ABSTRACT
The genus Fictibacillus contains twelve species significant in the synthesis of cellulose-degrading enzymes and phenylalanine dehydrogenase, isolated mainly from marine sedimentary environments. Here, we report a new biosurfactant-producing strain, Fictibacillus nanhaiensis ME46, isolated from Daqing oil field in China. The biosurfactant extracted from Strain ME46 was determined as surfactin, one of the representative families of lipopeptide biosurfactants. The yield of the surfactin produced by strain ME46 was 0.62 g·L−1 as determined by high-performance liquid chromatography, and the critical micelle concentration (CMC) of the surfactin was estimated to be about 68 mg·L−1 and the surface tension at CMC was 35.1 mN·m−1. This study extended our knowledge about the role of the species Fictibacillus nanhaiensis in the ecosystem of natural environments such as the oil field.
Introduction
Biosurfactants are a class of amphiphilic surface-active molecules and about 2000 different amphiphilic structures of biological origin have been described [Citation1]. Biosurfactants have a broad spectrum of industrial applications due to their emulsifying, foaming, detergency, dispersing, wetting, penetrating and antimicrobial properties [Citation2]. Microbial lipopeptides, one of the representative biosurfactants, are structurally characterized by cyclic peptides acylated with fatty acids [Citation3]. Fifty-seven families of lipopeptides covering about 400 lipopeptide compounds have been reported for two decades [Citation4]. About sixteen species of Gram-negative Pseudomonas have the potential to synthesize lipopeptide since Pseudomonas syringae PV has been reported to produce lipopeptide Phytotoxins in 1997 [Citation5]. In addition, a lipopeptide with a critical micelle concentration of 300 mg·L−1 isolated from Gram-negative Acinetobacter junii B6 in 2018 [Citation6]. Besides, Gram-negative Serratia sp. was reported to produce Stephensiolides in 2018 [Citation7]. And the lipopeptide produced by Gram-negative Citrobacter freundii MG812314.1 isolated from Al-Rahawy drain sediment has ability to remove heavy metals in wastewater in 2019 [Citation8]. In addition to Gram-negative bacteria, lipopeptides can also be synthesized and secreted by Gram-positive bacteria, including Bacillus, Lactobacillus, and Streptomyces. The first discovered natural lipopeptide was surfactin produced by Bacillus subtilis in 1968 [Citation4]. Mass spectrometric analysis of the products of Gram-positive Paenibacillus polymyxa revealed as fusaricidin B and polymyxin D1 in 2012 [Citation9]. The lipopeptide produced by gram-positive Rhodococcus opacus R7 was classified as a hydrophobic peptide in 2022 [Citation10]. And biosurfactants isolated from Streptomyces bikiniensis HD-087 were identified as surfactins, iturins, and fengycins in 2022 [Citation11]. Among them, surfactin is a cyclic heptapeptide attached to a β-OH (lactone) fatty acid chain of 11–16 carbon atoms lengths [Citation12–14]. Surfactin has received increasing attention as an antibacterial agent in the pharmaceutical industry, oil displacing agents in microbial enhanced oil recovery (MEOR), pesticides in agricultural control, and functional reagents in bioremediation [Citation15]. Although the biotechnological production of microbial biosurfactants has already been established so far, the number of effectively applicable biosurfactant-producing strains is still limited.
The genus Fictibacillus are gram-positive, motile, rod-shaped, endospore-forming bacteria [Citation16]. The earliest reported species of Fctibacillus was Fctibacillus barbaricus, which was previously classified as Bacillus barbaricus in 2003 [Citation17]. Glaeser et al. (Citation2013) further proposed the genus Fictibacillus and reclassified B. nanhaiensis, B. barbaricus, B. arsenicus, B. rigui, B. macauensis and B. gelatini as F. nanhaiensis, F. barbaricus, F.arsenicus, F.rigui, F.macauensis and F.gelatini, respectively [Citation18]. Until now, twelve species of Fctibacillus have been reported, namely F.phosphorivorans (2013) [Citation18], F. enclensis (2014) [Citation19], F. halophilus (2016) [Citation20], F. aquaticus (2018) [Citation21], F. iocasae (2018) [Citation22], F. marinisediminis (2022) [Citation23] and so on. F. nanhainensis JSM 082006T was first isolated on Naozhou Island in the South China Sea in 2011 [Citation24]. In 2016, a marine microorganism F. nanhaiensis DSF-15A2 with high salt activity and organic solvent heat-resistant phenylalanine dehydrogenase activity was reported [Citation25]. F. nanhaiensis LG2 with excellent cellulose degrading activity was isolated from coastal mangrove sediments [Citation26]. F. barnaricus DR4.0 reported in 2015 can synthesize bioemulsifier with an emulsification index of 63% [Citation27]. F. phosphorivorans R3 isolated in 2015 can use petroleum hydrocarbons to produce biosurfactants [Citation28]. However, only a limited number of biosurfactant compounds have been isolated, identified and characterized from the genus Fictibacillus.
In this study, a novel biosurfactant-producing bacterium, F. nanhaiensis ME46, was isolated from Daqing oil field, China. The composition, chemical structure and the surface activity of the surfactin produced by the strain F. nanhaiensis ME46 were analyzed using a combination of highly performance liquid chromatography and mass spectrometry. Fictibacillus species and in vitro experiments with synthetic biosurfactants improve understanding of the role of Fictibacillus species in natural ecosystems.
Material and methods
Materials
All the chemicals and biochemicals used were from Shanghai Meryer Chemical Technology Co., Ltd, Shanghai, China. Acetonitrile and methanol (HPLC-grade) were from Shanghai Boer Chemical reagents Co., Ltd, Shanghai. Acetonitrile and methanol (LC-MS grade) were from Shanghai Macklin Biochemical Co., Ltd, Shanghai, China. Genomic DNA Miniprep Kit was from Axygen Bioscience Co., Ltd, Union City, CA, USA. Produced water was collected from the Daqing Oilfield in China for microorganism isolation.
Screening for biosurfactants-producing microorganisms
Medium: Mineral Salt Medium (MSM) [Citation29] contains sucrose (20 g) as carbon source: Yeast extract 0.5 g, NaCl 1.1 g, NaNO3 15 g, KH2PO4 3.4 g, MgSO4·7 H2O 0.5 g, KCl 1.1 g, FeSO4·7 H2O, Na2HPO4 4.4 g per litre. Luria–Bertani (LB) medium: peptone 10.0 g, NaCl 10.0 g, yeast extract 5.0 g per litre The pH of the medium was adjusted to 7.2 after the preparation was completed. The medium sterilization condition was set at 115°C for 30 min.
Surface activity analysis: The sample used for surface tension analysis is the strain culture fluid after centrifugation to remove cell. The surface tension test instrument adopts a fully automatic tensiometer JK99B (Shanghai Wangxu Electric Co., Ltd., China). The tensiometer conditions of the surface tension were set as the measurement temperature: 25°C, the pH of the culture solution: 7.2, and all the test data were averaged three times in parallel [Citation30].
Phylogenetic tree construction
The AxyPrep™ Bacterial Genomic DNA Miniprep Kit (Axygen Bioscience, Union City, CA, USA) was used to extract total bacterial DNA. Polymerase Chain Reaction (PCR) Procedure: denaturation temperature: 95 °C, annealing temperature: 53 °C, extension temperature: 72 °C. The PCR procedure selected universal bacterial primers 27F (5'-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5'-GGTTACCTTGTTACGACTT-3’) [Citation31]. The phylogenetic tree reference database was selected from the National Centre for Biotechnology Information (NCBI) database. MEGA7.0 32-set arithmetic mean (UPGMA) and bootstrap's unweighted pairwise method were used to construct the phylogenetic tree [Citation32]..
Extraction of biosurfactant by acid precipitation
Acid precipitation method [Citation30]: the fermentation of the strain was centrifuged at 5,000×g for 25 mins to remove the cell. Because of the difference in solubility of lipopeptide amino acids under different pH conditions, 6M HCl was added to the cell-free fermentation broth to adjust the pH of the final fermentation to 2.0, and the crude product of lipopeptide was precipitated in a 4-degree refrigerator for 12 h. The biosurfactant extract is obtained by extracting the previously obtained biosurfactant crude product with ethyl acetate for three times and combining ethyl acetate. Ethyl acetate was removed by rotary evaporation to obtain a purified biosurfactant extract [Citation33].
Electrospray mass spectrometry: the extracted biosurfactant extract was dissolved in methanol, and the analytical instrument was electrospray mass spectrometry ESI-MS (Micromass Co., UK). The mass spectrometry programme was set as: negative ion and positive ion modes were selected for mass spectrometry analysis model, the ionization source temperature was 80 °C, the electrolyte voltage was 100 V, and the inlet temperature was 120 °C.
Reverse-phase HPLC analysis: HPLC system (LC 100) was equipped with HiQ sil C18 W column (KYA TECH, Japan; 5 μm, 250 × 21.2 mm). Reversed-phase HPLC analysis procedure: column temperature 30 °C, elution mode selection gradient elution, mobile phase 0.1% (v/v) formic acid added to acetonitrile solution and 0.1% (v/v) formic acid is dissolved in water, the flow rate of the mobile phase is 1 ml·min−1, and the detector is a UV spectroscopic detector with a wavelength of 205nm [Citation34]. All test data were averaged three times in parallel.
Tandem mass spectrometry: 5 mg of biosurfactant extract was dissolved in 1 ml of acetonitrile and analyzed by LC-MS/MS (Thermo Q-Exactive plus). The tandem mass spectrometry programme sets the collision energy of the mass spectrometer system to 30 V, and the mass spectrometer selects the target hydrogen ionized molecules to obtain ionized fragments generated by the fragmentation of hydrogen ionized molecules [Citation35]..
Results and discussion
Biosurfactant-producing microorganism
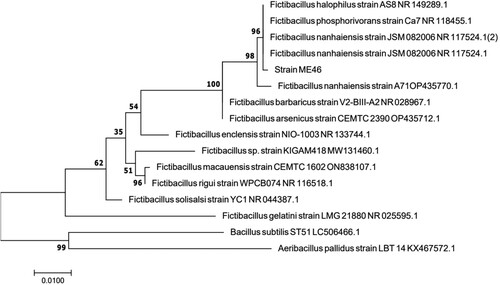
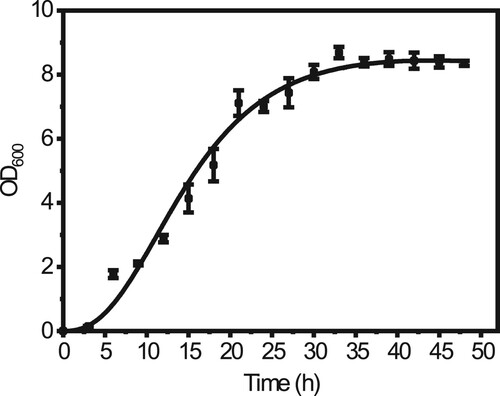
We isolated a novel strain ME46 which could reduce the surface tension of the medium to 32.8 mN·m−1 (The surface tension of sterile medium was 67.5 mN·m−1) from the Daqing oil field, China. The hemolytic assay test and the droplet collapse test were positive. The phylogenetic tree anlysis based on the sequences of the 16S rRNA-coding gene suggests that the strain belongs to F. nanhaiensis (). This is also the first report that the endospore forming bacterium, F. nanhaiensis ME46 isolated from oil reservoir has the ability to synthesize biosurfactants. The optical density value of F. nanhaiensis ME46 was measured every three hours during the 37 °C culture in LB medium. is the growth curve of strain ME46, and its maximum bacterium optical density value can reach 8.48 ± 0.21. In the first three hours, strain ME46 was in the lag phase then in the logarithmic phase from three hours to thirty hours, and enters the stationary phase after thirty hours. The synthesis of biosurfactants is basically accompanied by the sporulation process, so the synthesis of biosurfactants usually occurs after bacterial growth enters the stationary phase [Citation36].
The surface activity
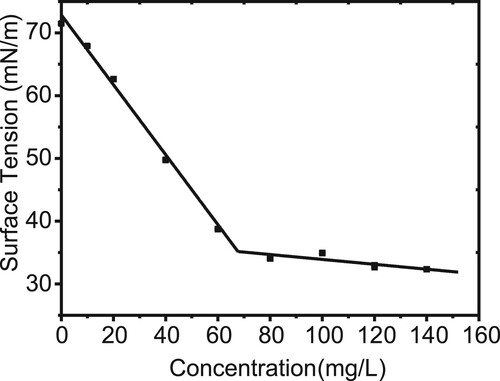
The surface activity of the biosurfactant extracted from F. nanhaiensis ME46 was measured by a surface tensiometer. The surface tension of the solution decreased rapidly and then remained stable with the concentration of the biosurfactant. As shown in , the turning point of the surface tension curve corresponds to the critical micelle concentration of the biosurfactant at 68 mg·L−1, which corresponds to a surface tension of 35.1 mN·m−1.
Figure 3. Surface tensions of the purified biosurfactant at different concentrations. The black dots represent the measured surface tension of the purified biosurfactant at different concentrations. The intersection of the two black lines represents the CMC, which is estimated to be approximately 68 mg·L−1.
Structure and composition of biosurfactants
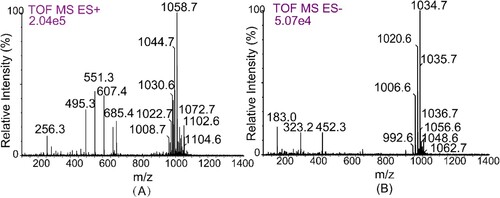
Electrospray mass spectrometry provided molecular weight information for the biosurfactants produced by F. nanhaiensis ME46 mixture. As show in A, six major peaks (m/z 1008.7, 1022.6, 1044.7, 1058.7, 1072.7, 1102.6, and 1104.6) were detected for positive ionization. The mass-to-charge ratio difference between each peak was fourteen, which corresponds exactly to the CH2 residues. The results of mass spectrogram comparison showed that the purified biosurfactant was a mixture of surfactants. Negative ionization detected peaks with different m/z values 992.6, 1006.6, 1020.6, 1034.7, 1048.6, and 1062.7 (B), which had mass-to-charge ratios of exactly one [Na+] molecular weight less than those detected by positive ionization. Thus, the biosurfactant produced by strain ME46 was identified as a surfactin variant.
Figure 4. ESI-MS of the biosurfactant produced by F. nanhaiensis ME46. (A) is positive mode and (B) is negative mode.
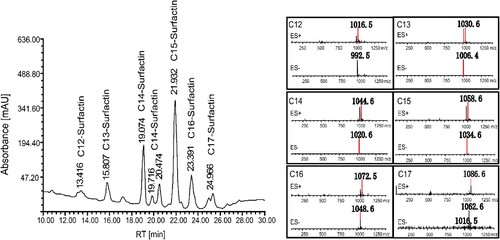
The biosurfactant extract of F. nanhaiensis ME46 was separated by HPLC into ten peaks with retention times ranging from 10 min to 30 min. As shown in , the chromatographic peak with a retention time of 21.932 min corresponding to mass-to-charge ratios of 1034.6 and 1058.6 was identified as C15-surfactin, and the peak with a retention time of 23.391 min was identified as C16-surfactin. According to the mass-to-charge ratio obtained by liquid chromatography-mass spectrometry, it was confirmed that the incompletely resolved doublet with a retention time of 24.966 min represents C17-surfactin, and its corresponding mass-to-charge ratio was determined to be 1062.6 and 1086.6. In addition, the peak of C12-surfactin appeared at 13.416 min, corresponding to mass-to-charge ratios of 992.5 and 1030.6. And the three peaks with retention times of 19.074, 19.761 and 20.474 min respectively represented C14-surfactin with mass-to-charge ratios of 1020.6 and 1044.6 Active variants. The yield of the strain ME46 was determined to be 0.62 g·L−1 by using the surfactin sample preserved in our laboratory as a standard [Citation35]. The proportions of surfactin variants were C12-surfactin (4.6%), C13-surfactin (6.3%), C14-surfactin (27.2%), C15-surfactin (41.7%), C16-surfactin (14.3%) and C17-surfactin (5.8%), respectively.
Figure 5. Composition of the biosurfactant produced by F. nanhaiensis ME46. The mass-to-charge ratios of positive and negative ion mass spectra of extracted ion mass spectrometry correspond to the relevant liquid chromatographic peaks.
The ability of synthesizing phenylalanine dehydrogenase and cellulolytic enzymes by F. nanhaiensis has attracted attention [Citation16, Citation25]. However, there are rare reports about the lipopeptide produced by Fictibacillus species. The main factor determining biosurfactants biosynthesis is the genetic makeup of the producer organisms and the induction by environmental factors is also essential (such as oil reservoirs) [Citation37]. Genome analysis of F. marinisediminis KIGAM418T revealed the presence of type III Polyketide Synthases (PKS) and Nonribosomal Peptide Synthase (NRPS) clusters which are necessary genes responsible for lipopeptide synthesis [Citation23]. Generally, endospore-forming bacteria of the genera Bacillus, Enterobacter, and Pseudomonas are commonly reported as producers of lipopeptides [Citation38]. In this study, we report a new biosurfactant-producing strain, F. nanhaiensis ME46, isolated from the oil field. The structure of biosurfactants produced by F. nanhaiensis ME46 was identified as surfactin with six variants. ESI-MS analysis of Bacillus velezensis SK product reveals a set of protonated molecular ion peaks [M + Na]+ with m/z ratios 1043.58, 1057.59 and 1071.61 corresponding to three component surfactin variants (C14-surfactin, C15-surfactin, C16-surfactin) [Citation39]. In fact, most lipopeptide-producing bacteria mainly synthesize surfactin with fatty acid chain lengths of fourteen, fifteen and sixteen carbon atoms, and only a few have been reported to synthesize surfactin with fatty acid chain lengths of seventeen carbon atoms or surfactin with fatty acid chain lengths of twelve carbon atoms. Synthesis of lipopeptides usually accompanies sporulation and is dominated by clusters of nonribosomal peptide synthetases. Microbes will adjust their internal growth and metabolic processes in response to environmental pressure, and the composition and component ratio of surfactin can reflect the internal self-regulation of microorganisms to a certain extent degree [Citation36]. Therefore, Genetic analysis of Fictibacillus species and in vitro experiments with synthetic biosurfactants improve understanding of the role of Fictibacillus species in natural ecosystems. In 2023, the surfactin with seventeen carbon atoms fatty acid chain was showed a very low critical micelle concentration (CMC) of 4.02 × 10−6 mol·L−1 [Citation33]. In general, the longer the fatty acid chain of surfactin, the lower the critical micelle concentration [Citation40]. Therefore, different ratios of surfactin variants correspond to different critical micelle concentrations, and the greater the proportion of long-chain surfactin variants, the greater the contribution to low critical micelle concentration. Naturally, the evaluation of the surface activity of biosurfactants is the key question to the industrial application.
Conclusions
In this study, a new endospore-forming bacteria, F. nanhaiensis ME46, with biosurfactant-producing ability, was isolated from Daqing oil field, China. The critical micelle concentration of the biosurfactants was calculated as 68 mg·L−1 and the corresponding surface tension was 35.1 mN·m−1. The structure of biosurfactants produced by F. nanhaiensis ME46 was identified as surfactin with six variant·s and the yield of the surfactins produced by this strain ME46 was 0.62 g·L−1.
Author contributions
Shi-Zhong Yang, and Bo-Zhong Mu. conceptualized and supervised the overall work; Jia-Yi Li carried out the experiments and validate the data; Jia-Yi Li, Shi-Zhong Yang, and Bo-Zhong Mu. writing original draft preparation; Yi-Fan Liu, Lei Zhou, Hong-Ze Gang, Jin-Feng Liu, Gang-Zheng Sun, and Wei-Dong Wang writing, review and editing; Jia-Yi Li, Yi-Fan Liu, Lei Zhou, Hong-Ze Gang, Jin-Feng Liu, Gang-Zheng Sun, Wei-Dong Wang, Shi-Zhong Yang, and Bo-Zhong Mu. All authors have read and agreed to the published version of the manuscript.
Data accessibility statement
The data underlying this article are available in [the GenBank Nucleotide Database] at [NCBI], and can be accessed with [Genbank accessions number OP999719].
Acknowledgment
The authors thank the Research Programme of the State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, for providing facilities and financial support. The authors are also thankful to the School of Chemistry and Molecular Engineering, East China University of Science and Technology.
Disclosure statement
Author Jin-Feng Liu is employed by Daqing Huali Biotechnology Co., Ltd. Author Gang-Zheng Sun and Wei-Dong Wang are employed by Research Institute of Petroleum Engineering and Technology, Shengli Oilfield Company, Sinopec, Dongying, China. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Funding
References
- Gayathiri E, Prakash P, Karmegam N, et al. Biosurfactants: Potential and eco-friendly material for sustainable agriculture and environmental safety-a review. Agronomy. 2022;12(3):662. doi:10.3390/agronomy12030662
- Singh P, Patil Y, Rale V. Biosurfactant production: emerging trends and promising strategies. J Appl Microbiol 2019;126(1):2–13. doi:10.1111/jam.14057
- Twigg MS, Baccile N, Banat IM, et al. Microbial biosurfactant research: time to improve the rigour in the reporting of synthesis, functional characterization and process development. Microb Biotechnol. 2021;14(1):147–170. doi:10.1111/1751-7915.13704
- Li JY, Wang L, Liu YF, et al. Microbial lipopeptide-producing strains and their metabolic roles under anaerobic conditions. Microorganisms. 2021;9(10):2030. doi:10.3390/microorganisms9102030
- Hutchison ML, Gross DC. Lipopeptide phytotoxins produced by pseudomonas syringaepv.syringae: comparison of the biosurfactant and Ion channel-forming activities of syringopeptin and syringomycin. Mol Plant Microbe Interact 1997;10(3):347–354. doi:10.1094/MPMI.1997.10.3.347
- Ohadi M, Dehghannoudeh G, Forootanfar H, et al. Investigation of the structural, physicochemical properties, and aggregation behavior of lipopeptide biosurfactant produced by Acinetobacter junii B6. Int J Biol Macromol 2018;112:712–719. doi:10.1016/j.ijbiomac.2018.01.209
- Ganley JG, Carr G, Ioerger TR, et al. Discovery of antimicrobial lipodepsipeptides produced by aserratiasp. within mosquito microbiomes. ChemBioChem. 2018;19(15):1590–1594. doi:10.1002/cbic.201800124
- Gomaa EZ, El-Meihy RM. Bacterial biosurfactant from Citrobacter freundii MG812314.1 as a bioremoval tool of heavy metals from wastewater. Bull Nat Res Cent. 2019;43:1–14. doi:10.1186/s42269-019-0088-8
- Quinn GA, Maloy AP, McClean S, et al. Lipopeptide biosurfactants from Paenibacillus polymyxainhibit single and mixed species biofilms. Biofouling. 2012;28(10):1151–1166. doi:10.1080/08927014.2012.738292
- Zampolli J, De Giani A, Di Canito A, et al. Identification of a novel biosurfactant with antimicrobial activity produced by Rhodococcus opacus R7. Microorganisms. 2022;10(2):475. doi:10.3390/microorganisms10020475
- Liu W, Wang J, Zhang H, et al. Xanthoepocin, a photolabile antibiotic of Penicillium ochrochloron CBS 123823 with high activity against multiresistant gram-positive bacteria. Microb Cell Fact 2022;21(1):1–11. doi:10.1186/s12934-021-01718-9
- Yang SC, Wang YH, Ho CM, et al. Targeting formyl peptide receptor 1 withanteiso-C13-surfactin for neutrophil-dominant acute respiratory distress syndrome. Br J Pharmacol 2023. doi:10.1111/bph.16073
- Liu XY, Yang SZ, Mu BZ. Production and characterization of a C15-surfactin-O-methyl ester by a lipopeptide producing strain Bacillus subtilis HSO121. Process Biochem. 2009;44(10):1144–1151. doi:10.1016/j.procbio.2009.06.014
- Li Y, Zou AH, Ye RQ, et al. Counterion-induced changes to the micellization of surfactin-C16 aqueous solution. J Phys Chem B. 2009;113(46):15272–15277. doi:10.1021/jp9062862
- Brown LR. Microbial enhanced oil recovery (MEOR). Curr Opin Microbiol. 2010;13(3):316–320. doi:10.1016/j.mib.2010.01.011
- Chen Y, Wang W, Zhou D, et al. Biodegradation of lignocellulosic agricultural residues by a newly isolated Fictibacillus sp. YS-26 improving carbon metabolic properties and functional diversity of the rhizosphere microbial community. Bioresour Technol 2020;310:123381. doi:10.1016/j.biortech.2020.123381
- Täubel M, Kämpfer P, Buczolits S, et al. Bacillus barbaricus sp. nov., isolated from an experimental wall painting. Int J Syst Evol Microbiol 2003;53(3):725–730. doi:10.1099/ijs.0.02304-0
- Glaeser SP, Dott W, Busse HJ, et al. Fictibacillus phosphorivorans gen. nov., sp. nov. and proposal to reclassify Bacillus arsenicus,Bacillus barbaricus,Bacillus macauensis,Bacillus nanhaiensis,Bacillus rigui,Bacillus solisalsiand Bacillus gelatiniin the genus Fictibacillus. Int J Syst Evol Microbiol 2013;63(8):2934–2944. doi:10.1099/ijs.0.049171-0
- Dastager SG, Mawlankar R, Srinivasan K, et al. Fictibacillus enclensis sp. nov., isolated from marine sediment. Antonie Van Leeuwenhoek. 2014;105(3):461–469. doi:10.1007/s10482-013-0097-9
- Sharma A, Kohli P, Singh Y, et al. Fictibacillus halophilus sp. nov., from a microbial mat of a hot spring atop the Himalayan Range. Int J Syst Evol Microbiol 2016;66(6):2409–2416. doi:10.1099/ijsem.0.001051
- Pal D, Bhardwaj A, Kaur N, et al. Fictibacillus aquaticus sp. nov., isolated from downstream river water. Int J Syst Evol Microbiol 2018;68(1):160–164. doi:10.1099/ijsem.0.002474
- Wang HL, Zhang J, Sun L. Fictibacillus iocasae sp. nov., isolated from the deep-sea sediment in Pacmanus, Manus Basin. Arch Microbiol 2018;200(7):1123–1128. doi:10.1007/s00203-018-1527-x
- Cho ES, Hwang CY, Jung DH, et al. Fictibacillus marinisediminis sp. nov., a nitrate-reducing bacterium isolated from marine sediment in Hupo Basin, Republic of Korea. Arch Microbiol 2022;204(8):1–8. doi:10.1007/s00203-022-03067-6
- Chen YG, Zhang L, Zhang YQ, et al. Bacillus nanhaiensis sp. nov. Bacillus nanhaiensis sp. nov., isolated from an oyster. Int J Syst Evol Microbiol 2011;61(4):888–893. doi:10.1099/ijs.0.022889-0
- Jiang W, Sun D, Ren H, et al. Isolation, purification and characterization of a salt-active and organic-solvent-thermostable phenylalanine dehydrogenase from Bacillus nanhaiensis DSF-15A2. J Mol Catal B: Enzym. 2016;133:12–19. doi:10.1016/j.molcatb.2016.07.009
- Pramono H, Mariana A, Ryandini D, et al. Short communication: diversity of cellulolytic bacteria isolated from coastal mangrove sediment in Logending Beach, Kebumen, Indonesia. Biodiversitas. 2021;22(4):1869–1878. doi:10.13057/biodiv/d220433
- Couto CRdA, Alvarez VM, Marques JM, et al. Inhibitory effect of zingiber officinale towards Streptococcus mutans virulence and caries development: in vitro and in vivo studies. BMC Microbiol 2015;15(1):1–17. doi:10.1186/s12866-014-0320-5
- Pandey R, Sharma P, Rathee S, et al. Isolation and characterization of a novel hydrocarbonoclastic and biosurfactant producing bacterial strain: Fictibacillus phosphorivorans RP3. 3 Biotech. 2021;11(2):1–11. doi:10.1007/s13205-021-02655-5
- Ahsani Arani Y, Noormohammadi Z, Rasekh B, et al. Evaluation of rhamnosyl transferase enzyme expression in the bio surfactant production pathway of Pseudomonas aeruginosa bacteria in the presence of SDS-based iron nanostructure. New Cell Mol Biotechnol J. 2022;13(49):3–10. http://ncmbjpiau.ir/article-1-1474-en.html
- Modabber G, Akhavan Sepahi A, Yazdian F, et al. Evaluation of production of lipopeptide biosurfactants and surfactin micelles by native Bacillus of Iran, for a broader application range. J Surfactants Deterg. 2023;26(1):3–11. doi:10.1002/jsde.12626
- Harirchi S, Sar T, Ramezani M, et al. Bacillales: from taxonomy to biotechnological and industrial perspectives. Microorganisms. 2022;10(12):2355. doi:10.3390/microorganisms10122355
- Jin W, Zhang Y, Su X, et al. First report of apple latent spherical virus infecting Angelica sinensisin China. Plant Dis. 2023;107(4):1252. doi:10.1094/PDIS-08-22-1756-PDN
- Qin WQ, Fei D, Zhou L, et al. A new surfactin-C 17 produced by Bacillus subtilis TD7 with a low critical micelle concentration and high biological activity. New J Chem. 2023;47(16):7604–7612. doi:10.1039/D3NJ00123G
- Liu JF, Yang J, Yang SZ, et al. Effects of different amino acids in culture media on surfactin variants produced by Bacillus subtilis TD7. Appl Biochem Biotechnol 2012;166:2091–2100. doi:10.1007/s12010-012-9636-5
- Li JY, Liu YF, Zhou L, et al. Structural diversity of the lipopeptide biosurfactant produced by a newly isolated strain, Geobacillus thermodenitrifcans ME63. ACS Omega. 2023;8:22150–22158. doi:10.1021/acsomega.3c02194
- Ishikawa F, Konno S, Uchida C, et al. Chemoproteomics profiling of surfactin-producing nonribosomal peptide synthetases in living bacterial cells. Cell Chem Biol. 2022;29(1):145–156.e8. doi:10.1016/j.chembiol.2021.05.014
- Kubicki S, Bollinger A, Katzke N, et al. Marine biosurfactants: biosynthesis, structural diversity and biotechnological applications. Mar Drugs. 2019;17(7):408. doi:10.3390/md17070408
- Chafale A, Kapley A. Biosurfactants as microbial bioactive compounds in microbial enhanced oil recovery. J Biotechnol 2022. doi:10.1016/j.jbiotec.2022.05.003
- Barale SS, Ghane SG, Sonawane KD. Purification and characterization of antibacterial surfactin isoforms produced by Bacillus velezensis SK. Amb Express. 2022;12(1):1–20. doi:10.1186/s13568-022-01348-3
- Zhou Y, Liu F, Liu L, et al. Effects of length and type of the alkyl chain on the micellization behavior of mixed systems of HS15 with fatty acids. Food Chem. 2022;397:133830. doi:10.1016/j.foodchem.2022.133830