Abstract
Distromatic foliose blades of the algal genus Ulva are notoriously difficult to identify due to their simple morphologies and few diagnostic characteristics that often exhibit intraspecific variation and interspecific overlap. Hence, species differentiation is difficult and diversity estimates are often inaccurate. Two major goals of this study were to assess the diversity of distromatic Ulva spp. in the Great Bay Estuarine System (GBES) of New Hampshire and Maine, USA, and to compare historical and present day records of these species. Molecular analysis (using ITS sequences) of field-collected specimens revealed four distinct taxa: Ulva lactuca, U. rigida, U. compressa, and U. pertusa. Prior to molecular screening, Ulva lactuca was the only distromatic Ulva species reported for the GBES. Ulva pertusa and the foliose form of U. compressa are newly recorded for the Northwest Atlantic, and the range of U. rigida has been extended. Molecular analysis of historical herbarium voucher specimens indicates that U. rigida, U. pertusa, and the foliose form of U. compressa have been present in the GBES since at least 1966, 1967, and 1972, respectively. The distromatic morphotype of U. compressa is found only in low salinity areas, which suggests that salinity may influence its morphological development. Molecular and morphological evaluations are critical if we are to distinguish between cryptic taxa, accurately assess biodiversity, and effectively monitor the spread of non-indigenous macroalgae.
Introduction
Distromatic foliose blades of Ulva L. (Ulvales, Chlorophyta) are notoriously difficult to identify to species level due to their simple morphologies and few diagnostic characteristics that often exhibit intraspecific variation and interspecific overlap (Bliding, Citation1968; Tanner, Citation1979, Citation1986; Blomster et al., Citation1999; Malta et al., Citation1999; Tan et al., Citation1999). The primary morphological features used for species differentiation include cell alignment in surface view, cell shape and size, chloroplast structure, pyrenoid numbers, and organization of starch grains (Bliding, Citation1968; Koeman & van den Hoek, Citation1981; Hoeksema & van den Hoek, Citation1983). Even so, several investigators (Titlyanov et al., Citation1975; Tanner, Citation1979, Citation1986; Malta et al., Citation1999) have shown that these features may be phenotypically variable. Further complications in differentiating between species based on morphology result from the ability of some individuals within the same species to exist as either a foliose blade or hollow tube, both of which are characteristic of the genus (Tan et al., Citation1999; Blomster et al., Citation2002; Hayden et al., Citation2003). Due to the unreliability of morphological characteristics for species identification in the genus Ulva, molecular analysis (the nuclear genome, nuclear ribosomal ITS sequences and the chloroplast rbcL gene) has become useful for differentiating taxa (Blomster et al., Citation1998, Citation1999; Coat et al., Citation1998; Malta et al., Citation1999; Tan et al., Citation1999; Hayden & Waaland, Citation2002, Citation2004; Hayden et al., Citation2003; Shimada et al., Citation2003; Loughnane et al., Citation2008). However, even when molecular analyses are used, specimens can still be misidentified if the determination is based on comparison to sequences posted on databases like GenBank (National Institutes of Health) and not validated against nomenclatural type material.
The challenge of identifying distromatic Ulva spp. from morphological characters has in some cases resulted in inaccurate biodiversity estimates. Molecular data have been used to resolve taxonomic issues in the genus, which in turn has led to more accurate biodiversity estimates. For example, in Europe, using a combination of morphological and molecular characters, Maggs et al. (Citation2007) synonymized U. scandinavica Bliding and U. armoricana P. Dion, B. de Reviers & G. Coat with U. rigida C. Agardh. As a result, the number of distromatic Ulva taxa in Ireland and southern Britain is seven rather than nine (Loughnane et al., Citation2008). Furthermore, the Asian taxon U. pertusa Kjellman has recently been reported from the Iberian Peninsula (Spain), albeit present and identified as another taxon as far back as 1990 (López et al., Citation2007), thus increasing biodiversity estimates for the region. Biodiversity assessments of macroalgal communities are important for examining community structure and ecological shifts that may coincide with environmental changes including abiotic factors such as water quality and climate, and biotic factors such as the introduction of non-native species.
Water quality in coastal environments throughout the world has declined from anthropogenic pollution, particularly nutrient runoff (Smith, Citation2002). Eutrophic, or high nutrient, conditions in shallow embayments can result in the formation of extensive blooms of macroscopic green algae or ‘green tides’ (Fletcher, Citation1996). Initially, green tides were assumed to consist of a single species (Fletcher, Citation1996), but recent investigations have shown that they can often comprise a mixture of species (Malta et al., Citation1999; Hiraoka et al., Citation2004). Ulva species are able to form green tides due to their high surface area to volume ratios and efficient nutrient uptake mechanisms (Rosenberg & Ramus, Citation1984; Taylor et al., Citation1998). In the Great Bay Estuarine System of New Hampshire and Maine, USA (GBES), Ulva blooms occur in many parts of the system during summer months. Historically, Ulva lactuca L. was the only distromatic species recorded in the GBES (Mathieson & Hehre, Citation1986), but molecular screening of Ulva populations in other geographic regions has indicated that their diversity is often underestimated (Loughnane et al., Citation2008; Heesch et al., Citation2009). As a result, the goals of the present study were to assess the biodiversity of distromatic Ulva spp. within the GBES using a combination of morphological and molecular tools for species identification, and to compare past and recent collections of these taxa throughout the GBES in order to document changes in distributions and approximate dates of any introductions.
Materials and methods
Site descriptions
The GBES is located on the border of New Hampshire and Maine, USA (). It consists of two major bays (Great Bay and Little Bay), plus the Piscataqua River, Portsmouth Harbor, and eight tidal tributaries. The system contains over 160 km of shoreline and its substratum is dominated by mud but becomes rockier near the open coast (Hardwick-Witman & Mathieson, Citation1983; Mathieson & Hehre, Citation1986). Temperature, salinity, and water clarity vary throughout the estuary. Generally, temperature and salinity are more variable in inner than outer estuarine locations (Emerich Penniman et al., Citation1985). For example, the seasonal temperature ranges at Dover Point (−2°C to 24°C) and Adams Point (−2°C to 27°C), which are 13–19 km inland (; ), are wider than those on the open coast (Fort Stark: −1°C to 19°C). In addition, salinity varies greatly at Dover Point (1–30 psu) and Adams Point (7–31 psu), whereas it remains fairly constant at Fort Stark (27–32 psu). Water clarity also varies spatially and temporally, being low in inland sites and increasing near the open coast.
Fig. 1. Map of the Great Bay Estuarine System, New Hampshire/Maine, USA, showing locations of 11 study sites (filled circles). Dashed white line indicates border between the two states along the Piscataqua River. Filled stars represent two low salinity estuarine sites where Ulva compressa (tubular form) was reported by Mathieson & Hehre (Citation1986).
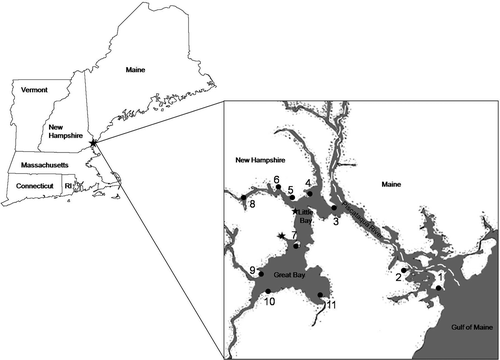
Table 1. Collection sites and distribution of distromatic Ulva taxa within the GBES and on the adjacent open coast.
Sample collection, processing and analysis
Ulva specimens were collected at eleven sites (); location, latitude and longitude coordinates, distance inland from the open coast and a short site description for each are summarized in . Ulva collections were made on low tides during the summer and fall of 2008 (). Specimens were chosen haphazardly, and care was taken to collect specimens exhibiting a range of morphologies in order to increase the probability of collecting several different taxa from the same site. Collections were placed in plastic bags and kept on ice during transport to the University of New Hampshire (UNH). In the laboratory, specimens were rinsed in artificial seawater prepared with Instant Ocean Sea Salt (Spectrum Brands Inc., Atlanta, Georgia, USA), sorted by general morphology, and examined under a compound light microscope (Olympus BX40F4, Olympus Optical Co. Ltd, Tokyo, Japan) for cell shape, chloroplast structure, numbers and organization of starch grains, and numbers of pyrenoids. Photomicrographs were taken using an Olympus PM-30 camera (Olympus Optical Co. Ltd). A small sample (∼2 cm2) of each specimen was preserved in silica gel for molecular analysis (see below), while the remainder of each blade was pressed as an herbarium voucher specimen and deposited in the Hodgdon Herbarium at the University of New Hampshire (NHA). Average cell size was measured on rehydrated herbarium specimens; measurements were taken of the longest and shortest parts of each cell. Historical specimens of GBES material from the 1950–1970s in NHA were also evaluated morphologically and with molecular data. Eighty-six freshly collected specimens and 43 specimens from pre-existing herbarium collections were genetically screened for identification.
DNA was extracted from preserved seaweed samples (i.e. in silica gel) using a Puregene Tissue Extraction Kit (Qiagen Inc., Valencia, CA, USA) and amplified using a modification of the methods detailed by Bray et al. (Citation2006). Polymerase Chain Reaction (PCR) analysis was done using 4 µl DNA and 0.25 µl Taq Polymerase (Go-Taq Flexi, Promega, Madision, WI) in a final volume of 50 µl per reaction. Amplification was conducted in an Eppendorf Mastercycler 5333 (Hamburg, Germany). As outlined in , the primers and PCR reaction profile used for amplification of the rDNA internal transcribed spacers (ITS1 and ITS2) and 5.8 S gene were identical to those used by Hayden et al. (Citation2003) and Blomster et al. (Citation1998). The ITS regions were used as molecular markers because attempts with rbcL primers were not as successful as ITS, especially for old herbarium specimens. However successful rbcL sequence identifications were always in agreement with the ITS markers.
Table 2. Primers used for ITS sequence amplification and sequencing based upon Blomster et al. (Citation1998) and Hayden et al. (Citation2003)
Purification of amplified DNA was conducted on a 0.8% UltraPure Low Melting Point Agarose (Invitrogen Corporation, Carlsbad, CA, USA) gel in 0.5x nTBE Buffer (54 g l−1 Trisbase, 27.5 g l−1 boric acid, 0.2% 0.5 M solid EDTA at pH 8.0). Quantification of DNA was conducted via fluorometry using an Invitrogen Qubit Fluorometer (Turner BioSystems, Sunnyvale, CA, USA) and reagents from a Quant-iT dsDNA BR assay kit (Invitrogen Corporation). Sequencing was done at the UNH Hubbard Center for Genome Studies using Applied Biosystems BigDye Terminator Cycle Sequencing Kits (v1.1 and v3.1) and an ABI 3130 DNA Analyzer.
The DNA sequences were trimmed using Chromas 2.22 (Technelysium Pty Ltd, Tewantin, Queensland, Australia), assembled in SeqMan Pro 7.2.1 and aligned based on primary structure using the ClustalW method in Megalign 7.2.1 (both from DNASTAR, Lasergene Inc, Madison, WI, USA). Sequences obtained in this study were aligned with those retrieved from GenBank (National Institutes of Health) using the Blast feature in Megalign 7.2.1; the GenBank sequences used for comparison are shown in . Sequence divergence percentages were calculated using the sequence distance calculation feature in Megalign. Sequence comparisons were made between all taxa found at all GBES collection sites.
Table 3. GenBank ITS sequences that were identical to ITS sequences of Ulva spp. collected from the GBES.
Results
Initial morphological observations of distromatic Ulva specimens from the GBES showed a range of thallus and cellular characters that suggested the presence of multiple species. Molecular analysis of both ITS regions plus the 5.8 S rDNA gene confirmed the presence of four distinct ribotypes, which matched the sequences identified on Genbank as U. lactuca, U. pertusa, U. rigida, and U. compressa (). Ulva pertusa is a new record for the Northwest Atlantic, while U. rigida is a new record for New Hampshire. Although the tubular morphology of U. compressa is common in the Northwest Atlantic, the presence of its distromatic form represents a new record for this geographical area. Within each of the four taxa, there was no sequence variation among specimens collected from GBES. The sequence divergence values between the four taxa for the ITS regions are shown in . Ulva lactuca and U. rigida had the highest percentage sequence divergence, followed by U. pertusa and U. rigida (19.5 and 18.8% divergence, respectively), whereas U. pertusa and U. lactuca had the lowest sequence divergence (4.8%).
Table 4. Percent sequence divergences within the ITS regions for the four Ulva taxa found in the GBES. None of the species showed any intraspecific sequence divergence.
The ITS sequences obtained from Ulva rigida specimens matched exactly a sequence in GenBank identified as U. scandinavica, but differed by 5 base pairs (bp) from a sequence identified as U. rigida. Despite the 5-bp difference, the name U. rigida was used for the specimens collected from the GBES based on the current nomenclature (). Distromatic Ulva compressa specimens collected from GBES were identical to ITS sequences listed in GenBank as this taxon, but also matched a single sequence identified as U. pseudocurvata (). The latter GenBank sequence is thought to have been misidentified (; see Discussion), hence the use of the name U. compressa rather than U. pseudocurvata for specimens collected in the present study.
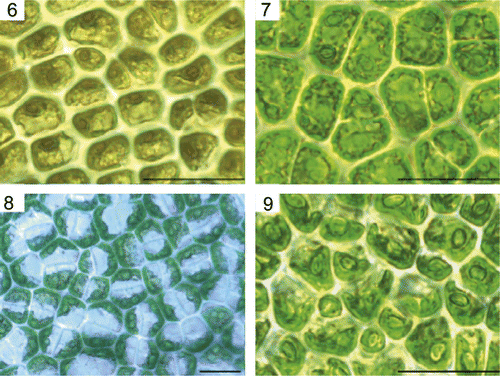
Although somewhat variable, the blade and cellular characteristics of the specimens generally agreed with the molecular delineation of the four taxa. Ulva lactuca specimens generally had ruffled margins and were often rounder in shape than other taxa (); cells typically had a lobed parietal chloroplast and a single pyrenoid, but multiple pyrenoids per cell were also observed (). Ulva rigida thalli were generally irregularly shaped, dark green in colour, and with a wrinkled texture that was especially visible upon drying (). The cells of U. rigida contained a parietal chloroplast with multiple pyrenoids (). The species’ characteristic marginal ‘teeth’ were rarely observed. In surface view, Ulva pertusa looked similar to U. lactuca as its chloroplasts were confined to one side of the cell, but most cells had multiple pyrenoids (). Ulva pertusa blades were often extensively perforated, especially in older specimens. Reproductive thalli of U. pertusa were common and easily recognized by their wide, pale margin (). The morphology of distromatic Ulva compressa was extremely variable () and specimens were frequently difficult to distinguish from U. rigida. These U. compressa specimens exhibited a wrinkled texture upon drying similar to that of U. rigida. The chloroplasts of U. compressa were hooded but sometimes filled the entire cell, and they contained a single large pyrenoid (). Cell measurements of rehydrated herbarium material showed that U. compressa cells were noticeably smaller (9.8 × 5.2 µm) than those of the other three taxa, which all had similar cell measurements (12–14 × 8 µm). Ulva lactuca and U. rigida cells were often rectangular in shape, while U. compressa and U. pertusa cells were polygonal. The arrangement of cells into rows in surface view was not observed as a distinguishing characteristic for any of the taxa.
Figs 2–5. Representative blade morphologies of the four Ulva taxa found in the GBES. . Ulva lactuca. . Ulva rigida. . Ulva pertusa. . Ulva compressa. Scale bar: 4 cm.

Figs 6–9. Representative cell morphologies (from fresh material) of the four Ulva taxa found in GBES. . Ulva lactuca . Ulva rigida. . Ulva pertusa. . Ulva compressa. Scale bars: 20 µm.
Spatial distribution within GBES varied among the species (). Ulva rigida was most ubiquitous, occurring at nine of the eleven sites; eight of the nine sites where U. rigida occurred were more than 16 km inland from the open coast. The species was also found rarely at South Mill Pond, a highly polluted site only 5 km from the open coast. Ulva lactuca and U. pertusa had broad estuarine distributions and were always found together; they occurred at four sites ranging from the open coast to the inner estuary (0–20 km inland; ). The distromatic foliose form of U. compressa was found at five inner estuarine sites (17.3–24.8 km inland; ) where it was always found with U. rigida. In general, U. rigida and the foliose form of U. compressa tended to occupy inner estuarine sites with relatively low salinities, while U. lactuca and U. pertusa were more often found closer to the open coast in relatively high salinities.
Molecular analysis of specimens from GBES collected during the 1960s and 1970s indicated that all three newly reported taxa (U. rigida, U. pertusa, and distromatic U. compressa) had been previously collected and misidentified as U. lactuca. The three taxa have been present in the GBES since at least 1966, 1967, and 1972, respectively. It is possible that these were present prior to the 1960s, but to date, useable DNA sequences have not been obtained from older specimens. The species distributions in recent and historical collections were similar, except that one historical collection (1968) of Ulva pertusa from an inner estuarine site (#8) was not confirmed during recent collections.
Discussion
Many investigators have noted that several morphological features in foliose distromatic forms of Ulva are unreliable for species identification due to phenotypic plasticity (Coat et al., Citation1998; Malta et al., Citation1999; Woolcott & King, Citation1999; Shimada et al., Citation2003; Loughnane et al., Citation2008). By contrast, several investigators (Tan et al., Citation1999; Hayden et al., Citation2003; Shimada et al., Citation2003; López et al., Citation2007) have suggested that molecular characters are more dependable. The use of ITS sequences, in combination with morphological characterization, allowed us to successfully differentiate four Ulva taxa from the GBES, where previously only one species (U. lactuca) was known (Mathieson & Hehre, Citation1986; Mathieson & Penniman, Citation1986). Ulva rigida, U. pertusa, and the foliose form of U. compressa are new records for the GBES, and the latter two are new records for the Northwest Atlantic.
Ulva rigida is a common species on the east coast of North America, extending from Newfoundland, Canada to Long Island Sound, New York, USA. Despite extensive morphological surveys previously conducted on macroalgal species throughout the GBES, U. rigida had not been reported from Massachusetts or New Hampshire (Mathieson & Hehre, Citation1986; Sears, Citation2002). In the present study the species was found at several outer and inner estuarine sites within the GBES. The ITS sequences of U. rigida reported here were identical to U. scandinavica (AB097659) and differed by 5 bp from U. rigida sequences submitted by Hayden et al. (Citation2003) and Tan et al. (Citation1999). Previous studies have shown analogous differences between the ITS sequences of U. armoricana, U. scandinavica, and U. rigida. Coat et al. (Citation1998) found a 6-bp difference between U. armoricana and U. rigida. Loughnane et al. (Citation2008) and Malta et al. (Citation1999) found few differences in rbcL and ITS sequences between the three taxa and noted that they were very closely related. Maggs et al. (Citation2007) reported that the intraspecific variation in ITS sequences was similar to the interspecific variation between the three groups, and they considered the three taxa synonymous, with the name U. rigida having precedence. Therefore, the name U. rigida was used for specimens collected here in light of the conspecifity of the taxa reported by Maggs et al. (Citation2007) and Loughnane et al. (Citation2008).
Ulva pertusa specimens contained conspicuous holes that distinguished them from other Ulva species. The holes result from morphological development rather than grazing, which is common in other Ulva taxa (Shimada et al., Citation2003; López et al., Citation2007). Ulva pertusa is an Asiatic species, occurring in the North and South Pacific, plus the coasts of South Africa and Europe (López et al., Citation2007; Guiry & Guiry, Citation2009). Its means of introduction to the North Atlantic is unknown, but it is assumed to have been transferred with Asian oyster aquaculture (Verlaque, Citation2001; Verlaque et al., Citation2002; López et al., Citation2007). The species was reported in 1994 off the coast of Brittany, France and in the Mediterranean Sea, where it was previously misidentified as U. rotundata (cf. Coat et al., Citation1998; Verlaque et al., Citation2002). In 2007 it was recorded from the Iberian Peninsula (Spain), but it was thought to have been there since at least 1990 based on herbarium collections (López et al., Citation2007). Subsequently, it has been reported from the Netherlands (Stegenga et al., Citation2007). Molecular analysis of herbarium specimens from the GBES indicates that U. pertusa has been present in the Northwest Atlantic since at least 1967, which is earlier than the report by López et al. (Citation2007). The present study provides the first report of U. pertusa from the Northwestern Atlantic. Mathieson et al. (Citation2008) documented the presence of 20 introduced seaweeds in this geographic region, including the Asiatic green alga Codium fragile subsp. fragile (as subsp. tomentosoides). The presence of U. pertusa in the GBES confirms 21 introduced species, three of which are green algae. Ulva pertusa was found at four sites in the GBES, always in association with U. lactuca. Ulva pertusa and U. lactuca were even found growing together as epiphytes on the same Chondrus crispus Stackhouse thallus at one site; hence, the two species can coexist and share analogous habitats. While U. pertusa has a wide tolerance of estuarine salinities, it does not seem to be outcompeting U. lactuca based on frequent collections of both species together. Future studies of competitive interaction between species should determine if there is potential for U. pertusa to outcompete native Ulva taxa. Additional systematic collections should be made to monitor the distribution and potential spread of U. pertusa throughout the Northwest Atlantic.
The tubular form of Ulva compressa is common within the GBES (Mathieson & Hehre, Citation1986), but its foliose distromatic morphotype has not been previously identified. Its foliose morphology is found in other geographical areas, including the Northeast Atlantic and the Pacific Northwest coast of North America (mistaken as U. pseudocurvata: Tan et al., Citation1999; Hayden et al., Citation2003), and it is apparently not as rare as formerly thought (F. Mineur, pers. comm.). It also appears that distromatic U. compressa was previously confused with U. pseudocurvata due to similarities in morphological and cellular characteristics. As a result, U. compressa was believed to be conspecific with U. pseudocurvata (Tan et al., Citation1999; Hayden et al., Citation2003; Hayden & Waaland, Citation2004). However, despite morphological similarities, molecular evidence indicates that U. pseudocurvata and the foliose form of U. compressa are not closely related (Maggs et al., Citation2007). Therefore, it is likely that the ITS sequence identified as U. pseudocurvata in GenBank (accession number AJ234312) that is identical to the sequence identified as U. compressa (AJ550765) is actually a sequence from a distromatic specimen of U. compressa (F. Mineur, pers. comm.).
Intraspecific morphological plasticity has also been reported for Ulva intestinalis L., which generally has a tubular form. Blomster et al. (Citation2002) reported the occurrence of Monostroma–like macroalgal sheets in green tide blooms on the west coast of Finland in the Baltic Sea. Their ITS sequences revealed that they were actually a monostromatic morphotype of U. intestinalis (reported as Enteromorpha intestinalis). They stated that the ITS sequences were identical to those of attached tubular U. intestinalis from southwestern Finland. The existence of a monostromatic foliose U. intestinalis in the Baltic Sea plus a distromatic form of U. compressa in the GBES provides two examples of Ulva spp. exhibiting both tubular and foliose morphologies. In their tubular form the two species can appear very similar (Blomster et al., Citation1998), whereas in their foliose forms U. intestinalis is a monostromatic blade and U. compressa forms a distromatic blade. The three different types of morphology exhibited by these two species further illustrates that morphological development in Ulva can be extremely variable and complicates the identification and classification of the genus Ulva (Hayden et al., Citation2003).
The factors controlling the development of Ulva compressa into variable morphologies are uncertain. Blomster et al. (Citation1998) reported that the morphologies of U. compressa and U. intestinalis (reported as Enteromorpha spp.) varied depending on the salinity in their environment. Low salinities or salinity shocks were correlated with branch development in U. intestinalis, which complicated differentiation between the two species because the tubular form of U. compressa is also branched. Further evidence for the influence of salinity on morphological development in Ulva spp. is provided by Tan et al. (Citation1999) who suggested that salinity might influence morphological development in U. compressa. They reported that U. compressa specimens (as U. pseudocurvata) occurred in the upper Ythan Estuary (Scotland) near a freshwater source, whereas tubular U. compressa occurred in more coastal and outer estuarine sites. In the present study, distromatic U. compressa was found at sites more than 16 km inland from the coast, which had relatively low salinities compared to open coastal sites. Because no blade-forming U. compressa specimens were collected from outer estuarine sites with higher salinities, the results support a possible link between low salinity and blade formation in U. compressa.
Mathieson & Hehre (Citation1986) and Mathieson and Penniman (Citation1986) reported that Ulva compressa (as Enteromorpha compressa) was a widely distributed species within the GBES and on the adjacent open coast. In the present study the species was recorded at two inner estuarine sites (; ) with highly variable salinities and extensive freshwater discharges. We found distromatic U. compressa at sites with similar hydrographies, and through molecular screening we documented its presence as far back as 1977. Due to the fact that U. intestinalis can exhibit branching under low salinity conditions (Blomster et al., Citation1998), it is possible that the specimens identified as U. compressa from inner estuarine sites (Mathieson & Hehre, Citation1986; Mathieson & Penniman, Citation1986) were actually branched forms of U. intestinalis. Molecular investigation of NHA herbarium specimens morphologically identified as U. compressa from one low salinity site in the GBES (# 8) has not confirmed the presence of tubular U. compressa, but rather has revealed these specimens as other Ulva species, including U. intestinalis (unpublished data). As a result, there is no molecular confirmation that the tubular and blade form of U. compressa coexist at site 8, which is consistent with Tan et al.'s (Citation1999) hypothesis that there may be reversible genetic switches controlling morphological development in Ulva that are influenced by salinity.
A combination of eutrophic conditions and low salinities may influence the presence of blade and tubular morphologies of Ulva taxa. Valiela et al. (Citation1997) suggested that changes in macroalgal morphology under eutrophic conditions may be a response to alter surface area:volume ratios and subsequently nutrient uptake. Morphologies that increase nutrient uptake efficiency provide an ecological advantage, perhaps facilitating bloom formation (Poole & Raven, Citation1997). The monostromatic blades of U. intestinalis reported by Blomster et al. (Citation2002) from Olkiluodonvesi, Finland, were from a eutrophic, microtidal site with a salinity of 4–5 psu. Attached U. intestinalis specimens with tubular morphologies did not coexist with monostromatic blades at Olkiluodonvesi. Hence, the eutrophic conditions and low salinity at Olkiluodonvesi may have contributed to the unique morphological development of monostromatic foliose U. intestinalis (Blomster et al., Citation2002).
In summary, it is clear that the diversity of Ulva species in the GBES and within the Northwest Atlantic as a whole has been underestimated because of morphological plasticity and the previous lack of molecular tools. In conjunction with several other studies (Malta et al., Citation1999; Tan et al., Citation1999; Loughnane et al., Citation2008), our results provide further evidence that morphological characteristics are phenotypically variable, making definitive species identifications of distromatic Ulva thalli difficult. Without accurate species identifications, it is impossible to adequately understand ecological systems and evaluate temporal changes. Thus, molecular data combined with morphological observations are essential for ensuring correct species identification, accurate biodiversity assessments, and effective monitoring of introduced species. In order to make molecular data most significant, every effort should be made to sequence nomenclatural type material or at least samples from type locations. Otherwise, GenBank sequences can be incorrect based upon misidentifications. The names used for the taxa here were based on sequences from GenBank that have not been authenticated against nomenclatural type material; therefore we cannot be completely sure of their accuracy. The problem introduced by misidentified sequences on GenBank is a clear indication that sequencing holotype material, when available and intact, is an important pursuit that should be continued to alleviate taxonomic confusion.
Acknowledgements
We thank Frédéric Mineur and Chris Maggs for their constructive comments regarding GenBank records of various Ulva taxa. We also thank the University of New Hampshire College of Life Sciences for a Summer TA Fellowship and the National Estuarine Research Reserve System for a Graduate Research Fellowship (NOAA award number NA08NOS4200285). This paper is Scientific Contribution Number 2401 from the New Hampshire Agricultural Experiment Station, as well as Contribution Number 485 from the Jackson Estuarine Laboratory and Center for Marine Biology.
References
- Bliding , C . 1968 . A critical survey of European taxa in Ulvales, II: Ulva, Ulvaria, Monostroma, Kornmannia . Bot. Notiser , 121 : 535 – 629 .
- Blomster , J , Bäck , S , Fewer , DP , Kiirikki , M , Lehvo , A , Maggs , CA and Stanhope , MJ . 2002 . Novel morphology in Enteromorpha (Ulvophyceae) forming green tides . Am. J. Bot. , 89 : 1756 – 1763 .
- Blomster , J , Maggs , CA and Stanhope , MJ . 1998 . Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles . J. Phycol. , 34 : 319 – 340 .
- Blomster , J , Maggs , CA and Stanhope , MJ . 1999 . Extensive intraspecific morphological variation in Enteromorpha muscoides (Chlorophyta) revealed in molecular analysis . J. Phycol. , 35 : 575 – 586 .
- Bray , TL , Neefus , CD and Mathieson , AC . 2006 . Morphological and molecular variability of Porphyra purpurea (Roth) C. Agardh (Rhodophyta, Bangiales) from the Northwest Atlantic . Nov. Hedwig. , 82 : 1 – 22 .
- Coat , G , Dion , P , Noailles , M-C , De Reviers , B and Fontaine , J-M . 1998 . Ulva armoricana (Ulvales, Chlorophyta) from the coasts of Brittany (France). II. Nuclear rDNA ITS sequence analysis . Eur. J. Phycol. , 33 : 81 – 86 .
- Emerich Penniman , C , Mathieson , AC , Loder , TC , Daly , MA and Norall , TL . 1985 . Nutrient and hydrographic data for the Great Bay Estuarine System New Hampshire-Maine part III . Jackson Estuarine Laboratory Contribution , : 151
- Fletcher , RL . 1996 . “ The occurrence of 'green tides': a review ” . In Marine Benthic Vegetation: Recent Changes and the Effects of Eutrophication , Edited by: Schramm , W and Nienhuis , PH . 7 – 43 . Berlin : Springer .
- Guiry, M.D. & Guiry, G.M. (2009). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; acessed on 22 May 2009.
- Hardwick-Witman , MN and Mathieson , AC . 1983 . Intertidal macroalgae and macroinvertebrates: Seasonal and spatial abundance patterns along an estuarine gradient . Estuar. Coast. Shelf Sci. , 16 : 113 – 129 .
- Hayden , HS , Blomster , J , Maggs , CA , Silva , PC , Stanhope , MJ and Waaland , JR . 2003 . Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera . Eur. J. Phycol. , 38 : 277 – 294 .
- Hayden , HS and Waaland , JR . 2002 . Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences . J. Phycol. , 38 : 1200 – 1212 .
- Hayden , HS and Waaland , JR . 2004 . A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific . Phycologia , 43 : 364 – 382 .
- Heesch , S , Broom , JES , Neill , KF , Farr , TJ , Dalen , JL and Nelson. , WA . 2009 . Ulva, Umbraulva, and Gemina: genetic survey of New Zealand taxa reveals diversity and introduced species . Eur. J. Phycol. , 44 : 143 – 154 .
- Hiraoka , M , Ohno , M , Kawaguchi , S and Yoshida , G . 2004 . Crossing test among floating Ulva thalli forming ‘green tide’ in Japan . Hydrobiologia , 512 : 239 – 245 .
- Hoeksema , BW and van den Hoek , C . 1983 . The taxonomy of Ulva (Chlorophyceae) from the coastal region of Roscoff (Brittany, France) . Bot. Mar. , 26 : 65 – 86 .
- Koeman , RPT and van den Hoek , C . 1981 . The taxonomy of Ulva (Chlorophyceae) in the Netherlands . Br. Phycol. J. , 16 : 9 – 53 .
- López , SB , Fernandez , IB , Lozano , RB and Ugarte , JC . 2007 . Is the cryptic alien seaweed Ulva pertusa (Ulvales, Chlorophyta) widely distributed along European Atlantic coasts? . Bot. Mar. , 50 : 267 – 274 .
- Loughnane , CJ , McIvor , LM , Rindi , F , Stengel , DB and Guiry , MD . 2008 . Morphology, rbcL phylogeny and distribution of distromatic Ulva (Ulvophyceae, Chlorophyta) in Ireland and southern Britain . Phycologia , 47 : 416 – 429 .
- Maggs , CA , Blomster , J , Mineur , F and Kelly , J . 2007 . “ Ulva. ” . In In Green Seaweeds of Britain and Ireland , Edited by: Brodie , J , Maggs , CA and John , DM . Dunmurry , , Northern Ireland : The British Phycological Society, Dataplus Print and Design .
- Malta , EJ , Draisma , SGA and Kamermans , P . 1999 . Free-floating Ulva in the southwest Netherlands: species or morphotypes? A morphological, molecular and ecological comparison . Eur. J. Phycol. , 34 : 443 – 454 .
- Mathieson , AC and Hehre , EJ . 1986 . A synopsis of New Hampshire seaweeds . Rhodora , 88 : 1 – 139 .
- Mathieson , AC and Penniman , CA . 1986 . Species composition and seasonality of New England seaweeds along an open coastal-estuarine gradient . Bot. Mar. , 29 : 161 – 176 .
- Mathieson , AC , Pederson , JR , Neefus , CD , Dawes , CJ and Bray , TL . 2008 . Multiple assessments of introduced seaweeds in the Northwest Atlantic . ICES J. Mar. Sci. , 65 : 730 – 741 .
- Poole , LJ and Raven , JA . 1997 . The biology of Enteromorpha . Prog. Phycol. Res. , 12 : 1 – 148 .
- Rosenberg , G and Ramus , J . 1984 . Uptake of inorganic nitrogen and seaweed surface area: volume ratios . Aqua. Bot. , 19 : 65 – 72 .
- Sears , JR . 2002 . NEAS Keys to Benthic Marine Algae of the Northeastern Coast of North America from Long Island Sound to the Strait of Belle Isle, , 2nd , Fall River (MA) , , USA : Northeast Algal Society .
- Shimada , S , Hiraoka , M , Nabata , S , Iima , M and Masuda , M . 2003 . Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva . Phycol. Res. , 51 : 99 – 108 .
- Smith , VH . 2002 . Eutrophication of freshwater and coastal marine ecosystems–a global problem . Environ. Sci. Polut. Res. , 10 : 126 – 139 .
- Stegenga , H , Karremans , M and Simons , J . 2007 . Zeewieren van de voormalige oesterputten bij Yerseke . Gorteria , 32 : 125 – 142 .
- Tan , IH , Blomster , J , Hansen , G , Leskinen , E , Maggs , CA , Mann , DG , Sluiman , HJ and Stanhope , MJ . 1999 . Molecular phylogenetic evidence for a reversible morphogenetic switch controlling the gross morphology of two common genera of green seaweeds, Ulva and Enteromorpha . Mol. Biol. Evol. , 16 : 1011 – 1018 .
- Tanner, C. E. (1979). The Taxonomy and Morphological Variation of Distromatic Ulvaceous Algae (Chlorophyta) From the Northeast Pacific. PhD thesis. University of British Columbia, Vancouver, Canada.
- Tanner , CE . 1986 . Investigations of the taxonomy and morphological variation of Ulva (Chlorophyta): Ulva californica Wille . Phycologia , 25 : 510 – 520 .
- Taylor , RB , Jeroen , TA , Peek , AT and Rees , V . 1998 . Scaling of ammonium uptake by seaweeds to surface area:volume ratio: geographical variation and the role of uptake by passive diffusion . Mar. Ecol. Prog. Ser. , 169 : 143 – 148 .
- Titlyanov , EA , Geebova , NT and Kotlyarova , LS . 1975 . Seasonal changes in structure of the thalli of Ulva fenestrata P. et R . Ehkologiya , 6 : 320 – 324 .
- Valiela , IJ , McClelland , J , Hauxwell , J , Behr , PJ , Hersh , D and Foreman , K . 1997 . Macroalgal blooms in shallow estuaries: controls and ecophysiological and ecosystem consequences . Limnol. Oceanogr. , 42 : 1105 – 1118 .
- Verlaque , M . 2001 . Checklist of the macroalgae of Thau Lagoon (Herault, France), a hot spot of marine species introduction in Europe . Oceanol. Acta , 24 : 29 – 49 .
- Verlaque , M , Belsher , T and Deslous-Paoli , JM . 2002 . Morphology and reproduction of Asiatic Ulva pertusa (Ulvales, Chlorophyta) in Thau Lagoon (France, Mediterranean Sea) . Cryptogamie Algologie , 23 : 301 – 310 .
- Woolcott , GW and King , RJ . 1999 . Ulva and Enteromorpha (Ulvales, Ulvophyceae, Chlorophyta) in eastern Australia: comparison of morphological features and analysis of nuclear rDNA sequence data . Aust. Syst. Bot. , 12 : 709 – 725 .