Abstract
Molecular phylogenies inferred from rbcL sequences including 39 representative members of the Laurencia complex confirm the four genera currently recognised within the complex: Laurencia sensu stricto, Osmundea, Chondrophycus and the recently described genus Palisada. Furthermore, Palisada poiteaui was resolved as a fifth independent lineage suggesting that the complex is actually composed of five rather than four genera. Palisada poiteaui is the type species of the subgenus Yuzurua, and elevation of this subgenus to generic rank is proposed. This new genus allied strongly with Laurencia s.s. However, the other intergeneric relationships were not well supported, suggesting that rbcL sequences may not have sufficient signal to clarify infrageneric relationships fully within the Laurencia complex.
Introduction
The genus Laurencia was erected by Lamouroux in Citation1813; thereafter its taxonomic history has been convoluted, and here we discuss only the major changes that have occurred; the reader is invited to refer to Saito (Citation1967), McDermid (Citation1988), Furnari & Serio (Citation1995) and Furnari et al. (Citation2001) for a more comprehensive history. Lamouroux included eight species in the original description of Laurencia. Subsequently, Schmitz (Citation1889) recognised Laurencia obtusa (Hudson) J.V. Lamouroux as the ‘typische Species’ for the genus and this species is currently considered the generitype. Thorough anatomical studies during the last four decades (e.g. Saito, Citation1967; Nam et al., Citation1994; Garbary & Harper, Citation1998; Nam, Citation1999, Citation2006) have revealed that Laurencia is a highly diverse genus, encompassing species that display distinctive features usually diagnostic at the generic level. The genus has therefore been referred to the Laurencia complex and, three additional genera, Osmundea Stackhouse, Chondrophycus (Tokida & Y. Saito) Garbary & Harper, and Palisada (Yamada) Nam have been proposed successively to reflect its morphological diversity. Saito (Citation1967) was the first to divide Laurencia into two subgenera, Laurencia and Chondrophycus, based on the occurrence of secondary pit connections between epidermal cells, and the type of tetrasporangial arrangement. The genus Osmundea (Stackhouse, Citation1809), which had been placed in synonymy with Laurencia (nom. cons., see Papenfuss, Citation1947), was resurrected by Nam et al. (Citation1994) to accommodate taxa that exhibit a filament-type spermatangial development rather than a trichoblastic-type, and tetrasporangial initials arising from a random epidermal cell rather than a particular pericentral cell. Osmundea currently includes 18 species mostly reported from temperate waters (Nam et al., Citation2000; McIvor et al., Citation2002; Guiry & Guiry, Citation2008).
Nam & Saito (Citation1995) showed that the number of pericentral cells in Laurencia subgenera (four in subgenus Laurencia and two in subgenus Chondrophycus) were different, and they also noted other distinctive characters, such as the presence/absence of additional terasporangium-bearing pericentral cells, the position of pericentral cells bearing tetrasporangia, and the number of pericentral cells of the procarp-bearing segment. The taxonomic status of Chondrophycus had been a matter of debate for more than 20 years (e.g. Furnari & Serio, Citation1993) and the subgenus was elevated to generic rank by Garbary & Harper in Citation1998. Nam (Citation1999) highlighted further diversity within this genus, in both reproductive and vegetative features, and proposed an infrageneric classification including four subgenera: Chondrophycus, Kangjaewonia, Palisada and Yuzurua.
Finally, Nam (Citation2006) proposed elevating Palisada to generic rank to accommodate members of Chondrophycus that have, among other features, the first pericentral cell located underneath the trichoblast rather than on the side, and tetrasporangial axes with one sterile pericentral cell rather than two. Nineteen species were transferred to the genus Palisada (Nam, Citation2006), leaving Chondrophycus with 17 species. However, the generic name Palisada was only validated the following year with the publication of the Latin diagnosis of this genus (Nam, Citation2007). Recently, Senties & Díaz-Larrea (Citation2008) transferred Chondrophycus corallopsis Montagne to Palisada, so the two genera currently include 16 and 20 species, respectively.
Contrasting with Lamouroux's initial concept of the genus Laurencia, Laurencia s.s. (i.e. Laurencia sensu Garbary & Harper, Citation1998) presently includes more than 140 species and the Laurencia complex encompasses almost 200 species (Guiry & Guiry, Citation2008), which are distributed from temperate to tropical waters (McDermid, Citation1988).
The phylogeny of the Laurencia complex has been studied mainly from anatomical and developmental perspectives, and Garbary & Harper (Citation1998), Nam et al. (Citation2000), and Nam (Citation2006), inferred the interspecific relationships among members of the Laurencia complex based on cladistic analyses. Molecular studies were initiated with analyses of sequences of the plastid-encoded, large subunit of RuBisCO (rbcL) to infer interspecific relationships within Osmundea (Nam et al., Citation2000; McIvor et al., Citation2002). Fujii et al. (Citation2006) have published a molecular phylogeny including sequences from Osmundea, Laurencia and Palisada (as Chondrophycus) species, and confirmed the monophyly of the three genera previously inferred from morphological characters, although with restricted sampling. Abe et al. (Citation2006) inferred a molecular phylogeny of the Laurencia complex from rbcL sequences and confirmed the monophyly of Osmundea, however Palisada (as Chondrophycus) and Laurencia were both resolved as non-monophyletic. Finally, Díaz-Larrea et al. (Citation2007) published a molecular phylogeny inferred from a data set built to assess species boundaries between Palisada poiteaui (J.V. Lamouroux) Nam (as Chondrophycus poiteaui (J.V. Lamouroux) Nam) and Palisada gemmifera (Harvey) Sentíes, Fujii & Díaz (as Chondrophycus gemmiferus (Harvey) Garbary & Harper) and concluded that they were conspecific. To the best of our knowledge, the delimitation of Palisada and Chondrophycus has not been tested using phylogenies inferred from molecular data. The aim of the present study was therefore to assess the generic boundaries of the four genera currently recognised within the Laurencia complex using rbcL sequences.
Materials and methods
Specimen collection
Specimens included in molecular analyses are listed in , along with their valid names and GenBank accession numbers (NCBI GenBank). Newly sequenced specimens were collected by SCUBA in the Western Pacific in the vicinity of New Caledonia (158–169°E and 18–23°S) except for Laurencia pyramidalis, Osmundea osmunda and O. hybrida, which were collected at low tide along the French Atlantic coast of Brittany. A part of each sample was stored in 5% buffered formalin in seawater, and the rest was dried as herbarium specimens and deposited in the herbaria of NOU-IRD (Phycological Herbarium, Institut de Recherche pour le Développement, Nouméa, New Caledonia) and PC Herbarium (abbreviations are in accordance with the Index Herbariorum (Holmgren et al., Citation1990), sciweb.nybg.org/science2.IndexHerbariorum.asp).
Table 1. List of species used in this study for phylogenetic analyses, with their collection data and GenBank accession numbers.
Morphological characters for specimen identification were observed using an Olympus BH2 compound microscope (Olympus Optical Co. Ltd., Tokyo, Japan). Most of our specimens were from New Caledonia rather than type localities and given the putative cryptic diversity (including sibling species) within the Laurencia complex, we treated the species identification cautiously and used the Latin-derived word ‘confer’ before the specific epithet of these specimens.
Extraction, amplification and sequencing
Total cellular DNA was extracted from herbarium specimens using the DNeasy Plant Mini Kit (QIAGEN, Valencia, California, USA). Between 0.5 × 10−3 to 10−3 mg of proteinase K was added to the lysis buffer to improve the DNA yield (Hughey et al., Citation2001). Both the rbcL coding region (1,647 base pairs [bp]) and rbcL–rbcS spacer (∼200 bp) were amplified using the following combinations of primers (): F-rbcLstart × R-753 (Freshwater & Rueness, Citation1994) for the 5′ end, rbcLFC × 1011R (Nam et al., Citation2000) or F-577 × R1381 (Freshwater & Rueness, Citation1994) for the middle fragment, and F-993 × R-rbcS start (Freshwater & Rueness, Citation1994) for the 3′ end. The protocol used for PCR amplifications was modified from Nam et al. (Citation2000) and Lin et al. (Citation2001). Sequence-amplifications were performed by PCR in a final 30 µl volume. For the primer pair rbcLFC × 1011R the cycle was 5 min of initial denaturation at 94°C followed by 40 cycles of 60 s at 94°C, 60 s at 52°C and 60°C at 72°C and a final extension at 72°C during 5 min. Conditions for amplifications with other primer pairs were: 4 min at 96°C for denaturation, followed by 35 cycles of 60 s at 94°C, 60 s at temperatures varying from 42°C to 50°C, and 90 s at 72°C, with a final 8 min extension at 72°C. The resulting PCR products were purified and used as templates for cycle sequencing reaction with the same primers as for the initial amplifications. These steps were performed by Genoscope (www.genoscope.fr, Evry, France).
Table 2. Primers used for amplification and sequencing in this study.
Sequence alignments and phylogenetic analyses
Sequences were obtained for both DNA strands and assembled and corrected using Sequencher™ 4.1 (Gene Codes Corporation, Ann Arbor, Michigan). Twenty-six sequences from GenBank were included to broaden the taxonomic range of the phylogeny, but only after carefully checking that they (i) belonged to the tribe Laurenciae, (ii) covered at least 70% of the full length of the rbcL gene and (iii) were associated with a published manuscript. Three of these selected sequences displayed suspicious phylogenetic affinities: Chondrophycus translucidus (Fujii & Cordeiro-Marino) Garbary & Harper (AY585408), Laurencia complanata (Suhr) Kützing (AF465813) and Palisada flagellifera (J. Agardh) Nam (EF061647). We therefore performed distance analyses partitioning the rbcL gene into two equivalent-sized fragments (∼702 bp) or three fragments (coinciding with the regions bordered by the universal red algal primers F-rbcLstart × R-753, F-577 × R1381 and F-993 × R-rbcS start). It appeared that the position of the three taxa changed considerably depending on the partition used, suggesting that they were likely to be chimeric, and therefore we excluded them from our analyses. Out-group species (see ) were chosen from the Rhodomelacean sequences available in GenBank. Alignments were performed with MEGA version 3.1 (Kumar et al., Citation2004) using the CLUSTAL algorithm (Thompson et al., Citation1994).
Phylogenetic analyses of rbcL
Maximum parsimony (MP) analyses were performed using PAUP* version 4.0b10 (Swofford, Citation2003) and used a heuristic search with 1000 random additions, unordered and unweighted characters, with tree bisection-reconnection branch swapping in effect. The program Modeltest version 3.7 (Posada & Crandall, Citation1998) was used to determine the model that best fit our data for maximum likelihood (ML) and Bayesian inference (BI) analysis, using the Akaike Information Criterion (AIC). ML analysis was carried out with PhyML 2.4.4 (Guindon & Gascuel, Citation2003) using a BioNJ starting tree, and parameter values estimated during the run. ML and MP analyses were subjected to bootstrap resampling (1000 replicates and 1000 replicates with 10 random additions, respectively) to estimate robustness (Felsenstein, Citation1985). MrBayes version 3.1 (Huelsenbeck & Ronquist, Citation2001) was used to complete BI, with the sequence data analysed as a single partition (an analysis using codon partitioning was also run, but topologies were unaffected therefore the simplest partition scheme was selected). Analyses were run with four heated Monte-Carlo Markov Chains for 2,000,000 generations. Output trees and data were sampled every 100 generations. Appropriate burn-in for each run was determined by plotting the overall likelihood against generations prior to estimating the posterior probability distribution. In all analyses, likelihood values were stable after the first 200,000 generations, and the final results were based on the pooled samples from the stationary phase of the two independent runs.
Results
Sampling and analyses of rbcL sequences
Complete rbcL (1467 bp) and rbcL–rbcS spacer sequences were successfully generated for 16 specimens (). In order to minimize missing data and, when combined with sequences available on GenBank, 59 bp at the 5′ end of the rbcL and the spacer region at the 3′ end were removed leading to a final alignment of 1404 characters. This alignment showed no gap and no stop codon (except for the final one), 545 (38.8%) sites were variable, and 138 were parsimony-informative. One hundred and one sites were variable and 67 (66%) were parsimony-informative at first codon positions; 35 sites were variable and 11 (31%) parsimony-informative at second codon positions; 409 sites were variable and 360 (88%) were parsimony-informative at third codon positions.
Phylogenetic analyses
Parsimony analysis of our dataset resulted in nine equally most parsimonious trees, which were 1954 steps long, with a consistency index of 0.4012 and a retention index of 0.6562. The best model selected under an Akaike Information Criterion was the general time reversible model (GTR) of nucleotide substitution, with the percentage of sites considered invariable and a gamma distribution of rates for variable sites (GTR + I + G).
All our phylogenetic analyses () strongly supported the monophyly of the Laurencia complex and resolved five, fully supported lineages. Laurencia s.s., Chondrophycus and Osmundea, were resolved as monophyletic genera. The two remaining lineages accommodated taxa currently assigned to the genus Palisada. Supra-generic relationships were not supported except for the relationship between the P. poiteaui group and Laurencia s.s. recovered in all our analyses ().
Fig. 1. Bayesian phylogram inferred from analyses of rbcL sequences for 39 Laurencia complex taxa and six outgroup species. Numbers above branches correspond to support values for Bayesian inference posterior probability/Maximum Likelihood bootstrap/and Maximum Parsimony bootstrap, respectively. Bold lines and % indicate a fully supported node in all the three analyses. Taxa marked in bold indicate newly determined sequences. Asterisks indicate generitypes, and the scale bar refers to the substitutions per site.
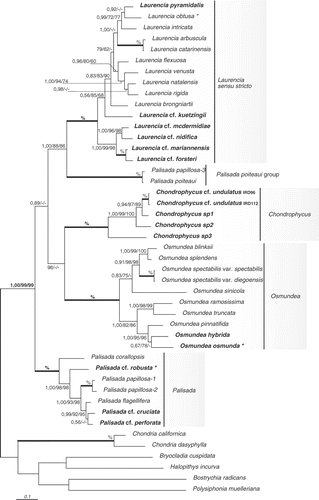
The genus Laurencia s.s. was resolved as a monophyletic lineage including the generitype L. obtusa, plus 14 other species. Infrageneric relationships within Laurencia s.s. lacked resolution, however two lineages appeared strongly supported. The first included specimens from New Caledonia (L. cf. mcdermidiae, L. cf. nidifica, L. cf. forsteri and L. cf. mariannensis), and the second, fully supported, encompassed L. arbuscula and L. catarinensis, two specimens from South America. Two other lineages were moderately supported; the first contained cosmopolitan species (L. pyramidalis, L. obtusa and L. intricata), and the second included taxa mainly from the Indian and Pacific oceans (L. flexuosa, L. venusta, L. natalensis and L. rigida).
One of the lineages in our analysis contained five specimens from New Caledonia. Two specimens were identified as Chondrophycus cf. undulatas, based on descriptions by Yamada (Citation1931) and Nam (Citation1999). The other three were difficult to identify to species due to the lack of reproductive structures and problematic tissue rehydration. Specimens were therefore referred to as sp1, sp2, and sp3. These specimens formed small, dark red clumps, cartilaginous in texture, with compressed fronds, and irregularly ramified axes with short wart-like ultimate branchlets. Epidermal cells were neither palisade-like in transversal section, nor secondarily pit connected, and lenticular thickenings were not observed in medullary cells. In longitudinal section, we observed projecting, domed epidermal cells near the branchlet in Chondrophycus sp3, a character mentioned by Nam (Citation1999) for some species of Chondrophycus. Based on our observations and molecular results, we assigned this clade to the genus Chondrophycus.
Taxa belonging to the genus Osmundea, including the type species O. osmunda, formed a fully supported and well-resolved lineage. Two lineages within this genus were moderately supported: one containing the North-East Pacific species (O. blinksii, O. splendens, O. spectabilis and O. sinicola) and the second containing species from North Atlantic coasts (O. ramosissima, O. truncata, O. pinnatifida, O. hybrida and O. osmunda). Osmundea was resolved as sister to Chondrophycus with high support in BI.
Phylogenetic analyses resolved specimens assigned to Palisada in two distinct lineages. One, fully supported in all analyses, included a specimen from New Caledonia, which presented the morpho-anatomical features of P. robusta, the generitype of Palisada; we therefore assigned this lineage to Palisada. The second lineage, fully supported in all analyses, included P. poiteaui and P. papillosa-3, two specimens with only three differences in their sequences (divergence 0.2%).
Discussion
Monophyly of the Laurencia complex
In our molecular phylogenetic analyses, the four genera forming the Laurencia complex are resolved as a strongly supported, monophyletic group, confirming previous studies based on molecular and morpho-anatomical characters (Garbary & Harper, Citation1998; Nam et al., Citation2000; McIvor et al., Citation2002; Abe et al., Citation2006; Fujii et al., Citation2006 and Díaz-Larrea et al., Citation2007). These results suggest that the main morpho-anatomical features (apical cells sunk in apical pits of branchlets, a recognisable axial cell row only near the apical cell and an extensive cortex (Kylin, Citation1956; Saito & Womersley, Citation1974; Womersley, Citation2003; Nam, Citation2006) used to distinguish members of the Laurencia complex from the remaining Rhodomelaceae are phylogenetically significant. However, using cladistic analyses of morphological characters, Nam & Choi (Citation2001) and Nam (Citation2006) resolved Laurencia clavata Sonder (an Australian endemic) as sister to the remaining species of the complex, and suggested that L. clavata may link the Laurencia complex and the genus Chondria C. Agardh. On the one hand, L. clavata has basally constricted branches, many refractive discoid starch grains in medullar and sub-cortical cells, which are characters shared with Chondria. On the other hand, it lacks specialized plates in its spermatangial structure, which is a characteristic feature of the tribe Chondriae, and has four pericentral cells, epidermal cells with secondary pit connections and unrecognizable axial cell rows in mature thalli, characters attributed to the Laurencia complex. Based on these observations, Nam (Citation2006) proposed resurrecting the genus Corynecladia J. Agardh to accommodate L. clavata. Similarly, Laurencia flexilis Setchell, which also has basally constricted branches (Masuda et al., Citation2006), was resolved by Abe et al. (Citation2006) as an early divergence in the Laurencia complex. Because Abe et al.'s sequences are not deposited in any public database, it was not possible to include their sequence of L. flexilis in the present study. Further analyses including both C. clavata and L. flexilis are needed to improve circumscription of the Laurencia complex, and to determine the evolutionary relationships of these two enigmatic species.
Interspecific relationships within the Laurencia complex
The five lineages resolved in our analyses are fully supported and correspond respectively to the genera Laurencia s.s., Chondrophycus, Osmundea, Palisada, and to a lineage encompassing P. poiteaui and P. papillosa-3. Palisada is divided into two unrelated lineages and is therefore polyphyletic. One of these two lineages () corresponds to Palisada since it includes a specimen that has the morpho-anatomical features of P. robusta, the generitype of Palisada. In addition, all the species of this group have epidermal cells with a conspicuous palisade structure (Martin-Lescanne, personal observation; see also ). The second lineage, the ‘P. poiteaui group’ (), includes two specimens identified morphologically as P. papillosa and P. poiteaui, although their rbcL sequences only differed by three base pairs (uncorrected p distance value = 0.2%). Two other specimens identified as P. papillosa were resolved within the Palisada lineage rather than the P. poiteaui group. Consequently, specimens assigned to P. papillosa did not form a monophyletic taxon. Palisada poiteaui and specimens referred to as P. papillosa-3 were both collected in the Gulf of Mexico (Florida) (McIvor et al., Citation2002; Senties & Diaz-Larrea, Citation2008), are similar in habit and difficult to distinguish (Littler & Littler, Citation2000), especially when they are fertile. We believe that P. papillosa-3 may have been misidentified and is probably P. poiteaui; however, the voucher specimen should be re-examined to confirm this hypothesis.
Table 3. Diagnostic characters used to identify the different genera of the Laurencia complex.
The P. poiteaui group was resolved as sister to Laurencia s.s. with moderate (ML, MP) to full (BI) support. Interestingly, P. poiteaui and P. gemmifera (which is now considered as conspecific with P. poiteaui, Díaz-Larrea et al., Citation2007) were two species transferred to Palisada by Nam (Citation2006), which lack palisadic epidermal cells. Both species also differ from other Palisada species by having five rather than four pericentral cells in procarp-bearing segments (Fujii et al., Citation1996). Nam (Citation2006) resolved these taxa as sister to the remaining species of Palisada based on morphological characters. The P. poiteaui group and Laurencia s.s. also have secondary pit connections between epidermal cells, a feature exhibited by most representatives of Laurencia s.s. but not of Palisada. Palisada poiteaui should either be included in Laurencia s.s. and this genus emended to accommodate it, or placed in a separate genus. Since P. poiteaui has two pericentral cells, whereas Laurencia s.s. has four, and lacks corps en cerise, in contrast to other Laurencia s.s. species, it seems preferable to accommodate it in a distinct genus (). This is consistent with the sequence divergences obtained in molecular analyses, P. poiteaui being as distant from Laurencia s.s. species (9–11%) as Chondrophycus species are from Osmundea (10–11%), and distances between P. poiteaui and Palisada species also ranged from 10–11%. Nam (Citation1999) proposed the subgenus Yuzurua to accommodate P. poiteaui, as well as Palisada parvipapillata (C.K. Tseng) Nam (as Chondrophycus parvipapillatus (C.K. Tseng) Garbary & Harper), Palisada iridescens (Wynne & Ballantine) Nam (as C. iridescens (Wynne & Ballantine) Garbary & Harper) and C. gemmiferus (now included in P. poiteaui, see above). Nam (Citation1999) designated P. poiteaui (as C. poiteaui) as the type species of the subgenus. We propose elevating Yuzurua to generic rank. At this time, we formally transfer P. poiteaui to Y. poiteaui, however we refrain from including P. parvipapillatus and P. iridescens in the new genus because of the lack of molecular data and the unknown state of the key characters in these species.
Species belonging to the genus Laurencia s.s. form a fully supported monophyletic cluster in which all species sequenced in the present study display Laurencia s.s. features: four pericentral cells per vegetative axial segment, longitudinally oriented secondary pit connections between contiguous superficial cortical cells and, when fresh material was available, corps en cerise within superficial cortical and trichoblast cells (). As in previous molecular and morpho-anatomical phylogenies (Garbary & Harper, Citation1998; Abe et al., Citation2006; Fujii et al., Citation2006), general infrageneric relationships were poorly resolved, and only a few interspecific nodes were well supported (). Since interspecific nodes were well supported in other genera, the absence of supported relationships within Laurencia s.s. could be due to either a weak phylogenetic signal (maximum divergence 0.05%) or insufficient taxon sampling. As mentioned in our results, some geographic structuring in our trees was observed but wider geographic sampling is needed to discuss any biogeographical scenarios.
Our molecular data largely echo the morpho-anatomical features used to delineate genera and sub-genera within the Laurencia complex. This suggests that molecular tools are helpful for assigning unidentified specimens of the Laurencia complex to a genus, especially when distinguishing morpho-anatomical characters cannot be observed, as it is often the case for reproductive features.
Despite the fact that the genera of the Laurencia complex are molecularly well-defined, relationships among them are currently poorly resolved and more molecular data and a larger taxon sampling are still necessary to further improve our understanding of this taxonomic complex in an evolutionary framework.
Taxonomic conclusion
Genus Yuzurua (Nam) Martin-Lescanne stat. nov. with the characters of the subgenus Yuzurua Nam ( Citation 1999 , Eur. J. Phycol . 34: 467)
TYPE SPECIES: Yuzurua poiteaui comb. nov. (J.V. Lamouroux) Martin-Lescanne.
BASIONYM: Fucus poiteaui J.V. Lamouroux (Citation1805, Ann. Mus. Hist. Nat. Paris 20: 63–64).
SYNONYMS: Chondrophycus poiteaui (J.V. Lamouroux) K.W. Nam (Citation1999, Eur. J. Phycol. 34: 463); Palisada poiteaui (J.V. Lamouroux) K.W. Nam (Citation2007, Algae 22: 54).
TYPE LOCALITY: Santo Domingo, Dominican Republic.
TYPE MATERIAL: CN, unnumbered.
SPECIES PRESENTLY INCLUDED IN THE GENUS: Yuzurua poiteaui.
Acknowledgements
We are grateful to Prof. Ki Wan Nam who generously provided us with specimens of Chondrophycus cartilagineus and Palisada robusta for morpho-anatomical study. This work was supported by the ‘Consortium National de Recherche en Génomique’, and the ‘Service de Systématique Moléculaire’ (IFR 101) of the Muséum national d’histoire naturelle. This research has benefited from funding allocated by the ANR BIODIVERSITE labelled BIONEOCAL granted to Philippe Grancolas. It is part of the agreement n°2005/67 between the Genoscope and the Muséum national d'histoire naturelle on the project ‘Macrophylogeny of life’ directed by Guillaume Lecointre. The travel and accommodation in New Caledonia for the first author was supported by the Institut de Recherche pour le Développement (IRD). New Caledonian samples were mostly collected during campaigns onboard ALIS vessel supported by IRD grants. The diving team of IRD Noumea is acknowledged for its kind help with field sampling. The first author benefited from a scholarship from the French ‘Ministère de l’éducation nationale, de l’enseignement supérieur et de la recherche’.
References
- Abe , T , Kurihara , A , Kawaguchi , S , Terada , R and Masuda , M . 2006 . Preliminary report on the molecular phylogeny of the Laurencia complex (Rhodomelaceae) . Coast. Mar. Sci. , 30 : 209 – 213 .
- Díaz-Larrea , J , Senties , A , Fujii , MT , Pedroche , FF and Oliveira , MC . 2007 . Molecular evidence for Chondrophycus poiteaui var. gemmiferus comb. et stat. nov. (Ceramiales, Rhodophyta) from the Mexican Caribbean Sea: implications for the taxonomy of the Laurencia complex . Bot. Mar. , 50 : 250 – 256 .
- Felsenstein , J . 1985 . Confidence limits on phylogenies: an approach using the bootstrap . Evolution , 39 : 783 – 791 .
- Freshwater , DW and Rueness , J . 1994 . Phylogenetic relationships of some European Gelidium (Gelidiales, Rhodophyta) species, on the basis of rbcL nucleotide sequence analysis . Phycologia , 33 : 187 – 194 .
- Fujii , MT , Collado-Vides , L and Cordeiro-Marino , M . 1996 . Morphological studies of Laurencia gemmifera and Laurencia poiteaui (Rhodomelaceae, Rhodophyta) from the Nichupté Lagoon System, Quintana Roo, Mexico . Bot. Mar. , 39 : 317 – 326 .
- Fujii , MT , Guimaranes , SMPB , Gurgel , CFD and Fredericq , S . 2006 . Characterization and phylogenetic affinities of the red alga Chondrophycus flagelliferus (Rhodomelaceae, Ceramiales) from Brazil based on morphological and molecular evidence . Phycologia , 45 : 432 – 441 .
- Furnari , G , Comarci , M and Serio , D . 2001 . The Laurencia complex (Rhodophyta, Rhodomelaceae) in the Mediterranean sea: an overview . Cryptogamie, Algol , 22 : 331 – 373 .
- Furnari , G and Serio , D . 1993 . The distinction of Laurencia truncata (Ceramiales, Rhodophyta) in the Mediterranean Sea from Laurencia pinnatifida . Phycologia , 32 : 367 – 372 .
- Furnari , G and Serio , D . 1995 . Progress in the taxonomy of the genus Laurencia (Ceramiales, Rhodophyta) up to the resurrection of the genus Osmundea . Giorn. Bot. Ital. , 129 : 185 – 188 .
- Garbary , DJ and Harper , JT . 1998 . A phylogenetic analysis of the Laurencia complex (Rhodomelaceae) of the red algae . Cryptogamie, Algol. , 19 : 185 – 200 .
- Guindon , S and Gascuel , O . 2003 . A simple, fast and accurate algorithm to estimates large phylogenies by maximum likelihood . Syst. Biol , 52 : 696 – 704 .
- GUIRY, M.D. & GUIRY, G.M. (2008). AlgaeBase version 4.2. World-wide electronic publication, National University of Ireland, Galway. Available at: http://www.algaebase.org, accessed August 2008
- Holmgren , PK , Holmgren , NH and Barnett , LC . 1990 . Index Herbariorum. Part 1, The Herbaria of the World, , 8th , Bronx, New York, (NY), , USA : New York Botanical Garden .
- Huelsenbeck , JP and Ronquist , FR . 2001 . MrBayes. Bayesian inference of phylogeny . Biometrics , 17 : 754 – 755 .
- Hughey , JR , Silva , PC and Hommersand , MH . 2001 . Solving taxonomic and nomenclature problems in Pacific Gigartinaceae (Rhodophyta) using DNA from type material . J. Phycol , 37 : 1091 – 1109 .
- Kumar , S , Tamura , K and Nei , M . 2004 . MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment . Brief. Bioinform. , 5 : 150 – 163 .
- Kylin , H . 1956 . Die Gattungen der Rhodophyceen , Lund, , Sweden : Gleerups .
- Lamouroux , JVF . 1805 . Dissertations sur plusieurs espèces de Fucus, peu connues ou nouvelles; avec leur description en latin et en français , Agen & Paris : de l'Imprimerie de Raymond Nouvel & Chez Treuttel et Würtz .
- Lamouroux , JV . 1813 . Essai sur les genres de la famille des thallasiophytes non articulées . Ann. Mus. Hist. Nat. Paris , 20 : 21 – 47 . 115–139, 267–293, pls 5–13
- Lin , SM , Fredericq , S and Hommersand , MH . 2001 . Systematics of the Delesseriaceae (Ceramiales, Rhodophyta) based on LSU rDNA and rbcL sequences, including the Phycodryoideae, subfam. nov . J. Phycol. , 37 : 881 – 899 .
- LITTLER, D.S. & LITTLER, M.M. (2000). Caribbean Reef Plants. An Identification Guide to the Reef Plants of the Caribbean, Bahamas, Florida and Gulf of Mexico. Offshore Graphics, Washington, (DC), USA
- Masuda , M , Tani , M and Kurihara , A . 2006 . New records of three marine red algae from Japan . Phycol. Res. , 54 : 244 – 254 .
- McDermid , KJ . 1988 . “ Section V. Laurencia (Rhodophyta, Rhodomelaceae) Introduction ” . In Taxonomy of Economic Seaweeds, with Reference to Some Pacific and Caribbean Species , Edited by: Abbott , IA . Vol. 2 , 221 – 229 . La Jolla, (CA), , USA : California Sea Grant College Program .
- McIvor , L , Maggs , CA , Guiry , MD and Hommersand , MH . 2002 . Phylogenetic analysis of the geographically disjunct genus Osmundea Stackhouse (Rhodomelaceae, Rhodophyta) . Constancea , 83 : 1 – 11 .
- Nam , KW . 1999 . Morphology of Chondrophycus undulata and C. parvipapillata and its implications for the taxonomy of the Laurencia (Ceramiales, Rhodophyta) complex . Eur. J. Phycol. , 34 : 455 – 468 .
- Nam , KW . 2006 . Phylogenetic re-evaluation of the Laurencia complex (Rhodophyta) with a description of L. succulenta sp. nov. from Korea . J. Appl. Phycol. , 18 : 679 – 697 .
- Nam , KW . 2007 . Validation of the Generic Name Palisada (Rhodomelaceae, Rhodophyta) . Algae , 22 : 53 – 55 .
- Nam , KW and Choi , HG . 2001 . Morphology of Laurencia clavata and L. elata (Ceramiales, Rhodophyta) in relation to generic circumscription in the Laurencia complex . Eur. J. Phycol , 36 : 285 – 294 .
- Nam , KW , Maggs , CA and Garbary , DJ . 1994 . Resurrection of the genus Osmundea with an emendation of the generic delineation of Laurencia (Ceramiales, Rhodophyta) . Phycologia , 33 : 384 – 395 .
- Nam , KW , Maggs , CA , Mcivor , L and Stanhope , MJ . 2000 . Taxonomy and phylogeny of Osmundea (Rhodomelaceae, Rhodophyta) in Atlantic Europe . J. Phycol. , 36 : 759 – 772 .
- Nam , KW and Saito , Y . 1991 . Laurencia similis (Ceramiales, Rhodophyta) a new species from Queensland, Australia . Br. Phycol. J. , 26 : 375 – 382 .
- Nam , KW and Saito , Y . 1995 . Vegetative and reproductive anatomy of some Laurencia (Ceramiales, Rhodophyta) species with a description of L. maris-rubri sp. nov. from the Red Sea . Phycologia , 34 : 157 – 165 .
- Papenfuss , GF . 1947 . Generic names of algae proposed for conservation: I . Madroño , 9 : 8 – 17 .
- Posada , D and Crandall , KA . 1998 . MODELTEST: testing the model of DNA substitution . Mol. Biol. Evol. , 23 : 615 – 625 .
- Saito , Y . 1967 . Studies on Japanese species of Laurencia, with special reference to their comparative morphology . Mem. Fac. Fish. Hokkaido Univ. , 15 : 1 – 81 .
- Saito , Y and Womersley , HBS . 1974 . The Southern Australian species of Laurencia (Ceramiales: Rhodophyta) . Aust. J. Bot. , 22 : 815 – 874 .
- Schmitz , F . 1889 . Systematische Übersicht der bisher bekannten Gattungen der Florideen . Flora (Jena) , 72 : 435 – 456 .
- Senties , A and Diaz-Larrea , J . 2008 . Proposals for Palisada poiteaui var. gemmifera comb. nov. and Palisada corallopsis comb. nov. (Rhodomelaceae, Rhodophyta) . Bot. Mar. , 51 : 69 – 70 .
- Stackhouse , J . 1809 . Tentamen marino-cryptogamicum, ordinem novum; in genera et species distributum, in Classe XXIVta Linnaei sistens . Mém. Soc. Imp. Nat. Moscou , 2 : 50 – 97 .
- Swofford , DL . 2003 . PAUP*. Phylogenetic Analyses Using Parsimony (*and Other Methods) , Sunderland, (MA), , USA : Sinauer Associates .
- Thompson , JD , Higgins , DG and Gibson , TJ . 1994 . CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice . Nucl. Acids Res. , 22 : 4673 – 4680 .
- Womersley , HBS . 2003 . The Marine Benthic Flora of Southern Australia. Part IIID , Canberra, , Australia : Australian Biological Resources Study, Canberra and the State Herbarium of South Australia. Pirion .
- YAMADA, Y. (1931). Notes on Laurencia, with special reference to the Japanese species. California: University of California Publications in Botany. Vol. 16, no. 7, pp. 185–310, 30 pIs.
- Zuccarello , GL and West , JA . 2006 . Molecular phylogeny of the subfamily Bostrychioideae (Ceramiales, Rhodophyta): subsuming stictosiphonia and highlighting polyphyly in species of Bostrychia . Phycologia , 45 : 24 – 36 .