Abstract
The genetic diversity of the green algal genus Ulva sensu lato in the New Zealand region was surveyed, examining rbcL sequences of 581 samples from a wide geographical range. Twenty-four genetically distinct taxa were discovered in New Zealand waters, belonging to three genera–Ulva (19 species), Umbraulva (four species) and Gemina (one species). Of the 19 species of Ulva reported here, 13 could be identified to the species level based on morphological and genetic data. The remaining six species cannot currently be assigned to known species groups due to a lack of close homology with sequences in GenBank. These species may include undescribed endemic taxa, recognised taxa for which rbcL sequences are not yet available, or may represent cryptogenic species. The genus Umbraulva is recorded for the first time for the New Zealand region and for the Southern Hemisphere. Of the four species distinguished, one is considered to be introduced to the region and the other three are undescribed indigenous taxa. Subantarctic samples provide the first evidence of the genus Gemina since its description in 1952: sequence data confirmed that Gemina is distinct from Ulva and Umbraulva. A number of the species identified in this study can be distinguished through a combination of growth form, morphological, ecological and distributional characters. However there remain considerable problems in distinguishing a number of other species by morphological characters alone. Based on information such as distribution in New Zealand (percentage of samples occurring in highly modified environments and/or areas with frequent vessel traffic), as well as the genetic similarity of New Zealand samples to material from overseas, we have concluded that at least five species have been introduced to the New Zealand region: Ulva armoricana, U. californica, U. flexuosa, U. lactuca and Umbraulva olivascens.
Introduction
The genus Ulva L. sensu lato (Ulvaceae, Ulvales, Chlorophyta) is found throughout the world, with many entities described: 548 names are listed in AlgaeBase of which 125 are considered current (Guiry & Guiry, Citation2007). Recent systematic studies employing molecular sequencing data have established that Ulva and Enteromorpha Link are paraphyletic with respect to each other: generic separation based on thallus form (placement in Ulva for sheet-like thalli and Enteromorpha for tubular thalli) does not reflect the phylogenetic relationships of species placed in these taxa (Malta et al., Citation1999; Blomster et al., Citation2002; Hayden & Waaland, Citation2002; Hayden et al., Citation2003). As a consequence they have been synonymised and species previously placed in Enteromorpha are now placed in the genus Ulva (Hayden et al., Citation2003).
Species of Ulva are found in a wide range of habitats and environments, and they are notoriously difficult to identify because of the morphological plasticity expressed by many members, and the few reliable characters available for differentiating taxa. In the New Zealand region, species of Ulva have been recorded from the subtropical Kermadec Islands through to the subantarctic islands, and from the high intertidal shore through to subtidal habitats of 40 m depth (Chapman, Citation1956; Adams, Citation1994). In the only full account of the New Zealand green algal flora published to date, Chapman (Citation1956) described many taxa in the Ulvaceae, including two new genera for ulvoid algae, Gemina V.J.Chapm. and Lobata V.J.Chapm. Within Ulva sensu lato Chapman (Citation1956) distinguished 67 separate entities including subspecific rankings (46 within Enteromorpha and 21 within Ulva), many of which subsequent authors have had difficulty recognising (e.g. Adams, Citation1994).
The capacity of many Ulva species to tolerate a wide range of conditions makes them ideal candidates for human-mediated dispersal; they are among the most notorious fouling organisms on ships’ hulls (Schaffelke et al., Citation2006). A number of ulvacean species are regarded as cryptogenic based on their cosmopolitan distribution, and may have been distributed in previous centuries by wooden sailing vessels. Shipping continues to be an important vector for the worldwide transport of macroalgal species (Schaffelke et al., Citation2006). Ulva species are also known for their potential for rapid, proliferous growth. A number of studies have focused on ‘green tide’ phenomena, where species of Ulva produce large amounts of biomass, apparently in response to human activities, such as eutrophication of the environment (e.g. Hawes & Smith, Citation1995; de Winton et al., Citation1998; Raffaelli et al., Citation1998; Malta et al., Citation1999; Taylor, Citation1999; Blomster et al., Citation2002; Nelson et al., Citation2003; Hiraoka et al., Citation2004 a).
An important element in assessing the risks posed by introduced species is the capacity to recognise introduced and potentially invasive taxa. Reliable taxonomy is a critical cornerstone for biosecurity management. Although it is clear that a thorough taxonomic investigation of Ulva in New Zealand is required in order to recognise potentially introduced species, the confused nomenclatural history and taxonomy, and the paucity of reliable morphological characters, mean that a revision based solely on traditional approaches is unlikely to be satisfactory. Molecular sequencing data have been shown to be very useful in untangling relationships and clarifying morphological plasticity along ecological and environmental gradients within Ulva (e.g., Tan et al., Citation1999; Hayden & Waaland, Citation2002; Shimada et al., Citation2003). Preliminary molecular sequencing data of samples from New Zealand indicated that some of the species concepts previously applied in New Zealand were likely to be incorrect (McIvor & Maggs, unpublished data, pers. comm.).
The objective of this study was to assess the genetic diversity of Ulva sensu lato in New Zealand from a range of habitats reflecting differing degrees of human modification. We explored whether such an approach would allow taxa to be distinguished, and would also provide data to enable the recognition of native and non-native taxa.
Materials and methods
Field work
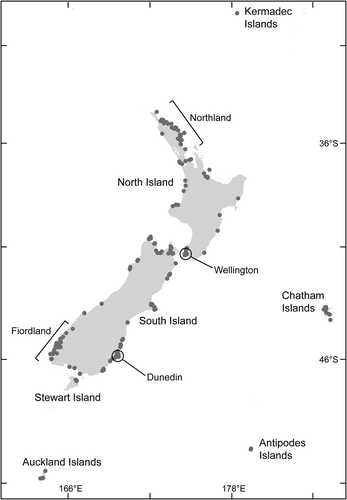
Collections of Ulva were obtained from populations throughout the New Zealand region (). In total 195 localities were visited (Kermadecs: 1; North Island: 75; South Island: 97; Chatham Islands: 11; Stewart Island: 5; Antipodes Islands: 2; Auckland Islands: 4). Full collection data are available in Heesch et al. (Citation2007). The sampling sites covered a wide geographic range and spectrum of habitats, including varying degrees of wave exposure and freshwater influence. Specimens were collected to represent populations of particular morphologies, as well as from particular habitat types e.g. high, mid and low intertidal pools, exposed vertical faces, on rocks experiencing sand abrasion, epiphytic/epiplithic growth forms. Both pristine and modified environments were specifically targeted. Seasonal collections were made in Wellington and Dunedin, re-sampling the same sites in spring (September–November), summer (December–February), autumn (March–May), and winter (June–August) (Wellington), and in spring, summer and autumn (Dunedin). Additional samples examined in this study were collected as part of baseline and targeted surveillance collecting studies of New Zealand ports and harbours for Biosecurity New Zealand.
Collections of specimens with comparable morphology were placed in individual plastic bags and transported to the laboratory in a chilled container for further sorting. If multiple samples showing similar morphologies were collected from the same site, a representative of these samples was chosen. Collections containing specimens of varying morphologies were separated into sub-samples. Collections that appeared to contain more than one Ulva species or were contaminated with other green algae (e.g. with epiphytes), and were not physically separable, were discarded.
Each sample that underwent further processing was allocated a unique number. Pieces of tissue, either approximately 1–2 cm x 1–2 cm from large thalli, or whole smaller thalli, or groups of very small individuals of the same morphology, were quickly dried in desiccant silica gel for subsequent DNA extraction; the remainder of each sample was prepared as a herbarium voucher lodged at the herbarium of the Museum of New Zealand Te Papa Tongarewa (WELT–Holmgren et al. Citation1990). Some samples were initially preserved in formalin/seawater before preparation of voucher specimens.
Molecular biology
DNA was extracted using the Chelex method of Goff & Moon (Citation1993) and diluted (1 : 10 or 1 : 100) with TrisEDTA buffer (10 mM Tris, 0.1 mM EDTA pH 8.0) for PCR amplification. A portion of the rbcL gene approximately 1200 bp in length was amplified using published primers RH1 and 1385R (Manhart, Citation1994) or primers designed by us: SH F1 and SH R4 ().
Table 1. Primers used for amplification and sequencing of the partial rbcL gene.
PCR reactions of 25 µl volume containing 1 µl diluted DNA extract, 2.5 µl 10x reaction buffer, 50 nmol MgCl2, 10 nmol of each dNTP, 25 pmol of each primer and 0.1 U Platinum ® Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, USA). Amplifications were performed in a Robocycler Gradient 96 thermocycler (Stratagene Corporation, La Jolla, USA) using an initial denaturation step (94°C, 2 min), followed by 35 cycles of denaturation (94°C, 30 s), annealing (40–47°C, 1 min), and extension (72°C, 2 min), with a final extension of 10 min at 72°C. PCR products were assessed by agarose gel electrophoresis, purified by PEG precipitation and sequenced on an ABI 377 automated sequencer according to standard methods.
An iterative approach was taken to allow simultaneous identification of known species and of new genetic entities. An initial phylogenetic dataset was constructed in PAUP* 4.0.b10 (Swofford, Citation2002) containing sequences of both bladed and tubular Ulva entities retrieved from GenBank. Incoming sequences from New Zealand specimens were added to this dataset and were subjected to neighbor joining phylogenetic analyses, placing them in relationship to the existing sequences.
Specimens generating sequences that formed a well-supported monophyletic group with GenBank sequences, were identified as belonging to the species named in GenBank. Sequences that were not significantly similar to GenBank sequences, i.e. those from species without representatives in GenBank, were retained in the dataset. Sequences that formed a monophyletic group with those already in the dataset were removed to make phylogenetic analyses tractable. This framework tree served as a means of evaluating incoming sequences: sequences could be quickly identified as either novel or belonging to an existing clade, whether this clade was based on sequences from New Zealand or from elsewhere. Each clade was distinguished either by a species name or a unique clade number.
For the phylogenetic analysis reported here, we assembled a dataset containing representatives of New Zealand clades and related sequences from GenBank (). Abbreviations for species authorities follow the International Plant Name Index (www.ipni.org/index.html). Sequences were aligned by eye in Se-Al version 2.0 a 11 (Rambaut, Citation1996). The dataset included 27 sequences of Ulvaceae from other parts of the world, with an outgroup consisting of six representatives of the Kornmanniaceae Golden & K.M.Cole (Kornmannia leptoderma [Kjellm.] Bliding and Blidingia minima [Nägeli ex Kütz.] Kylin), Gomontiaceae De Toni (Monostroma nitidum Wittr.), the Ulotrichaceae Kütz. (Pseudoneochloris marina S.Watan., A.Himizu, L.A.Lewis, G.L.Floyd & P.A.Fuerst) and the Dasycladales Pascher (Bornetella nitida [Harv.] Munier-Chalmas and Batophora oerstedii J.Agardh). One representative was retained from sets of identical New Zealand sequences. The final dataset contained 59 taxa: 35 GenBank sequences including the outgroup species, and 24 sequences from New Zealand specimens.
Table 2. Details of rbcL sequences used in phylogenetic analyses.
ModelTest version 3.06 (Posada & Crandall, Citation1998) was used to find the model of sequence evolution that best fitted the dataset. The model selected was the General Time Reversible (GTR) model allowing for invariant sites and with rate heterogeneity modelled by four gamma-distributed rate classes (GTR + I + Γ). This model was used for all distance and likelihood calculations. The dataset was analysed using maximum parsimony (MP) and maximum likelihood (ML) optimality criteria, and Bayesian analysis. MP trees were estimated with PAUP*4.0b10. MP analyses were conducted using an heuristic search strategy with 10 replicates of random-order sequence addition followed by tree bisection reconnection (TBR) branch swapping. Bootstrap support was estimated with 1000 replicates, each of 10 replicates of random order sequence addition followed by TBR branch swapping. Maximum likelihood analyses were conducted using PHYML v2.4.4 (Guindon & Gascuel, Citation2003) under the GTR + I + G model of sequence evolution, with concurrent estimation of parameters for invariant sites and gamma-modelled rate heterogeneity. Support was estimated using 500 bootstrap replicates.
Bayesian analyses were carried out using MrBayes v3.1.1 (Ronquist & Huelsenbeck, Citation2003) to run four chains of Metropolis-coupled Markov chain Monte Carlo (MCMC) iterations (one cold and three incrementally heated, temperature parameter = 0.2). Two independent MrBayes analyses were run under the GTR + I + G model of sequence evolution for 2,000,000 generations. Model parameters were treated as unknown and were estimated in each analysis. Chains were initiated with random starting trees and trees were sampled every 100 generations. Appropriate burn-in values were determined by inspection of plots of log-likelihood against generation time for each run. The likelihood scores stabilised at approximately 60,000 generations, and thus the first 600 trees were discarded as burn-in. The remaining trees were used to calculate a 50% majority rule consensus tree, in which each clade posterior probability (PP) value is represented by the proportion of trees containing that clade.
Results
Molecular data and phylogenetic analyses
The 581 samples of Ulva sensu lato collected in the New Zealand region could be separated into 24 distinct monophyletic entities on the basis of rbcL sequence data. Of these, 19 were identified as belonging to the genus Ulva, four to Umbraulva Bae & I.K.Lee and one to Gemina ().
Table 3. List of entities, numbers of samples per entity in collections analysed, and distribution of taxa by habitat (open coast vs harbour/embayment), and geographic distribution in selected regions (from north to south: Kermadec Islands to subantarctic islands).
Fourteen of the entities were resolved with sequences from GenBank in the phylogenetic analyses. The names attached to sequences in GenBank were treated with care: the accuracy of the identification of material that has been sequenced and for which there are data lodged in GenBank is critical. When selecting species names for clades found in New Zealand we sought to use GenBank entries based on material from the region of the type locality of the taxon. Based on similarity with GenBank sequences species names were allocated to 13 entities (). A total of 11 entities did not match any named sequences in GenBank. Of these, two were tentatively identified on the basis of morphology as Gemina letterstedtioidea V.J.Chapm. and Ulva species 5 “U. ralfsii ”, while the other nine remain without species names.
The partial alignment of the rbcL sequences comprised 1144 base pairs. Some sequences did not cover the whole length of the alignment. Missing data were represented by IUPAC code N. The average base composition of the reduced dataset was A: 0.28755; C: 0.16531; G: 0.21547; T: 0.33167. The maximum likelihood tree inferred from the analyses is presented in . In the phylogenetic analyses, the genus Ulva was well supported as a monophyletic clade, with Ulva species 4 as a sister group to the rest of the genus. Gemina and Umbraulva were resolved with weak support on a polytomy with Percusaria percursa (C.Agardh) Rosenv. and Ulvaria obscura var blyttii (Aresch.) Bliding, this group was resolved as a sister group to Ulva. Support for Umbraulva as a monophyletic group was weak.
Fig. 2. Maximum likelihood phylogram (rbcL sequences) showing entities of Ulva sensu lato from New Zealand identified in this study (shaded in two tones for clarity). ML/MP bootstrap support values are shown above, and Bayesian PP values below, each node. Only nodes that are supported under two methods of analysis are labelled (> 60% for ML and MP, PP > 0.70). The discontinuity drawn in the branch from the outgroups indicates that this branch only is not drawn to scale.
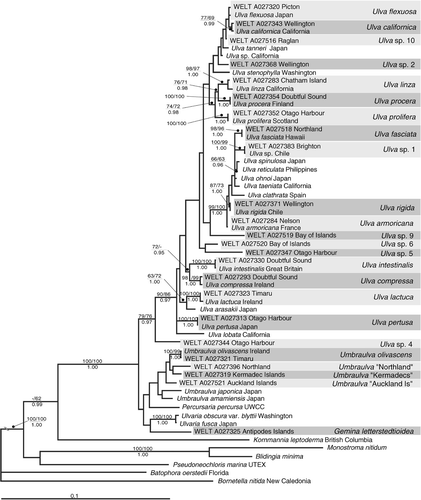
The topology of the ML tree was essentially the same as that of the tree inferred from the Bayesian analysis, as well as the 16 equally most parsimonious trees produced by the MP analysis, based on 298 parsimony-informative characters (length of MP trees: 1016 steps; scores: Consistency Index: 0.519, Retention Index: 0.702, Rescaled Consistency Index: 0.364) (calculated using PAUP* 4.0b10). lists the maximal distances among sequences from New Zealand within entities and, where appropriate, the absolute distance of the representative New Zealand sequence to the GenBank sequence.
Table 4. Maximum base-pair distances between rbcL sequences from New Zealand specimens within single entities and absolute distance to the GenBank sequence of the representative sequence included in the reduced dataset.
Species distributions
Approximately equal numbers of samples came from open coast sites (282) as from sites in harbours and embayments (299) (). There were only 36 subtidal collections, in which 11 species were recorded (G. letterstedtioidea, Umbraulva olivascens [P.J.L.Dang.] Bae & I.K. Lee, Um. “Kermadecs”, Um. “Auckland Is”, Um. “Northland”, Ulva armoricana Dion, de Reviers & Coat, Ulva fasciata Delile, U. linza L., U. pertusa Kjellm., U. rigida C.Agardh, Ulva species 1). Of these, the Umbraulva species and U. fasciata were found solely in subtidal habitats. Seventeen collections were made from freshwater streams or drains, from which six taxa were identified (U. compressa L., U. intestinalis L., U. pertusa, Ulva species 1, Ulva species 5 “U. ralfsii ”, Ulva species 6). Of these species all but Ulva species 6 are known from other habitat types: Ulva species 6 is currently known from a single collection site.
In the distribution of species in selected regions (outlying island groups in the north and south, as well as from northern, central, and southern parts of the main islands) is presented. Four taxa (Ulva flexuosa Wulfen, U. lactuca L., U. “species 10” and Um. olivascens) were found in regions other than those listed. Although we found a greater diversity of taxa in the northern North Island (12 taxa, ) than in other regions, at most northern sites Ulva spp. were not conspicuous in terms of population size or biomass, in either intertidal or subtidal communities. Intertidal populations were considerably more conspicuous in more southern locations, for example, at both the Chatham Islands and in Fiordland seven taxa were recorded. Some taxa were widespread (e.g. Ulva species 1) whereas other taxa found in this study were from single regions/island groups (e.g. Umbraulva spp., Ulva species 9, Ulva species 10). Species composition was found to vary seasonally and between sites in both the Wellington (14 sites, 9 taxa) and Dunedin (16 sites, 10 taxa) regions. In both the Wellington and Dunedin samples analysed, the species U. compressa, U. pertusa and Ulva species 1 were present throughout the sampling period. These were also the most geographically widespread species in the New Zealand region.
The collecting strategy employed in this study–to sample a wide geographic range as well as to obtain seasonal collections–was constrained by the fact that the field work spanned a single year. Most samples were collected in the intertidal zone, and a relatively small number were from subtidal habitats. Thus the geographical ranges of entities that are mainly or always subtidal, such as Umbraulva species or Ulva fasciata, may be under-sampled.
In terms of habitat modification, three sites (at the Antipodes and Kermadec Islands) were scored as pristine and a further seven sites were recorded as having low volume of marine traffic (sites at Chatham, Stewart and Auckland Islands). From these sites a total of 16 collections were made, belonging to six taxa (G. letterstedtioidea, Umbraulva “Kermadecs”, Umbraulva “Auckland Is”, U. compressa, U. pertusa, and Ulva species 1). Seventy-seven collections were made from sites that were classified as highly modified (wharf pilings, harbour installations). At these sites, nine taxa were recorded (U. armoricana, U. californica Wille in Collins, Holden & Setch., U. compressa, U. lactuca, U. pertusa, U. rigida, Ulva species 1, Ulva species 2, Um. olivascens). A further 95 samples were analysed from sites adjacent to ports or harbour structures, where an additional seven taxa were found (U. flexuosa, U. intestinalis, U. procera (K.Ahlner) H.S.Hayden et al., U. prolifera O.F.Müll., Ulva species 5 “U. ralfsii”, Ulva species 6, Ulva species 9).
Discussion
Prior to this study, the taxonomy of the Ulvaceae in the New Zealand region was highly problematic: previous treatments of the group were considered unreliable and there has been difficulty in assigning names to even commonly occurring taxa. It is generally agreed that Chapman (Citation1956) recognised too many taxa particularly at infra-specific rank, and it has been difficult to gauge the true diversity within this family in New Zealand. In this study 24 genetically distinct taxa were distinguished in New Zealand waters, including 19 species of Ulva sensu lato, four species of Umbraulva (the first record of this genus for the Southern Hemisphere) and the confirmation of Gemina from the subantarctic region.
When sequence data are being used to define species or clarify species concepts, type specimens–or at least material collected at a species’ type locality–should provide the sequence in GenBank. However these data are not always available, particularly when type specimens are very old, or lost, and/or type localities are unknown, as is, for example, the case with most of the Ulva type specimens described by Linnaeus (Citation1753) (Womersley, Citation1984). The names used in this study based on GenBank entries need to be used with the understanding that sequence data based on type or topo-type material is still not available for most taxa in the Ulvales.
Our phylogenetic analysis supports the monophyly of the Ulvaceae, and the monophyly of Ulva itself. Support for Umbraulva as a monophyletic group is low, however this may be due to the nature of our taxon sampling, which was focused on a survey of New Zealand taxa. Wider taxon sampling within the Ulvaceae, including representatives of Letterstedtia Aresch., Ruthnielsenia O’Kelly, Wynsor & Bellows and Ochlochaete Thwaites may clarify the relationships of genera within the Ulvaceae, and confirm or rebut the monophyly of Umbraulva.
Within Ulva, branch lengths are generally low. This reflects the limited amount of variability between these taxa at the rbcL locus. More variable markers will be required to resolve relationships within the genus more clearly. This was outside the scope of the present study, since our purpose was to discriminate taxa in the New Zealand flora.
Some nodes in our analysis receive high support from Bayesian analysis but only low support under MP and ML. This probably reflects the tendency of Bayesian analysis to resolve polytomies with inappropriately strong support, which has been identified as a property of Bayesian analyses (Lewis et al. Citation2005). These high PP values should therefore be interpreted with some caution.
Although the diversity found within the Ulvaceae considered in this study may be regarded as high (e.g. compared with southern Australia with 14 taxa in Ulva sensu lato, Womersley Citation1984), we consider this number of taxa to be a conservative assessment, as it is based solely on collections from a single year with many sites visited on only one occasion. As the New Zealand region extends from 29–53°S, encompassing subtropical to subantarctic conditions, warm and cool water taxa are present in the flora. Moreover, solely rbcL sequence comparisons have been used here, and the status of morphologically distinguishable but genetically similar groups has not been further investigated in this account. The results of this study parallel the results of equivalent studies on the diversity of the red algal family, Bangiaceae, in the New Zealand region (Nelson et al., Citation2006). In both the Ulvaceae and Bangiaceae the morphological simplicity of the taxa, and their apparent capacity for phenotypic plasticity as well as cryptic speciation, create particular difficulties when distinguishing taxa, and in both instances molecular sequence data provide a valuable source of data for comparative and phylogenetic analyses.
Four species of Umbraulva were discovered in the course of this study, of which three are undescribed (“Kermadecs”, “Northland”, “Auckland Is”) and one, Um. olivascens, we consider to be introduced to the New Zealand region. Prior to this study, Um. olivascens was only known from Europe and the Canary Islands, while Um. amamiensis Bae & I.K.Lee and Um. japonica (Tanaka) Bae & I.K.Lee have so far been reported only from South East Asia (Bae & Lee, Citation2001). The rbcL sequences of the undescribed New Zealand Umbraulva species are distinct from Ulva, and group with sequences of the three known species of Umbraulva from Japan and Ireland. Although these three New Zealand Umbraulva species were each collected only once, their occurrence in areas with low or no human influence suggests that they are native in their respective habitats, and additionally, are probably endemic species. The genus Umbraulva was separated from Ulva based on DNA sequence comparisons and the presence of an additional accessory pigment, siphonaxanthin. This pigment gives the thalli an olive-green tinge and apparently allows them to survive in deeper waters (Yokohama, Citation1981; Bae & Lee, Citation2001). The specimens from Northland had a distinct olive-green colour, and all Umbraulva specimens were collected subtidally. Whether they contain siphonaxanthin as an accessory pigment has yet to be confirmed. Further material of these taxa will be required before formal descriptions can be prepared and names assigned.
The present study provides the first evidence of the genus Gemina since its description in 1952. This genus was originally described from samples collected in New Zealand, from Stewart Island and the subantarctic islands (Chapman, Citation1952). Although it is considered current in AlgaeBase (Guiry & Guiry, Citation2007), its status as a genus separate from Ulva has been doubted (Papenfuss, Citation1960; Adams, Citation1994). Although Gemina specimens were macroscopically similar to Ulva and Umbraulva specimens, they were microscopically different. The presence of paired cells enveloped by a mother cell wall and their palisade-like appearance in transverse sections agree well with the description given by Chapman (Citation1952) for the genus Gemina. The collection of six samples from distant sites (such as the Antipodes and Auckland Islands) with identical or nearly identical rbcL sequences confirms the presence of a single species. Further research is required to understand better the relationships and distinguishing morphological, anatomical and ecological characteristics of this taxon.
The assessment of the native/non-native status of the 19 species of Ulva recorded in this study was based on a range of criteria: on the numbers of collections as well as types of sites where an entity was observed, and the genetic distance between sequences from New Zealand and overseas specimens. The existence of few collections, in combination with a restricted distribution or close proximity to sites with high frequency of ship traffic, such as harbours and marinas, was considered indicative of an introduced species, especially if the rbcL sequences were identical or nearly identical to sequences obtained from samples collected outside New Zealand. The status of an entity as native or introduced was difficult to assess for those sequences with no significant close homology to existing sequences in GenBank (e.g. species 4, species 6, species 9 and species 10).
Based on the criteria used in this study, we have concluded that of the 19 species of Ulva, at least four are probable introductions (U. armoricana, U. californica, U. flexuosa and U. lactuca). A further five species are known from only one or two collections (U. linza, species 4, species 6, species 9, species 10). These species may be naturally rare–for example, either seasonally ephemeral or highly restricted in geographic range–or they may be introduced. Further research will be required to resolve questions about the identity of these taxa and to provide sufficient material for detailed analyses of their morphology, as well as improved understanding of their distribution and ecology.
As a consequence of the present study, some of the species concepts previously applied to the genus Ulva in New Zealand need to be amended. This is especially evident in the case of the three most common species in New Zealand, previously thought to be U. lactuca, U. rigida and U. clathrata (Roth) C. Agardh (including taxa currently regarded as its synonyms, i.e. Enteromorpha muscoides (Clemente) Cremades, E. ramulosa (Sm.) Carmich. and E. acanthophora Kütz. (Guiry & Guiry, Citation2007). Adams (Citation1994, p.24) states: “... the name, U. lactuca, has been widely and uncritically used in New Zealand.” In this study we found that U. lactuca had a very limited distribution, at three sites all of which have a high volume of vessel traffic, suggesting that this species is not native but has been introduced to New Zealand. The entity it is most likely to have been confused with is Ulva species 1, which clustered with an unnamed Chilean species in the phylogenetic analysis, and which was found to be the most abundant entity in New Zealand. Ulva species 1 may be synonymous with Ulva laetevirens Aresch. described from South Australia. If so, this would confirm Adams' observations (1994, p.24) that “the Ulva species on our shores have probably more in common with those found in southern Australia”. Similarly, U. rigida has a limited distribution in New Zealand, while the species previously mis-identified under this name, U. pertusa, was the second most abundant Ulva species among the samples collected.
Species that have been regarded as native but that may be introduced species include Ulva species 1 and U. pertusa, both of which occurred in habitats with high human influence through to areas where there has been little human-mediated change, as well as U. compressa and U. intestinalis, which have been recorded from New Zealand from the early period of European exploration (e.g. Hooker & Harvey, Citation1845). Third in the list of most abundant Ulva species in New Zealand, U. compressa is much more widespread than previously thought, and also more morphologically variable, including terete, much-branched Ulva specimens that until now may have been identified as U. clathrata (including its synonyms). The fourth most common species found in the current study was U. intestinalis and, in contrast to some other examples, this name appears to have been applied consistently in the past to this entity. Both U. intestinalis and U. compressa are known hull-fouling organisms; they are tolerant to a wide range of environmental conditions, and grow and reproduce rapidly, which may explain their cosmopolitan distribution (Blomster et al., Citation1998; Schaffelke et al., Citation2006). While the wide geographical spread of these taxa in New Zealand suggests that they are native, they may in fact have been naturalised some time ago. The origins of these taxa are not known and they should therefore be considered cryptogenic species.
The status of U. fasciata in New Zealand waters is not clear. This species is reported worldwide; in some regions it is considered cosmopolitan and in other regions, for example, in Mexico and Australia, regarded as an introduced taxon (Phillips, Citation1988; Anonymous, Citation2001; Aguilar-Rosas et al., Citation2005). Mature thalli of this species have a very distinct morphology. The specimens of U. fasciata collected in New Zealand were small and atypical. Whether they were immature or stunted by unfavourable environmental conditions will require further investigation. The rbcL sequences of the three New Zealand specimens were virtually identical to sequences from Japan, Australia and Hawaii, but as GenBank sequences are not available for material from the type locality in the Mediterranean, confirmation of the name U. fasciata for Pacific specimens is still required.
The present study was designed to provide information on the genetic diversity of the genus Ulva in New Zealand. Rather than focusing on an in-depth examination of phylogenetic and intrageneric relationships within Ulva sensu lato, it aimed to identify and better define entities within the New Zealand region and to enable native and alien taxa to be distinguished. Because of New Zealand's extensive coastline and the large numbers of samples to screen, our approach was to compare one gene in many specimens, rather than many genes in fewer specimens. The rbcL gene proved to be useful for this approach. However, this gene is not variable enough to separate levels below the rank of species. Distinct morphological groups were found within some taxa without any accompanying genetic distinctions using rbcL sequences (e.g. U. procera, U. compressa). Further molecular systematic studies could clarify the status of morphological variants observed within rbcL genetic entities, and extended field studies might confirm the presence of species so far only encountered in single samples, thus providing information about the geographic distribution of rarely collected species. This research has provided a platform for monographic studies of the family Ulvaceae in New Zealand, and for future biosecurity investigations of what has been a notoriously difficult group of algae.
Acknowledgements
Biosecurity New Zealand is acknowledged for funding this research (ZBS200408). We would like to thank the following people who have assisted us with aspects of this project: Rebecca Ansell (Victoria University) for assistance with database development and mapping; Roberta D’Archino (NIWA) for SCUBA collections and assistance with bibliographic databases; Darren Hart (University of Otago) for assistance in the field and for advice with sequencing and phylogenetic analyses; Mei Nee Lee and Ewen Cameron (Auckland Institute and Museum) for enabling access to the AK collections and for assistance with loans; Amonida Zadissa for translation of Swedish. We would like to thank Neill Barr for valuable advice and discussions about Ulva. A number of people assisted us in the field or provided collections for us, sometimes from very remote locations–the coverage we have achieved would not have been possible without their assistance: Paul Buisson, Clinton Duffy, Erin Green, Debbie Freeman, Lou Hunt, Helen Kettles, Peter de Lange, Anne McCrone (Department of Conservation); Sheryl Miller and Jeff Forman (NIWA); Chris Cornelisen, Rochelle Dewdney, Wilson Freshwater, Chris Hepburn, Rachel Law, Gabrielle Lockett, Abi Loughnan, Paul Meredith, Roland Pfisterer, Margaret Richards, Lisa Russell, Nick Shears, Franz Smith, Brett Stansfield, Mike Stuart, NIWA field staff for biosecurity collections (ZBS2000/04, ZBS2001/01, ZBS2005/18, ZBS2005/35).
References
- Adams , NM . 1994 . Seaweeds of New Zealand. An Illustrated Guide , Christchurch, , New Zealand : Canterbury University Press .
- Aguilar-Rosas , R , Aguilar-Rosas , LE and Pedroche , FF . 2005 . Ulva fasciata Delile (Ulvaceae, Chlorophycota): a species newly introduced into Pacific Mexico . Bot. Mar. , 48 : 46 – 51 .
- Anonymous . 2001 . Algae: Invasive Native. Ulva fasciata Delile 1813. , Botany : University of Hawaii at Manoa . Available at: www.hawaii.edu/reefalgae/invasive_algae/chloro/ulva_fasciata.htm, accessed July 2007
- Bae , EH and Lee , IK . 2001 . Umbraulva, a new genus based on Ulva japonica (Holmes) Papenfuss (Ulvaceae, Chlorophyta) . Algae , 16 : 217 – 231 .
- Blomster , J , Baeck , S , Fewer , DP , Kiirikki , M , Lehvo , A , Maggs , CA and Stanhope , MJ . 2002 . Novel morphology in Enteromorpha (Ulvophyceae) forming green tides . Am. J. Bot. , 89 : 1756 – 1763 .
- Blomster , J , Maggs , CA and Stanhope , MJ . 1998 . Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles . J. Phycol. , 34 : 319 – 340 .
- Blomster , J , Maggs , CA and Stanhope , MJ . 1999 . Extensive intraspecific morphological variation in Enteromorpha muscoides (Chlorophyta) revealed by molecular analysis . J. Phycol. , 35 : 575 – 586 .
- Chapman , VJ . 1952 . New entities in the Chlorophyceae of New Zealand . Trans. R. Soc. N. Z. , 80 : 47 – 58 .
- Chapman , VJ . 1956 . The marine algae of New Zealand. Part I. Myxophyceae and Chlorophyceae . J. Linn. Soc. Bot. , 55 : 333 – 501 .
- De Winton , MD , Hawes , I , Clayton , JS , Champion , PD and Smith , RK . 1998 . Sea lettuce dynamics and ecophysiology in Taruanga Harbour, Bay of Plenty. , NIWA Client Report BPR802, prepared for Environment B.O.P. Tauranga District Council and Western Bay of Plenty District Council .
- Goff , LJ and Moon , DA . 1993 . PCR Amplification of nuclear and plastid genes from algal herbarium specimens and algal spores . J. Phycol. , 29 : 381 – 384 .
- Guindon , S and Gascuel , O . 2003 . A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood . Syst. Biol. , 15 : 696 – 704 .
- Guiry , MD and Guiry , GM . 2007 . AlgaeBase version 4.2 , Galway : World-wide electronic publication, National University of Ireland . Available at: http://www.algaebase.org, accessed 13 September 2007
- Hawes , I and Smith , RA . 1995 . Effect of current velocity on the detachment of thalli of Ulva lactuca (Chlorophyta) in a New Zealand estuary . J. Phycol. , 31 : 875 – 880 .
- Hayden , HS , Blomster , J , Maggs , CA , Silva , PC , Stanhope , MJ and Waaland , JR . 2003 . Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera . Eur. J. Phycol. , 38 : 277 – 294 .
- Hayden , HS and Waaland , JR . 2002 . Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences . J. Phycol. , 38 : 1200 – 1212 .
- Hayden , HS and Waaland , JR . 2004 . A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific . Phycologia , 43 : 364 – 382 .
- Heesch , S , Broom , JE , Neill , K , Farr , TJ , Dalen , J and Nelson , WA . 2007 . Genetic diversity and possible origins of New Zealand populations of Ulva. , Biosecurity New Zealand Technical Paper No: 2007/01
- Hiraoka , M , Ohno , M , Kawaguchi , S and Yoshida , G . 2004a . Crossing test among floating Ulva thalli forming ‘green tide’ in Japan . Hydrobiologia , 512 : 239 – 245 .
- Hiraoka , M , Shimada , S , Uenosono , M and Masuda , M . 2004b . A new green-tide-forming alga, Ulva ohnoi Hiraoka et Shimada sp. nov. (Ulvales, Ulvophyceae) from Japan . Phycol. Res. , 52 : 17 – 29 .
- Holmgren , PK , Holmgren , NH and Branett , LC . 1990 . Index Herbariorum. Part. I: The Herbaria of the World , 8th Vol. 120 , Regnum Vegetabile
- Hooker , JD and Harvey , WH . 1845 . Algae Novae Zelandiae . London J. Bot , 4 : 521 – 551 .
- Lewis , PO , Holder , MT and Holsinger , KE . 2005 . Polytomies and Bayesian phylogenetic inference . Syst. Biol. , 54 : 241 – 253 .
- Linnaeus, C. (1753). Species Plantarum, Vol. 2. Sweden: Stockholm. pp. 561–1200 (+ 1–31)
- Malta , E-J , Draisma , SGA and Kamermans , P . 1999 . Free-floating Ulva in the southwest Netherlands: species or morphotypes? A morphological, molecular and ecological comparison . Eur. J. Phycol. , 34 : 443 – 454 .
- Manhart , J . 1994 . Phylogenetic analysis of green plant rbcL sequences . Mol. Phylogenet. Evol. , 3 : 114 – 127 .
- Nelson , TA , Lee , DJ and Smith , BC . 2003 . Are ‘green tides’ harmful algal blooms? Toxic properties of water-soluble extracts from two bloom-forming macroalgae, Ulva fenestrata and Ulvaria obscura (Ulvophyceae) . J. Phycol , 39 : 874 – 879 .
- Nelson , WA , Farr , TJ and Broom , JES . 2006 . Phylogenetic relationships and generic concepts in the red order Bangiales: challenges ahead . Phycologia , 45 : 249 – 259 .
- Papenfuss , GF . 1960 . On the genera of the Ulvales and the status of the order . J. Linn. Soc. Bot. , 56 : 303 – 318 .
- Phillips , J . 1988 . Field, anatomical and developmental studies on southern Australian species of Ulva (Ulvaceae, Chlorophyta) . Aust. Syst. Bot. , 1 : 411 – 456 .
- Posada , D and Crandall , K . 1998 . MODELTEST: testing the model of DNA substitution . Bioinformatics , 14 : 817 – 818 .
- Raffaelli , DG , Raven , JA and Poole , LJ . 1998 . Ecological impact of green macroalgal blooms . Ann. Rev. Mar. Biol. Oceanogr. , 36 : 97 – 125 .
- Rambaut , A . 1996 . Se-Al: Sequence Alignment Editor. . Available at: http://evolve.zoo.ox.ac.uk, accessed July 2007
- Ronquist , F and Huelsenbeck , JP . 2003 . MrBayes 3: Bayesian phylogenetic inference under mixed models . Bioinformatics , 19 : 1572 – 1574 .
- Schaffelke , B , Smith , JE and Hewitt , CL . 2006 . Introduced macroalgae–a growing concern . J. Appl. Phycol. , 18 : 529 – 541 .
- Shimada , S , Hiraoka , M , Nabata , S , Lima , M and Masuda , M . 2003 . Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva . Phycol. Res. , 51 : 99 – 108 .
- Swofford , DL . 2002 . PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods) , Sunderland, , USA : Sinauer Associates . Version 4.0b10
- Tan , IH , Blomster , J , Hansen , G , Leskinen , E , Maggs , CA , Mann , DG , Sluiman , HJ and Stanhope , MJ . 1999 . Molecular phylogenetic evidence for a reversible morphogenetic switch controlling the gross morphology of two common genera of green seaweeds, Ulva and Enteromorpha . Mol. Biol. Evol. , 16 : 1011 – 1018 .
- Taylor , R . 1999 . The green tide threat in the UK–a brief overview with particular reference to Langstone Harbour, south coast of England and the Ythan estuary, east coast of Scotland . Bot. J. Scot , 51 : 195 – 203 .
- Womersley , HBS . 1984 . The Marine Benthic Flora of Southern Australia. Part I , Adelaide, , Australia : Government Printer .
- Yokohama , Y . 1981 . Distribution of the green light-absorbing pigments siphonaxanthin and siphonein in marine green algae . Bot. Mar. , 24 : 637 – 640 .
- Zechman , FW . 2003 . Phylogeny of the Dasycladales (Chlorophyta, Ulvophyceae) based on analyses of rubisco large subunit (rbcL) gene sequences . J. Phycol. , 39 : 819 – 827 .