in terms of SeMet during intestinal digestion (Dhanjal et al., Citation2016; Cubadda et al., Citation2010). These Se-rich matrices provide an efficient reservoir for long-term shortage (Behne, Alber, & Kyriakopoulos, Citation2009), facilitating the availability of Se through catabolism of SeMet during the natural protein turn-over (Zeng, Botnen, & Jhonson, Citation2008). However, the ability of these Se-rich foods to mitigate expression of inflammatory genes while upregulating those proteins that are key in resolution of inflammation has not been demonstrated. In this study, we demonstrate the influence of bioavailable dietary Se in resolving the inflammatory responses through upregulation of expression of anti-inflammatory enzymes such as GPx-1 and H-PGDS, while downregulating pro-inflammatory enzymes (COX-2, mPGES-1 and iNOS) in SeDef and SeSup RAW264.7 macrophage cells.
Experimental
Extraction and quantification of selenium in wheat extract
The extraction of Se from wheat matrices (SeW and NSeW) was performed in duplicate with 1X PBS buffer. The samples were vortexed for 10 min and centrifuged at 4000 rpm for 30 min at 4°C. The supernatant was centrifuged at 16,000 rpm for 1 h at 4°C. The filtrate was collected under sterile conditions and passed twice through 0.22 μm sterile filters. The extracts were kept at −80°C until quantification was done. Se in wheat extract was oxidized in acid using microwave digestion (MARS 6 240/50:910905, CEM-NC) with nitric acid and perchloric acid (SD FineChem, India) followed by reduction with HCl (SD FineChem, India). The reduced Se (Se+4) was then allowed to complex with 2,3-diamino-naphtalene (Himedia, India) to obtain yellow coloured piazselenol complex in cyclohexane. The colour intensity was measured with a fluorescence spectrometer (Perkin Elmer LS-45) at emission spectrum of 520 nm corresponding to the amount of selenite present in the wheat extracts (SeW/NSeW). The emission spectrum of NIST-certified Selenium ICP standard solution (SRM-1349) was used for Se quantification (Dhanjal et al., Citation2016).
Cell culture and stimulation
RAW264.7 murine macrophage cell line was cultured in DMEM (Lonza) containing 10% defined FCS (Himedia), 4.5 g/L glucose, 2 mM L-Glutamine, 1 % v/v penstrep containing 100 U/mL of penicillin and 100 μg/mL streptomycin (Sigma), 0.5% amphotericin B solution (Sigma) at 37°C with 5% CO2 devoid of any Se source (SeDef). SeDef cells (1×105) were then cultured in three different Se-supplemented media (SeSup) for three days along with respective controls (). After 72 h, cells were stimulated with lipopolysaccharide (LPS) (Sigma) at 1 μg/mL. Cells were collected at different time intervals, washed twice with PBS (Lonza), scraped and centrifuged at 2000 rpm. Both SeDef and SeSup cells had similar growth rate and cell viability pattern.
Table 1. Different sources of Se supplementation with increasing three different Se concentrations (50, 100 and 500 nM) w.r.t. their controls. Control (C) represents cells devoid of any source of Se supplementation. In case of (NSeW)e and (NSeW)e + rMET, “e” represents the volume of non-seleniferous wheat extract taken equivalent to seleniferous wheat extract with 500 nM Se concentration.
Preparation of cell lysates
The pellets were re-suspended in 100 μL of CelLyticTM M (Sigma) protein extraction reagent containing PierceTM protease inhibitor (Thermo Scientific Life Sciences) and kept for 30 min at 4°C with intermittent vortexing. The cell lysate was centrifuged at 12,000 rpm for 30 min at 4°C and protein containing supernatant was collected. For mPGES-1 analysis, the pellet containing membranous proteins was re-suspended in extraction buffer and centrifuged at 16,000 rpm for 1 h at 4°C. Supernatant was collected and the protein concentration was estimated using the Bradford method (Alabi, Citation1979).
Western blot analysis
Along with prestained protein ladder (PUREgene Genetix Biotech), 30 μg of protein was separated on 15% SDS-polyacrylamide gel (Sigma) and transblotted onto the PVDF membrane (0.2 μm, Biorad). The membrane was blocked with TBST containing 10 mM Tris, pH 8.0 (Himedia), 150 mM NaCl (Sigma), 0.05% Tween-20 (Sigma) and 5% skim milk (Sigma) for 1 h at room temperature. The membrane was probed with primary antibody followed by an appropriate secondary antibody coupled to HRP. Anti-GPx-1 was procured from Abcam (Cat # ab108427); mouse anti-GAPDH (Cat # 437000) and HRP goat anti-mouse IgG (H + L) (Cat # 626520) were procured from Life Technologies; while goat anti-rabbit IgG HRP (Cat # 10004301), COX-2 (mouse) polyclonal antibody (Cat # 160126), H-PGDS polyclonal antibody (Cat # 160013) and mPGES-1 polyclonal antibody (Cat # 160140) were procured from Cayman Chemical Company. Anti-GAPDH was re-probed with blots for normalized protein loading. The immunoreactive bands were analysed densitometrically using the ImageJ software program (Schneider, Rasband, & Eliceiri, Citation2012), developed at the National Institute of Health (NIH).
Semi-quantitative reverse transcriptase PCR
Total RNA was extracted from SeDef and SeSup cells by TRIzol reagent (Life Technologies) according to the manufacturer’s instructions and quantified on the basis of UV absorbance values at 260 and 280 nm. To reduce any DNA contamination, RNA samples were treated with RNase-free DNase (Promega) for 15 min at 20–25°C. One microgram of total RNA was reverse-transcribed using Superscript® III First strand synthesis (Invitrogen). Equal amounts of DNA (100 ng) from each sample were polymerized using Applied Biosystems Veriti thermal cycler. Primers for GPx-1, COX-2, mPGES-1, H-PGDS, iNOS, GAPDH and β-Actin, as listed in , were used for PCR amplification and the product was analysed on 2% agarose gel (3B Black Bio Biotech India Ltd.). DNA bands were quantified on a GEL DOC XR system (Biorad) using Quantity One 1-D Analysis (Biorad) software program and the intensity of bands were analysed densitometrically using ImageJ software program (Schneider et al., Citation2012), developed by the National Institutes of Health.
Table 2. Primers used for pro-/anti-inflammatory genes in PCR amplification. One microgram of total RNA was reverse transcribed from each sample and amplified by PCR polymerization. Gene expression in SeSup and SeDef samples were then quantified densitometrically using ImageJ software program and normalized to the expression of GAPDH.
Nitrite assay
Nitric oxide (NO) release into cell-free medium was measured extracellularly from the culture media (as nitrite) taken from cells cultivated and stimulated with LPS under identical conditions. For standard curve, NaNO2 was used from scale 0 to 100 μM and incubated with Griess reagent (Sigma) for 15–20 min under dark conditions. Extracellular nitrite excretion was measured in a plate reader at 548 nm.
Results and discussion
Quantification of selenium wheat extract
The Se concentration of seleniferous wheat grain extract measured by fluorescence spectrometer was 7.1 ± 0.2 µg/ml. Earlier reports from our group showed the hyperaccumulation of Se in different cereal grains and their protein fractions from seleniferous belt of Punjab, India. The whole wheat grains accumulate up to 122.9 ± 0.6 µg/g Se than maize (26.5 ± 0.2 µg/g) and rice (19.7 ± 0.2 µg/g). In the in vitro gastrointestinal digestion studies, the intestinal accessibility of Se describes its potential bioavailability through dietary supplementation to the SeDef population (Dhanjal et al., Citation2016).
Dietary selenium transforms the expression profile of pro-inflammatory genes (COX-2, mPGES-1) into the anti-inflammatory genes (GPx-1, H-PGDS)
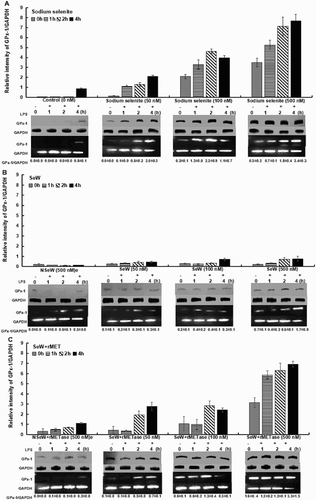
The expression profile of Se-dependent pro-/anti-inflammatory genes were measured in RAW264.7 cells. Cells were grown in various conditions as SeDef (Control, NSeW and NSeW + rMET) and Se-supplemented media (SS, SeW and SeW + rMET) with different Se concentrations (50, 100 and 500 nM). As GPx is a well-known marker for the Se status, the anti-inflammatory enzyme expression of Se-GPx-1 increased with increasing sodium selenite concentration (500 nM > 100 nM > 50 nM > 0 nM, 0 h) of SeSup cells with 3.5-fold higher expression as compared to SeDef cells ((A)). With oxidative stress upon LPS stimulation, control RAW264.7 cells that were devoid of Se also showed mild GPx-1 expression after 4 h ((A)) which clearly indicates the self-defense mechanism against the ROS generation. However, upon LPS stimulation up to 4 h, Se supplementation increased GPx-1 levels to a significant extent with increasing concentration (500 nM > 100 nM > 50 nM) w.r.t. control, i.e. from 2.4-fold at 50 nM to 9.0-fold at 500 nM. Increased expression of selenoproteins, including GPx1, is known to mitigate inflammation and bolster immunity that involves downregulation of COX-2 and mPGES-1, while upregulating H-PGDS to effect a process called “eicosanoid class switching mechanism” in Se-stimulated RAW264.7 cells (Yui, Imataka, Nakamura, Ohara, & Naito, Citation2015; Duntas, Citation2009; Vunta et al., Citation2007). Higher Se supplementation up to 2 μM in the form of sodium selenite is also reported to inhibit NFκB-dependent transcription (Nettore et al., Citation2017; Meplan et al., Citation2016; Youn et al., Citation2008; Maehira, Mivagi, & Eguchi, Citation2003). Under Se-deficient conditions, there is increased ROS generation that is closely associated with increased inflammatory genes, which results in augmentation of oxidative stress in cells. Thus, imbalances in oxidative stress exacerbate the modulation of repressor activator protein 1 (RAP1), which induces pro-inflammatory cytokines via activation of transcription factor NFκB (Cai et al., Citation2015). Prolonged NFκB activation results in neuro-inflammation followed by neuro-toxicity, as it is ubiquitously expressed in neurons (Shih, Wang, & Yang, Citation2015).
Figure 1. GPx-1 expression in RAW264.7 macrophages. Cells were treated with 50, 100 and 500 nM Se concentrations in different forms: (A) Sodium selenite (SS), (B) Seleniferous wheat extract (SeW) and (C) Seleniferous wheat extract in the presence of rMETase incubation (SeW + rMET) with respect to their controls, for 72 h and then inflamed with 1 μg/mL LPS for framed time interval up to 4 h. Densitometric values normalized to GAPDH are graphed with mean ± S.D. for protein expression and the values indicated below each panel of mRNA expression.
rMETase increases the bioavailability of selenium from cereal matrices
rMETase is the recombinant L-methionine γ-lyase or methioninase, a pyridoxal 5’-phosphate containing enzyme. It catalyses the α- and γ-elimination of methionine, sulphur-containing amino acids. Likewise, it is able to cleave C-Se and C-O bonds of selenium- and oxygen-containing amino acids (Fukumoto et al., Citation2012; Nakayama et al., Citation1984) and making them optically more active and bioavailable in functioning (Tanaka, Esaki, & Soda, Citation1985). Our results also showed the significant difference with rMETase application in comparison to controls demonstrating the facilitation of free Se from wheat matrices, and impacting the elevated trend of anti-inflammatory enzymes vis-à-vis downregulation of pro-inflammatory enzymes.
A marginally elevated trend of overall GPx-1 expression was observed when cells were incubated with ex-vivo dietary Se-supplementation in the form of seleniferous wheat extract (SeW) when compared to non-seleniferous wheat extract (NSeW). Even at highest concentration of SeW extract (500 nM), RAW264.7 cells showed 1.1-fold increase in GPx-1 expression at 4 h of LPS stimulation w.r.t. control (C) ((B)). The SS and SeW + rMET-incubated cells showed 9.0- and 8.0-fold higher GPx-1 expression, respectively, than SeW-supplemented cells at a similar concentration and time. This clearly demonstrates that Se bound to the wheat matrix was not completely bio-available and accessible to the cells. Hence, to facilitate the accessibility of Se and assess its efficacy, cells were incubated with rMETase (Gandhi et al., Citation2011) at 0.2 units/mL media along with SeW extract. The observations clearly indicated enhanced GPx-1 expression trending similar to SS-stimulated cells. In LPS-unstimulated (0 h) SeW + rMET-supplemented cells, the GPx-1 expression increased with increase in SeW + rMET extract concentration (500 nM > 100 nM > 50 nM) up to 3.2-fold in comparison to NSeW + rMET-incubated cells ((C)). Under similar conditions, the supplementation of SeW extract showed poor GPx-1 response of up to 1.1-fold ((B)). At 4 h of LPS stimulation, SeW + rMET supplementation (50 to 500 nM) w.r.t. NSeW + rMET enhanced the GPx-1 level from 2.5-fold to 6.2-fold, respectively ((C)).
Collectively, our data confirm the role of rMETase in facilitating the release of bio-available Se from Se-rich wheat grain (SeW + rMET) and its antioxidant expression in comparison to the SeW supplementation. Previous reports from our group showed that selenomethionine (SeMet) accounts for 72–85% of selenocompounds present in Se-rich wheat grains (Cubadda et al., Citation2010) and is the major form consumed by humans (Finley, Citation2006). It can be, thus, hypothesized that Se-rich natural dietary matrices facilitate sustained and slow release of Se through enzymatic cleavage of SeMet to be metabolized to bioactive forms that increase plasma Se levels with better antioxidant and anti-inflammatory capacity (Cao, Guo, Zhang, Dong, & Gong, Citation2014). SeMet supplementation in minute concentration of up to 3 μg/g abrogate the allergies caused due to these inflammatory responses under Se deficiency (Sakazaki et al., Citation2014).
Dietary selenium supplementation downregulates the COX-2 expression in comparison with SeDef macrophages
In contrast to GPx1, Se supplementation downregulated the expression of a well-known pro-inflammatory gene, COX-2. COX-2 is a marker of inflammation and is activated by diverse stimuli, including LPS. COX-2 levels were increased by 4.5-fold during the first 4 h in SeDef cells as compared to the SeSup group ((A)). However, SeW with rMETase treatment (SeW + rMET) showed a nearly similar trend to SS-incubated cells with moderately weaker expression of COX-2 after 4 h of LPS stimulation ((C)).
Figure 2. LPS-induced expression of COX-2 in RAW264.7 macrophages with same experimental conditions as for GPx-1. The graph shows the inverse relationship with GPx-1 upregulation with increased selenium concentration up to 4 h of LPS incubation. n = 3 is shown.
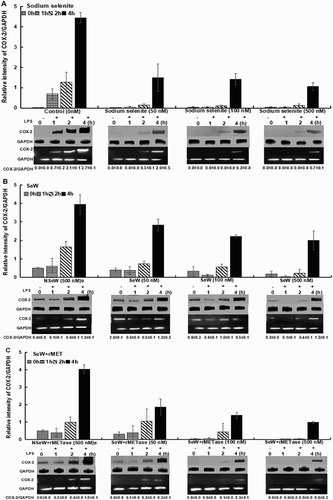
Upon SS supplementation, the COX-2 expression was downregulated within 2–4 h ((A)) and reduced by 3.0- to 4.3-fold in 4 h with 50 and 500 nM Se, respectively. Among SeW- and SeW + rMET-treated cells, COX-2 levels decreased from 1.4- to 2.0-fold and 2.2- to 4.0-fold with increasing dietary Se levels (500 nM > 100 nM > 50 nM) as compared to the NSeW- and NSeW + rMET-treated cells, respectively ((B,C)). Further at 500 nM, SeW + rMET, COX-2 expression significantly decreased by 4 h as compared to SeW-treated cells ((C)).
These results suggested that SeW extract with bio-available Se modulates the COX-2 expression leading to the downregulation of its downstream product mPGES-1 ((C)) and thus helps in the activation of other anti-inflammatory prostaglandin enzymes such as H-PGDS, which produces the precursor of anti-inflammatory cyclopentenone prostaglandins, D12-PGJ2 and 15d-PGJ2. With a delayed time lag of expression, levels of COX-2 and mPGES-1 correlate with each other and NF-κB transcription (Kirkbya et al., Citation2016) with an increase at the protein level under Se deficiency upon LPS stimulation (Davis et al., Citation2002). Such a functional coupling of COX-2 and mPGES-1 versus COX-1 and H-PGDS is known to drive the PG metabolism towards either pro-inflammatory or anti-inflammatory pathways under differential Se status (Gandhi et al., Citation2011; Vunta et al., Citation2007). The over-expression of COX-2 and mPGES-1 due to Se deficiency is thought to be one of the key underlying basis of inflammation that is seen in breast cancer (Jana et al., Citation2014), AIDS, (Ipp, Zemlin, Erasmus, & Glashoff, Citation2014) and asthma (Daham et al., Citation2014).
mPGES-1 expression is downregulated by dietary supplementation in LPS-stimulated RAW264.7 cells
To demonstrate the effect of Se on mPGES-1 expression as a downstream marker of COX-2 activity, cells were grown in SeDef and SeSup RAW264.7 cells with/without 1 μg/mL LPS stimulation from 0 to 12 h. Based on the literature (Davis et al., Citation2002; Vunta et al., Citation2007; Gandhi et al., Citation2011), a time course experiment was set up to check the expression of COX-2 and mPGES-1. In SeDef LPS-stimulated cells (control 0 nM), the mPGES-1 level was increased by 3.8-fold from 0 to 12 h as compared to the SeSup cells ((A)). However, an increasing trend in mPGES-1 expression over time was seen in all SeSup cells. In contrast to control (C, 12 h) SeW + rMET incubation of RAW264.7 cells clearly indicated a significant decreased mPGES-1 expression up to 12-fold at higher concentration (500 nM) as compared to the SeW cells (3.3-fold) ((B, C)).
Figure 3. mPGES-1 expression; selenium supplementation modulates the mPGES-1 levels when inflamed with 1 μg/mL LPS up to 12 h. Seleniferous wheat extract in the presence of rMETase incubation (SeW + rMET) shows maximum downregulation in comparison to sodium selenite and seleniferous wheat extract incubation. A noticeable observation of non-seleniferous wheat extract treated with rMETase (NSeW + rMET) showed significant decrease in mPGES-1 protein and mRNA levels (C). Results shown are representative of three independent experiments.
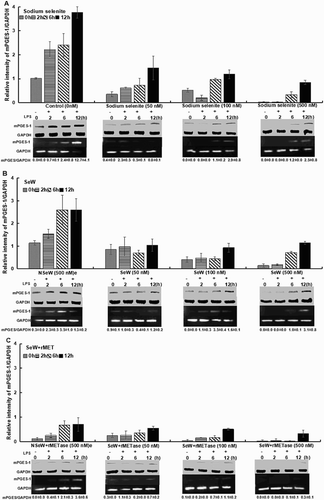
Interestingly, in LPS-stimulated NSeW + rMET-supplemented cells, rMETase co-treatment decreased mPGES-1 expression up to 5.9-fold over 12 h ((C)). This result highlights the role of rMETase in promoting the bioavailability of Se to plausibly skew the AA pathway. During resolution of inflammation, Se supplementation shifts AA metabolism from PGE2 towards PGD2 and its downregulated product 15d-PGJ2 through inactivation of NF-κB (Gandhi et al., Citation2011; Kaushal et al., Citation2014; Nelson, Lei, & Prabhu, Citation2011; Vunta et al., Citation2007; Davis et al., Citation2002). By virtue of being a Michael electrophile, 15d-PGJ2 covalently modifies key cysteine thiols in I Kappa B Kinase (IKK)-β (IKK2), which regulates the activation of NF-κB in LPS-treated macrophages (Cai et al., Citation2015; Gandhi et al., Citation2011).
Shunting of the AA pathway from mPGES-1 to H-PGDS by SeSup
Culturing of RAW.264.7 cells in LPS-stimulated SeDef and SeSup cells showed significant increase in H-PGDS expression. A 7.0-fold difference in H-PGDS in SeDef LPS-stimulated cells (control, C) was seen from 0 to 12 h ((A)). However, Se supplementation significantly increased the upregulation of H-PGDS with increasing concentration (500 nM > 100 nM > 50 nM) either in the form of SeW + rMET or SS as compared to the control ((A, C)).
Figure 4. H-PGDS expression; the increase in selenium level in any form (SS, SeW and SeW + rMET) increases the overall H-PGDS expression. A time course study was set up for up to 12 h of 1 μg/mL LPS stimulation, which shows a significant increase in case of SeW + rMET incubation. n = 3 shown with mean ± S.D. values.
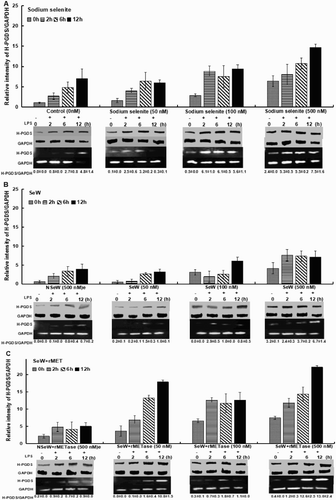
In addition, SS-supplemented cells without LPS stimulation (0 h) showed increasing levels of H-PGDS (up to 6.4-fold) with increase in Se concentration from 50 to 500 nM. A similar increasing trend was observed in SeW + rMET-treated cells, at a higher concentration (500 nM) with prominent hike in expression levels (4.4-fold) when compared to the SS (2.0-fold) and SeW (1.8-fold)-treated cells w.r.t. their controls, respectively (). Similar results were observed in earlier studies with sodium selenite stimulation of macrophages wherein enhanced levels of hematopoietic PGDS (H-PGDS) enzyme catalyse PGH2 to PGD2 and 15d-PGJ2 conversion for the downregulation of inflammation processes. 15d-PGJ2 acts as an endogenous ligand for peroxisome proliferator-activated receptor (PPAR)γ (Gandhi et al., Citation2011), a transcription factor that inhibits the expression of NF-kB-regulated genes, while activating the expression of various anti-inflammatory genes (Tyagi, Gupta, Saini, Kaushal, & Sharma, Citation2011).
iNOS expression
In addition to COX-2 and mPGES-1, expression of iNOS, a well-known NF-kB downstream target gene, was examined at the level of mRNA and product (as nitrite) cell-free supernatant. Upon LPS stimulation, iNOS expression was increased in all treatments. However, in SeDef (i.e. C, NSeW- and NSeW + rMET-treated cells), the mRNA expression was significantly higher than in SeSup cells ().
Figure 5. iNOS mRNA levels. Ex-vivo Se supplementation mitigates the iNOS mRNA expression levels by incubating RAW264.7 cells with 1 μg/mL post LPS stimulation for 0–24 h. Densitometry of iNOS and β-actin bands clearly shows that sodium selenite (500 nM) and dietary SeW + rMET (500 nM) reduces the oxidative stress by 3- and 10-fold, respectively. Representative of n = 3 shown and quantified by ImageJ software program.
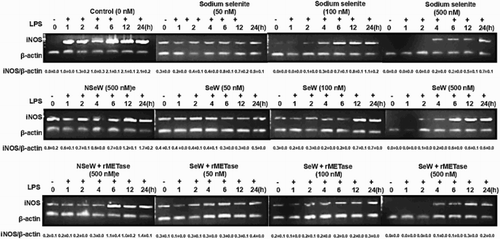
With increasing concentration of Se from 50 to 500 nM, iNOS expression was effectively inhibited within 4 h in all SeSup cells. Although among SS-, SeW- and SeW + rMET-stimulated cells, the SeW + rMET supplementation showed comparatively higher inhibition of iNOS expression (10-fold) as compared to the SS (3-fold) and SeW (3.5-fold)-stimulated cells w.r.t. control (C, 24 h). In SeDef cells, devoid of LPS stimulation, marginal expression of iNOS could be seen in NSeW (0.8-fold) and NSeW + rMET (0.2-fold) as compared to the control (C). It is known that LPS and IFNγ induction upregulates the iNOS expression to augment inflammatory responses in human U937 macrophages (Grkovich, Johnson, Buczynski, & Dennis, Citation2006). Our results corroborate with a previous report on the ability of Se supplementation to reduce iNOS expression by 3-fold as compared to SeDef LPS-unstimulated RAW 264.7 macrophages (Prabhu et al., Citation2002). Similar results were seen with the role of Aster Yomena (Kim et al., Citation2017), Eupartoium makinoi (Ahn et al., Citation2015), Siegesbeckia glabrescens (Lee, Kang, Hwang, & Kim, Citation2011) and lansai Cand D (Taechowisan, Wanbanjob, Tuntiwachwuttikul, & Liuin, Citation2010) in downregulation of iNOS transcription along with NO quenching which further decreases in COX-2 and PGE2 mRNA levels in LPS-induced macrophages.
The enzyme expression through protein levels of GPx-1 , COX-2 , mPGES-1 and H-PGDS showed consistent results in gene expression via mRNA levels. This demonstrates that SeSup regulates the AA metabolism at the transcriptional level. Thus, Se modulates the pro-inflammatory genes transcriptionally to reduced levels of COX-2 , mPGES-1 and iNOS expression. However, the antioxidant capacity of genes like GPx-1 and H-PGDS showed elevated mRNA expression with SeSup as compared to the control.
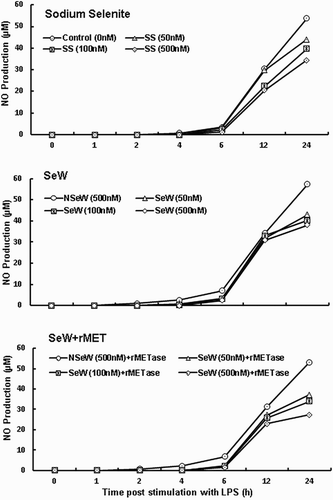
NO production
NO.production (measured as nitrite) was also analysed by collecting cell-free supernatants over time (). The SeDef (C, NSeW and NSeW + rMET) shows significantly higher NO level as compared to the SeSup cells (SS, SeW and SeW + rMET). The observations clearly indicate that upon LPS stimulation, Se deficiency results in exogenous NO production (Hoffmann, Citation2007), which is part of the inflammatory markers along with other cytokines such as TNF-α, IL-1β and IL-6 in LPS-induced J774.1 mouse macrophages (Safir, Wendel, Saile, & Chabraoui, Citation2003).
Conclusions
In the present study, we observed the enhanced anti-inflammatory role of bio-available Se facilitated through rMETase-treated seleniferous wheat extracts by inhibiting the expression of COX-2, mPGES-1, iNOS expression and nitrite production. This proves the regulatory effect of bio-available Se in induction of anti-inflammatory GPx-1 and H-PGDS protein levels through downregulation of pro-inflammatory protein levels. Our data highlight the role of rMETase enzyme in dietary Se supplementation in the form of wheat extract as nearly similar to that of sodium selenite supplementation in resolving inflammatory responses in the RAW264.7 macrophage model. To the best of our knowledge, this is the first report that demonstrates the influence of bio-available Se from Se-rich dietary matrices on inflammatory responses. Further studies with in-vivo models are to substantiate these observations on bioavailability and bio-activity of Se from Se-rich dietary matrices.
Disclosure statement
No potential conflict of interest was reported by the authors.
Notes on contributors
Noorpreet Inder kaur Dhanjal is a doctoral student at the Department of Biotechnology, Thapar working in the area of selenium biofortification and its bioactivity with specific reference to inflammation.
Dr. Siddharth Sharma, associated with Department of Biotechnology, Thapar University, India, is pursuing research on Pharmacogenetics of individualized cancer chemotherapy.
Dr. K. Sandeep Prabhu is associated with Pennsylvania State University, State College, USA, and has been exploring the role of selenium, selenoproteins, and redox homeostasis, macrophage biology; and pathways of inflammation and resolution.
Dr. N. Tejo Prakash is working in the area of selenium quantification, bioaccessibility and bioactivity, at School of Energy and Environment, Thapar University, Patiala, India.
ORCID
N. Tejo Prakash http://orcid.org/0000-0003-3422-5550
Additional information
Funding
References
- Ahn, S-I., Lim, S. J., Gu, G-J., Kim, J-S., Paek, J. H., Kim, S. Youn, H. S. (2015). Eupartoium makinoi suppresses lipopolysaccharide-induced inducible nitric oxide synthase and cyclooxygenase-2 expression. Food and Agricultural Immunology, 26(4), 496–503. doi: 10.1080/09540105.2014.968768
- Alabi, G. (1979). Bradford’s law and its application. International Library Review, 11, 151–158. doi: 10.1016/0020-7837(79)90044-X
- Behne, D., Alber, D., & Kyriakopoulos, A. (2009). Effects of long-term selenium yeast supplementation on selenium status studied in the rat. Journal of Trace Elements in Medicine and Biology, 23, 258–264. doi: 10.1016/j.jtemb.2009.05.001
- Bukkens, S. G., de Vos, N., Kok, F. J., Schouten, E. G., de Bruijn, A. M., & Hofman, A. (1990). Selenium status and cardiovascular risk factors in healthy Dutch subjects. Journal of the American College of Nutrition, 9, 128–135. doi: 10.1080/07315724.1990.10720361
- Cai, Y., Sukhova, G. K., Wong, H. K., Xu, A., Tergaonkar, V., & Vanhoutte, P. M. (2015). Rap1 induces cytokine production in pro-inflammatory macrophages through NFκB signaling and is highly expressed in human atherosclerotic lesions. Cell Cycle, 14, 3580–3592. doi: 10.1080/15384101.2015.1100771
- Cao, J., Guo, F., Zhang, L., Dong, B., & Gong, L. (2014). Pigs fed camelina meal increase hepatic gene expression of cytochrome 8b1, aldehyde dehydrogenase, and thiosulfate transferase. Journal of Animal Science and Biotechnology, 5, 1–7. doi:10.1186/2049-1891-5-1
- Conrado, A. B., D’Angelantonio, M., Torreggiani, A., Pecci, L., & Fontana, M. (2014). Reactivity of hypotaurine and cysteine sulfinic acid toward carbonate radical anion and nitrogen dioxide as explored by the peroxidase activity of Cu, Zn superoxide dismutase and by pulse radiolysis. Free Radical Research, 48, 1300–1310. doi: 10.3109/10715762.2014.951839
- Cubadda, F., Aureli, F., Ciardullo, S., D’Amato, M., Raggi, A., Acharya, R., … Nagaraja, T. P. (2010). Changes in selenium speciation associated with increasing tissue concentrations of selenium in wheat grain. Journal of Agricultural and Food Chemistry, 58, 2295–2301. doi: 10.1021/jf903004a
- Daham, K., James, A., Balgoma, D., Kupczyk, M., Billing, B., Lindeberg, A., … Dahlen, B. (2014). Effects of selective COX-2 inhibition on allergen-induced bronchoconstriction and airway inflammation in asthma. Journal of Allergy and Clinical Immunology, 134, 306–313. doi: 10.1016/j.jaci.2013.12.002
- Davis, F. Z., Ying, L. U., Thompson, J. T., Prabhu, K. S., Reddy, P. V., Sordillo, L. M., & Reddy, C. C. (2002). Nuclear factor-κB mediates over-expression of cyclooxygenase-2 during activation of RAW 264.7 macrophages in selenium deficiency. Free Radical Biology and Medicine, 32, 890–897. doi: 10.1016/S0891-5849(02)00775-X
- Dhanjal, N. I., Sharma, S., & Nagaraja, T. P. (2016). Quantification and in vitro bioaccessibility of selenium from Osborne fractions of selenium rich cereal grains. Cereal Chemistry Journal, 93, 339–343. doi: 10.1094/CCHEM-10-15-0199-R
- Duntas, L. H. (2009). Selenium and inflammation: Underlying anti-inflammatory mechanisms. Hormone and Metabolic Research, 41, 443–447. doi: 10.1055/s-0029-1220724
- Finley, J. W. (2006). Bioavailability of selenium from foods. Nutrition Reviews, 64, 146–151. doi: 10.1111/j.1753-4887.2006.tb00198.x
- Fukumoto, M., Kudou, D., Murano, S., Shiba, T., Sato, D., Tamura, T., … Inagaki, K. (2012). The role of amino acid residues in the active site of l-methionine γ-lyase from pseudomonas putida. Bioscience, Biotechnology, and Biochemistry, 76, 1275–1284. doi: 10.1271/bbb.110906
- Gandhi, U. H., Kaushal, N., Ravindra, K. C., Hegde, S., Nelson, S. M., Narayan, V., … Prabhu, K. S. (2011). Selenoprotein-dependent up-regulation of hematopoietic prostaglandin D2 synthase in macrophages Is mediated through the activation of peroxisome proliferator-activated receptor (PPAR) . Journal of Biological Chemistry, 286, 27471–27482. doi: 10.1074/jbc.M111.260547
- Gandhi, U. H., Nagaraja, T. P., & Prabhu, K. S. (2013). Selenoproteins and their role in oxidative stress and inflammation. Current Chemical Biology, 7, 65–73. doi: 10.2174/2212796811307010007
- Grkovich, A., Johnson, C. A., Buczynski, M. W., & Dennis, E. A. (2006). Lipopolysaccharide-induced cyclooxygenase-2 expression in human U937 macrophages is phosphatidic acid phosphohydrolase-1-dependent. Journal of Biological Chemistry, 281, 32978–32987. doi: 10.1074/jbc.M605935200
- Higgins, L. G., & Hayes, J. D. (2011). Mechanisms of induction of cytosolic and microsomal glutathione transferase (GST) genes by xenobiotics and pro-inflammatory agents. Drug Metabolism Reviews, 43, 92–137. doi: 10.3109/03602532.2011.567391
- Hoffmann, P. R. (2007). Mechanisms by which selenium influences immune responses. Archivum Immunologiae et Therapiae Experimentalis, 55, 289–297. doi: 10.1007/s00005-007-0036-4
- Huang, Z., Rose, A. H., & Hoffmann, P. R. (2012). The role of selenium in inflammation and immunity: From molecular mechanisms to therapeutic opportunities. Antioxidants and Redox Signaling, 16, 705–743. doi: 10.1089/ars.2011.4145
- Ipp, H., Zemlin, A. E., Erasmus, R. T., & Glashoff, R. H. (2014). Role of inflammation in HIV-1 disease progression and prognosis. Critical Reviews in Clinical Laboratory Sciences, 51, 98–111. doi: 10.3109/10408363.2013.865702
- Jana, D., Sarkar, D. K., Ganguly, S., Saha, S., Sa, G., Manna, A. K., … Mandal, S. (2014). Role of cyclooxygenase 2 (COX-2) in prognosis of breast cancer. Indian Journal of Surgical Oncology, 5, 59–65. doi: 10.1007/s13193-014-0290-y
- Kaushal, N., Kudva, A. K., Patterson, A. D., Chiaro, C., Kennett, M. J., Desai, D., … Prabhu, K. S. (2014). Crucial role of macrophage selenoproteins in experimental colitis. The Journal of Immunology, 193, 3683–3692. doi: 10.4049/jimmunol.1400347
- Kim, J. S., Kim, A. Y., Shin, H. M., Ahn, S. I., Shim, H. J., Nam, K. W., … Youn, H. S. (2017). Aster yomena suppresses LPS-induced cyclooxygenase-2 and inducible nitric oxide synthase expression. Food and Agricultural Immunology, 28, 202–210. doi: 10.1080/09540105.2016.1251395
- Kirkbya, N. S., Chanb, M. V., Zaissd, A. K., Vaze, E. G., Jiaod, J., Berglunde, L. M., … Mitchella, J. A. (2016). Systematic study of constitutive cyclooxygenase-2 expression: Role of NF-κB and NFAT transcriptional pathways. Proceedings of the National Academy of Sciences, 113, 434–439. doi: 10.1073/pnas.1517642113
- Lee, J. H., Kang, B. S., Hwang, K. H., & Kim, G. H. (2011). Evaluation for anti-inflammatory effects of siegesbeckia glabrescens extract in vitro. Food and Agricultural Immunology, 22, 145–160. doi: 10.1080/09540105.2010.549210
- Lu, J., Zhang, J., Jiang, C., Deng, Y., Ozten, N., & Bosland, M. C. (2016). Cancer chemoprevention research with selenium in the post-SELECT era: Promises and challenges. Nutrition and Cancer, 68, 1–17. doi: 10.1080/01635581.2016.1105267
- Maehira, F., Mivagi, I., & Eguchi, Y. (2003). Selenium regulates transcription factor NF-κB activation during the acute phase reaction. Clinica Chimica Acta, 334, 163–171. doi: 10.1016/S0009-8981(03)00223-7
- Meplan, C., Johnson, I. T., Polley, A. C. J., Cockell, S., Bradburn, D. M., Commane, D. M., … Hesketh, J. (2016). Transcriptomics and proteomics show that selenium affects inflammation, cytoskeleton, and cancer pathways in human rectal biopsies. The FASEB Journal, 30, 2812–2825. doi: 10.1096/fj.201600251R
- Nakayama, T., Esaki, N., Lee, W. J., Tanaka, I., Tanaka, H., & Soda, K. (1984). Purification and properties of L-methionine γ-lyase from aeromonas sp.. Agricultural and Biological Chemistry, 48, 2367–2369. doi: 10.1080/00021369.1984.10866502
- Nelson, S. M., Lei, X., & Prabhu, K. S. (2011). Selenium levels affect the IL-4–induced expression of alternative activation markers in murine macrophages. Journal of Nutrition, 141, 1754–1761. doi: 10.3945/jn.111.141176
- Nettore, I. C., De Nisco, E., Desiderio, S., Passaro, C., Maione, L., Negri, M., … Macchia, P. E. (2017). Selenium supplementation modulates apoptotic processes in thyroid follicular cells. Biofactors, 415–423. doi: 10.1002/biof.1351
- Onal, S., Naziroglu, M., Colak, M., Bulut, V., & Flores-Arce, M. F. (2011). Effects of different medical treatments on serum copper, selenium and zinc levels in patients with rheumatoid arthritis. Biological Trace Element Research, 142, 447–455. doi: 10.1007/s12011-010-8826-7
- Prabhu, K. S., Davis, F. Z., Stewart, J. B., Thompson, J. T., Sordillo, L. M., & Reddy, C. C. (2002). Selenium deficiency increases the expression of inducible nitric oxide synthase in RAW 264.7 macrophages: Role of nuclear factor-κB in up-regulation. Biochemical Journal, 366, 203–209. doi: 10.1042/bj20020256
- Premarathna, L., McLaughlin, M. J., Kirby, J. K., Hettiarachchi, G. M., Stacey, S., & Chittleborough, D. J. (2012). Selenate-enriched urea granules are a highly effective fertilizer for selenium biofortification of paddy rice grain. Journal of Agricultural and Food Chemistry, 60, 6037–6044. doi: 10.1021/jf3005788
- Rayman, M. P. (2012). Selenium and human health. The Lancet, 379, 1256–1268. doi: 10.1016/S0140-6736(11)61452-9
- Ricciotti, E., & FitzGerald, G. A. (2011). Prostaglandins and inflammation. Arteriosclerosis, Thrombosis, and Vascular Biology, 31, 986–1000. doi: 10.1161/ATVBAHA.110.207449
- Safir, N., Wendel, A., Saile, R., & Chabraoui, L. (2003). The effect of selenium on immune functions of J774.1 cells. Clinical Chemistry and Laboratory Medicine, 41, 1005–1011. doi: 10.1515/CCLM.2003.154
- Sakazaki, F., Arakawa, T., Shimizu, R., Ogino, H., Okuno, T., & Ueno, H. (2014). Allergies are aggravated by mild selenium deficiency and abrogated by supplementation with selenomethionine. Food and Agricultural Immunology, 25, 477–485. doi: 10.1080/09540105.2013.837866
- Schneider, C. A., Rasband, W. S., & Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675. doi: 10.1038/nmeth.2089
- Shih, R. H., Wang, C. Y., & Yang, C. M. (2015). NF-kappaB signaling pathways in neurological inflammation: A mini review. Frontiers in Molecular Neuroscience, 8, 3244–3248. doi: 10.3389/fnmol.2015.00077
- Taechowisan, T., Wanbanjob, A., Tuntiwachwuttikul, P., & Liuin, J. (2010). Anti-inflammatory effects of lansai C and D cause inhibition of STAT-1 and NF-κB activations in LPS-induced RAW 264.7 cells. Food and Agricultural Immunology, 21, 57–64. doi: 10.1080/09540100903419592
- Tanaka, H., Esaki, N., & Soda, K. (1985). A versatile bacterial enzyme: L-methionine γ-lyase. Enzyme and Microbial Technology, 7, 530–537. doi: 10.1016/0141-0229(85)90094-8
- Tyagi, S., Gupta, P., Saini, A. S., Kaushal, C., & Sharma, S. (2011). The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. Journal of Advanced Pharmaceutical Technology & Research, 2, 236–240. doi: 10.4103/2231-4040.90879
- Vunta, H., Davis, F., Palempalli, U. D., Bhat, D., Arner, R. J., Thompson, J. T., … Prabhu, K. S. (2007). The anti-inflammatory effects of selenium Are mediated through 15-deoxy- 12,14-prostaglandin J2 in macrophages. Journal of Biological Chemistry, 282, 17964–17973. doi: 10.1074/jbc.M703075200
- Youn, H. S., Lim, H. J., Choi, Y. J., Lee, J. Y., Lee, M. Y., & Ryu, J. H. (2008). Selenium suppresses the activation of transcription factor NF-κB and IRF3 induced by TLR3 or TLR4 agonists. International Immunopharmacology, 8, 495–501. doi: 10.1016/j.intimp.2007.12.008
- Yui, K., Imataka, G., Nakamura, H., Ohara, N., & Naito, Y. (2015). eicosanoids derived from arachidonic acid and their family prostaglandins and cyclooxygenase in psychiatric disorders. Current Neuropharmacology, 13, 776–785. doi: 10.2174/1570159X13666151102103305
- Zeng, H., Botnen, J., & Jhonson, L. (2008). A selenium-deficient caco-2 cell model for assessing differential incorporation of chemical or food selenium into glutathione peroxidase. Biological Trace Element Research, 123, 98–108. doi: 10.1007/s12011-008-8097-8