Abstract
Twenty-five specimens of the freshwater red alga Compsopogon were collected from locations in North America, South America, Europe, Asia, Australasia and Oceania, and from an aquarium, with the goal of determining genetic diversity among specimens and ascertaining the number of phylogenetic species. Specimens were morphologically identified as having either the ‘caeruleus’ morphology, with regular polyhedral cortical cells, or the ‘leptoclados’ morphology, with irregular cortical cells with rhizoidal outgrowths. The ‘leptoclados’ morphology has been used by some researchers to distinguish the genus Compsopogonopsis from Compsopogon, or at least to distinguish C. leptoclados from other Compsopogon species. Sequence data for the rbcL gene and cox1 barcoding region were obtained for most specimens. In addition, SSU and partial LSU (barcode) rDNA were explored for a few specimens, but all sequences were identical. For the 25 newly generated and eight previously published rbcL gene data, there were seven unique haplotypes, but the sequence divergence was very low (≤7 bp, ≤ 0.7%). One haplotype was widespread, represented by 21 specimens from diverse locations in all regions sampled. Likewise, the 22 new and one previously published cox1 barcode region sequences yielded seven unique haplotypes with little sequence divergence (≤13 bp, ≤ 2.0%). One haplotype was widespread, being shared among 16 specimens from all regions. The combined molecular and morphological data showed no genetic differentiation between the ‘caeruleus’ and ‘leptoclados’ morphologies. The ubiquitous distribution of Compsopogon in tropical/subtropical regions and its low genetic variation are probably facilitated by the alga's ability to tolerate a wide range of stream conditions and its propagation via asexual spores. Given the findings of previous culture-based studies, morphometric research and field observations, coupled with the results of our study, we conclude there is only a single monospecific genus worldwide and that the species is correctly called C. caeruleus, since this is the oldest validly published name; all other previously described species of Compsopogon and Compsopogonopsis are synonyms.
Introduction
The freshwater red algal genus, Compsopogon, is a member of the order Compsopogonales within the Class Compsopogonophyceae (Yoon et al., Citation2006; Müller et al., Citation2010). This order used to include only two genera: Boldia, with a monostromatic and hollow sac-like thallus without pit plugs; and Compsopogon, with a uniaxial to corticated multistromatic and usually compact thallus, with simple pit plugs, this last most likely representing a primitive state (Scott et al., Citation1988). Recently, a new genus has been described – Pulvinaster – which has prostrate-adherent filaments or free filaments composed of cells containing one or more discoid to spiral bluish-green plastids without pyrenoids; this alga is euryhaline (West et al., Citation2007).
Compsopogon has a simple cylindrical thallus with large inner cells covered by smaller cortical cells; in larger, more mature thalli the inner cells may disintegrate, leaving only the outer cortex (Kumano, Citation2002). No sexual reproduction is known and asexual reproduction is most commonly by triangular to circular monosporangia cut off from the cortical cells. More rarely, microsporangia are produced in clusters (Patel & Francis, Citation1969; Necchi & Dip, Citation1992). Compsopogon has a primarily tropical and subtropical distribution, with a few reports from temperate regions (Sheath & Hambrook, Citation1990). Within these climatic zones it appears to be globally distributed, having been often reported from North America, South America, Europe, Asia and Pacific Islands (Krishnamurthy, Citation1962; Starmach, Citation1977; Sheath & Hambrook, 1990; Kumano, Citation2002).
Since the establishment of the genus by Montagne (1846–1869), who recognized a single species, there has been much taxonomic research and debate about the morphological characters used to define species. In 1850, Montagne described three additional species from French Guiana, while Krishnamurthy (1962) recognized six species of Compsopogon and transferred C. leptoclados to a new genus, Compsopogonopsis. This monospecific genus was distinguished from Compsopogon primarily by rhizoidal cells arising from the cortical cells (Krishnamurthy, 1962). A morphometric study of specimens from North America and type specimens of 10 species concluded there were indeed two genera but only three distinct species, Compsopogon caeruleus, C. prolificus and Compsopogonopsis leptoclados (Vis et al., Citation1992). A morphometric study of specimens from Brazil, on the other hand, recognized only one genus and two species, Compsopogon caeruleus and C. leptoclados (Necchi & Dip, 1992).
Considerable phenotypic plasticity has been demonstrated in both culture studies and field research. For example, in a culture study of C. caeruleus, the morphological variability observed spanned the taxonomic characteristics of five other described species (Nichols, Citation1964). A study on a Compsopogon isolate from India showed morphological variation in many of the characters used to define species and concluded that most of the earlier described species are probably ecophenes of a single species (Shyam & Sarma, Citation1980). A one-year phenological study of C. caeruleus also showed much variation in taxonomic characters, which could be related to variation in physical and chemical parameters (Necchi et al., Citation1990). In addition, morphometric characters were evaluated in specimens from a tropical drainage basin by Necchi & Pascoaloto (Citation1995) and were demonstrated to be widely variable and therefore of little taxonomic value for species identification. Necchi & Pascoaloto suggested the use of qualitative characters (mode of cortex formation) for species delineation in Compsopogonaceae. Clearly, the morphology within the genus is highly plastic with regard to characteristics previously and currently used to define species, which makes a morphological determination of the number of taxa within the genus difficult, if not impossible.
To date, few molecular data have been generated for Compsopogon, with only 10 sequences for the ITS region, nine sequences for the rbcL gene, and fewer for other gene regions in GenBank (accessed 24 April 2012). The most comprehensive study provided data for rbcL from four locations, 18S rDNA gene from five locations, and ITS from 10 locations in North America and Hawaii (Rintoul et al., Citation1999). The major conclusion was that Compsopogon and Compsopogonopsis are congeneric; specimens used in the study showed little genetic variation in the rbcL gene. The research suggested that the systematics of the genus would benefit from a more global sampling.
Most recent systematic studies have concluded there are only one to three species within the genus (Necchi & Dip, 1992; Vis et al., 1992; Rintoul et al., 1999). However, Kumano (2002) still recognized nine species. Given these uncertainties and the limited sampling of the previous phylogenetic study (Rintoul et al., 1999), the current study was undertaken to provide DNA data for four species-level molecular markers from throughout the range of this genus to determine the extent of genetic variation and to ascertain the number of phylogenetic species.
Table 1. Morphology type, collection information, GenBank accession numbers and previous papers for rbcL and cox1 sequences of Compsopogon specimens used in this study. New sequences in bold
Table 2. Percent difference (lower left matrix) and base pair changes (upper right matrix) among the seven unique rbcL haplotypes for Compsopogon specimens. Location codes as in
Table 3. Percent difference (lower left matrix) and base pair changes (upper right matrix) among the seven unique cox1 barcode haplotypes for Compsopogon specimens. Codes as in
Table 4. Physical and chemical parameters of collection locations. Codes as in
Figs 1. World map showing locations of the new specimens sequenced for this study (stars) and previously sequenced specimens from GenBank (circles). Location codes as in .
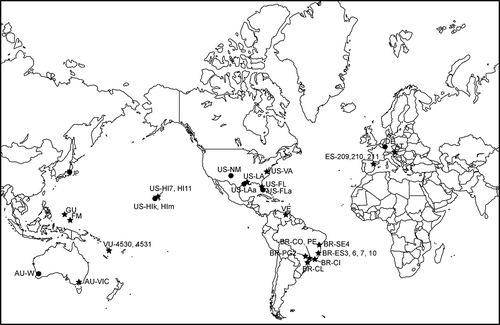
Figs 2. Maximum likelihood (ML) analysis tree (-Ln 11094.402668) showing the relationships within the Compsopogonales (with representatives of Compsopogon and Boldia) based on rbcL gene data, with the Erythropeltidales as the outgroup. Nodes with filled circles had statistical support ≥ 0.90 Bayesian posterior probability and 90 ML bootstrap values. Branches without values had statistical support < 0.70 posterior probability and 70 ML bootstrap. Haplotypes marked with * represent more than one sequence. Haplotypes AQUA and US-HI7 correspond to AF087115 and DQ308425, respectively: see and for information on haplotypes.
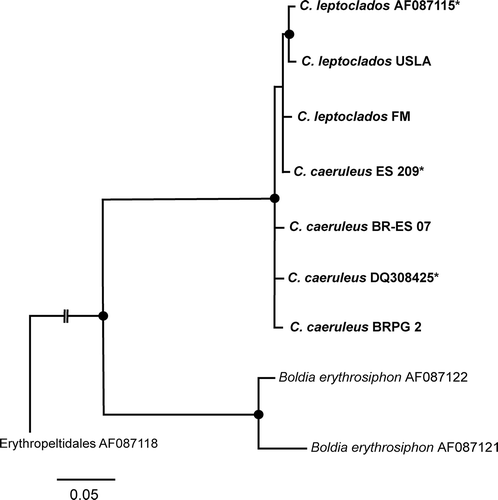
Figs 3–6. Compsopogon caeruleus: photomicrographs of important morphological features for both the ‘caeruleus’ and ‘leptoclados’ morphologies. Location codes as in Table 1. 3. Early cortication of ‘caeruleus’ morphology showing regular cortication. Specimen from BR-ES7. 4. Early cortication of ‘leptoclados’ morphology showing irregular cortication and rhizoidal cells (arrow-heads). Specimen from BR-CL. 5. Surface view of ‘caeruleus’ morphology showing regular (polygonal) cortical cells and monosporangia (arrowheads). Specimen from BR-ES3. 6. Surface view of ‘leptoclados’ morphology showing monosporangia (arrowheads), microsporangia (arrow) and rhizoidal cell (double arrowhead). Specimen from BR-CL. Scale bars = 50μm.
Necchi JR. Orlando
1Zoology and Botany Department, São Paulo State University, Rua Cristóvão Colombo, 2265-15054-000 São José do Rio Preto, SP, Brazil
Correspondence to: Orlando Necchi Jr. E-mail: [email protected]
Fo. Auro Silva Garcia
1Zoology and Botany Department, São Paulo State University, Rua Cristóvão Colombo, 2265-15054-000 São José do Rio Preto, SP, Brazil
Salomaki Eric D.
2Department of Environmental and Plant Biology, Ohio University, Porter Hall 315, Athens, OH 45701 USA
West John A.
3School of Botany, University of Melbourne, Parkville, Victoria 3010, Australia
Aboal Marina
4Laboratorio de Algología, Departamento de Biología Vegetal, Facultad de Biología, Universidad de Murcia, E-30100 Murcia, Spain
Vis Morgan L.
2Department of Environmental and Plant Biology, Ohio University, Porter Hall 315, Athens, OH 45701 USA
Materials and methods
Specimens were collected from 24 locations in North America, South America, Europe, Australia and Pacific Islands, as well as from an aquarium (). Morphological voucher specimens were either pressed on herbarium paper or preserved in 2.5% buffered glutaraldehyde and lodged at the herbaria BHO or SJRP (Thiers, Citation2012). Specimens for DNA analysis were cleaned of visible epiphytes and then either preserved in silica desiccant or cultured and later extracted.
Specimens were extracted for DNA by grinding a mortar and pestle with liquid nitrogen and using the NucleoSpin® extraction kit (Clontech, Mountainview, CA, USA) according to the manufacturer's protocols. A 1119 bp fragment of the plastid-encoded ribulose-1,5-bisphosphate carboxylase–oxygenase large-subunit gene (rbcL) gene was PCR amplified using the Comp1 and Comp2 primers (Rintoul et al., 1999). The 664 bp barcoding region near the 5′ end of the cox1 gene was PCR amplified using the GazF1 and GazR1 primers (Saunders, Citation2005). SSU rDNA and the LSU rDNA barcode (the D2–D3 region proposed as a secondary barcode for red algae by Saunders & McDevitt, Citation2012) were amplified using primers and cycles previously described (Milstein & Oliveira, Citation2005; Conklin et al., Citation2009). Three different amplification systems were used for PCR reactions for all markers as follows: (1) 25 μl AmpliTaqGold (Applied Biosystems, Carlsbad, CA, USA), 2.5 μl each primer, 17.5 μl dH2O, and 2.5 μl extracted DNA; (2) 25 μl Top Taq® Master Mix (Qiagen GmbH, Hilden, Germany), 2.0 μl each primer, 4.0 μl dH2O, and 2.0 μl extracted DNA; or (3) puReTaq® Ready-to-go PCR beads (GE HealthCare Life Sciences, Buckinghamshire, UK), 2.0 μl each primer, 19 μl dH2O, and 2.0 μl extracted DNA. The PCR protocols consisted of either (1) an initial denaturing at 95°C for 1 min followed by 35 cycles of 93°C for 30 s, 50°C for 30 s and 68°C for 1 min, and a final elongation period at 72°C for 10 min before holding at 4°C; or (2) an initial denaturing at 95ºC for 2 min followed by 35 cycles of 93ºC for 1 min, 47 ºC for 1 min and 72ºC for 4 min, and a final elongation period at 72ºC for 2 min before holding at 4ºC (Rintoul et al., 1999). The resulting PCR products of all markers were purified using the UltraClean™ PCR Clean-up DNA purification kit (Mo Bio, Carlsbad, CA, USA), Wizard SV Clean-up System (Promega, Madison, USA), or GFX® PCR (GE HealthCare Life Sciences, Buckinghamshire, UK), all according to manufacturer's protocols. The purified rbcL PCR products were sequenced using the amplification primers as well as the internal rbcL primers F650 and R897.comp (5′-GCTGGTAACTCAACATATTCTCG-3′) (Stewart & Vis, Citation2007), or F650 and R751 (5′-GCACGTTCGTACATATCTTCC-3′) (Rintoul et al., 1999). The PCR products from the cox1 barcode region were sequenced with both amplification primers. All sequencing was completed using the ABI PRISM Big DyeTM v3.1 Terminator Cycle Sequencing Ready Reaction Kit and the ABI PRISMTM 3130x1 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Sequence data were assembled using Sequencher™ version 4.10.1 (Gene Codes, Ann Arbor, MI, USA), Geneious Pro 5.5.6 (Drummond et al., Citation2012) or BioEdit version 6.0 (Hall, Citation1999). All new sequence data were submitted to GenBank (accession numbers in or in the Results section).
The internally transcribed spacer region (ITS) of the rRNA gene previously used by Rintoul et al. (1999) was evaluated as an additional nuclear marker. However, this region was determined not to be suitable because it was overly variable. For example, among the specimens examined in Rintoul et al. (1999), 55% of the bases in the final alignment were variable. In the present research, PCR amplification resulted in multiple PCR products, usually comprising more than six bands all of equal brightness.
Phylogenetic analyses of the rbcL data were conducted using Geneious (Drummond et al., 2012). The best-fit model of sequence evolution was determined using the Akaike Information Criterion (AIC) implemented in ModelTest 3.7 (Posada & Crandall, Citation1998), which is part of the PAUP*4.0 plugin for Geneious. Maximum likelihood (ML) topologies and bootstrap values from 1000 replicates were inferred using PhyML (Guindon & Gascuel, Citation2003) and Bayesian analysis (BS) was performed in MrBayes 3.1 (Huelsenbeck & Ronquist, Citation2001) with three runs of five chains of Metropolis coupled Markov Chain Monte Carlo for 4 × 106 generations. For these analyses the GTR + I model was determined to be the best-fit for the rbcL dataset with the following parameters: base frequencies A = 0.2898, C = 0.1685, G = 0.2242 and T = 0.3175, rate matrix A–C = 0.4672, A–G = 3.3152, A–T = 4.0077, C–G = 1.3739, C–T = 14.2762 and G–T = 1.0000, proportion of invariable sites = 0.5124.
Morphological characteristics of voucher specimens were examined using a Leica DM1000 microscope and photographed with a Leica DFC3200 digital camera (Leica Microsystems, Wetzlar, Germany). In particular, it was noted if the specimens had regular cortical cells corresponding to the caeruleus morphology or cells with rhizoidal outgrowths corresponding to the leptoclados morphology (). The presence of monosporangia and microsporangia was also recorded (Fig. 6).
Results
Molecular data
Twenty-five new rbcL, 22 cox1 barcode, seven SSU and five LSU sequences were generated for specimens from geographically distant locations (, ). The rbcL gene fragment was 1119 bp for all specimens with no indels among sequences. Likewise, the cox1 barcoding region was 664 bp with no indels.
For the rbcL gene region, the new sequences were compared with the nine previously published sequences in GenBank for Compsopogon caeruleus and synonymous taxa. Location information could be determined for eight of the accessions and are included in and . The location data (MT, Montana) for GenBank accession U04037 provided by Freshwater et al. (Citation1994) was an error and no location information is available for the culture isolate (LB 1553: http://web.biosci.utexas.edu/utex/algaeDetail.aspx?algaeID=3987) (D.W. Freshwater, personal communication). The alignment of the new and previously published sequences showed gaps in the aligned sequences of U04037, AF087120, AF460220. Since gaps are unlikely in the protein coding rbcL gene, these sequences were removed from the alignment and were not investigated further. Among the 25 new and six previously published sequences, there were seven unique haplotypes (). One haplotype was represented by 22 specimens, one by three, one by two, and four were unique to a single specimen from a single location. The sequence divergence and base pair changes among these unique haplotypes were very low (≤7 bp; ≤ 0.7%) ().
For the cox1 barcode region, there was one previously published sequence in GenBank from the Hawaiian Islands (, ). This sequence and the 22 new sequences were compared and resulted in 7 unique haplotypes (). One haplotype was shared by 16 specimens and one by two, and the other five were unique to a single specimen from a single location. Six of the haplotypes differed by only 1–3 bp (≤0.5%; ). The haplotype of US-LA differed from the others by 11–13 bp (1.7–2.0%). Based on the rbcL data, this result for US-LA seemed unusual and a new PCR reaction was therefore performed and the same sequence was obtained.
The seven new SSU sequences (JX511996–512002), as well as the five LSU barcode sequences (JX504701–504705) were all identical. The SSU sequences were 0–1 bp (0–0.1%) divergent from those in GenBank and reported in previous studies (AF087123–087128, Rintoul et al., 1999; AJ880416, Hoef-Emden et al., Citation2005). The LSU sequences were identical to the single sequence (HQ422322, C. caeruleus) in GenBank (Sherwood et al., Citation2010).
The rbcL data gave identical topologies from Bayesian and ML analyses (the ML tree is shown in ). There were two major clades representing the two genera (Compsopogon and Boldia), both with high support. However, only one of the internal nodes within Compsopogon clade had statistical support.
Correspondence between molecular and morphological data, and ecological characterization
Specimens from 12 locations had the ‘caeruleus’ morphology and eight locations had the ‘leptoclados’ morphology (). The ‘caeruleus’ morphology specimens had regular cortication consisting of polygonal cortical cells (Figs 3, 5). The ‘leptoclados’ morphology specimens had irregular cortication, consisting of cells with rhizoidal outgrowths covering other cells (Figs 4, 6). Monosporangia of specimens for both morphologies were oval to triangular and formed by uneven division of parental cell with an oblique wall formation (Figs 5, 6). Microsporangia produced in clusters were only observed in specimens from BR-CL (Fig. 6).
Combining the molecular data and morphological identification, there was no genetic differentiation between the ‘caeruleus’ and ‘leptoclados’ morphologies. Twelve specimens that were unequivocally identified as having the ‘caeruleus’ morphology and five with the ‘leptoclados’ morphology were identical in rbcL sequence (, , ). Likewise, seven ‘caeruleus’ morphology specimens and six ‘leptoclados’ specimens were identical in cox1 barcode region sequence (, ).
Temperature, pH and conductivity were measured at 18 sites (). Both pH and specific conductivity varied widely among locations, with pH ranging from slightly acidic to basic (pH 6.4–8.3) and conductivities corresponding to very low ion concentrations up to brackish waters (18–1766 µS.cm−1). The streams were warm (≥12.1 °C). Compsopogon specimens inhabited a wide range of ecoregions; for example, collections were made in Atlantic rain forest, Brazilian savannah and seasonal semi-deciduous forest in Brazil (T).
Discussion
Five molecular markers, SSU, LSU, rbcL, cox1 and ITS 1 and 2, all commonly employed in studies of red algae, were initially considered for inclusion in this research. However, only rbcL and cox1 proved to be suitable for examining relationships among all specimens. The SSU and LSU were quickly determined to show little or no variation in Compsopogon. Although ITS 1 and 2 have been employed previously to study Compsopogon species (Rintoul et al., 1999), our results and re-interpretation of previous results led us to the conclusion that this marker is not useful without considerably more research. In a reanalysis of the data from Rintoul et al. (1999) using MUSCLE and CLUSTALW through Geneious, no reliable alignment could be obtained, with sequences differing by over 50%. Therefore, it is likely that the ITS sequence data presented in Rintoul et al. (1999) may not represent homologous regions for phylogenetic comparison.
Compsopogon has a wide geographical range, being reported from North America, South America, Europe, Asia, Oceania and Australasia (Krishnamurthy, 1962; Starmach, 1977; Entwisle & Kraft, Citation1984; Necchi & Dip, 1992; Vis et al., 1992; Kumano, 2002). For our study, specimens were collected and analysed from the same regions, except Asia. Environmental data from previous research suggest that Compsopogon is not only geographically widespread, but also quite tolerant of a variety of stream conditions (Tomas et al., Citation1980; Yadava & Kumano, Citation1985; Sheath & Hambrook, 1990; Necchi & Dip, 1992; Vis et al., 1992; Necchi et al., 1999a, 1999b). For example, although it frequently occurs in alkaline waters (pH ≥ 7.5), it can also be found in more acidic streams (pH ≤ 6.5) (Tomas et al., 1980; Yadava & Kumano, 1985; Sheath & Hambrook, 1990; Necchi & Dip, 1992; Vis et al., 1992; Necchi et al., Citation1999a, Citation1999b), and Compsopogon has been reported from low ionic waters to near-brackish conditions (Tomas et al., 1980; Sheath & Hambrook, 1990; Vis et al., 1992). Similar tolerance to variation in pH and conductivity was found in the current study. Previous studies and the present research have recorded Compsopogon at water temperatures typically ≥ 10 °C, with most reports at temperatures ≥ 20 °C (Tomas et al., 1980; Yadava & Kumano, 1985; Sheath & Hambrook, 1990; Necchi & Dip, 1992; Vis et al., 1992; Necchi et al., 1999a, 1999b, this study). Fewer data are available for stream current velocity and nutrient content, but the limited information indicates that Compsopogon is found across wide ranges of these parameters as well (Tomas et al., 1980; Necchi & Dip, 1992; Vis et al., 1992; Necchi et al., 1999a, 1999b). This tolerance of a wide variety of stream conditions most likely contributes the ubiquitous distribution of Compsopogon in tropical–subtropical regions. Its preference for warm waters is underlined by the fact that two of the temperate locations (AT and US-VA) in this study were at the outflow of industrial waters with elevated temperature. A culture used in a previous molecular study was from a warm water factory outflow in Germany (Yoon et al., 2006), and Manny et al. (Citation1991) hypothesized that the Compsopogon they found growing at 21 m depth in the North American Great Lakes might have come from one of the many warm water discharges around Lake Huron.
It would therefore appear that Compsopogon disperses readily and over long distances, since the warm water outflows in the temperate zone are long distances from source populations in tropical and subtropical regions. To our knowledge, no empirical data on dispersal vectors have been published for Compsopogon, though several hypotheses have been put forward. Ship ballast water was suggested as a potential source of propagules for the Compsopogon in the Great Lakes (Manny et al., 1991). A recent discovery of Compsopogon on ‘extraordinarily isolated’ islands in Japan was attributed to transport by migratory birds (Kitayama, Citation2011). Others have suggested aquaria as sources (Rintoul et al., 1999; Sheath & Sherwood, 2002, and references therein), since Compsopogon is known as ‘Staghorn algae’ in the freshwater aquarium trade and there are numerous webpages devoted to its control and removal (e.g. http://www.aquahobby.com/articles/e_freshwater_algae.php); as well as indicating possible routes of dispersal, the nuisance caused by Compsopogon gives further support to the idea that it readily acclimates to new environments and grows quickly.
The discovery of Rintoul et al. (1999) that there is little genetic variation among specimens of Compsopogon from geographically distant locations and the extension of those findings by our study to pantropical locations strongly suggest that it is probably not a single vector that accounts for its wide distribution. In addition, the biology of this alga most likely plays a significant role in its global distribution. Compsopogon is known to reproduce predominantly by asexual monospores and occasionally microsporangia (Kumano, 2002). Furthermore, it has a very wide ecological niche (Tomas et al., 1980; Sheath & Hambrook, 1990; Necchi & Dip, 1992; Vis et al., 1992; Necchi et al., 1999b). It seems likely that the lack of sexual reproduction and the propagation via asexual spores coupled with its ecological plasticity has greatly contributed to the pattern seen in this study, of very low genetic diversity over a large geographical range.
The lack of genetic variation in Compsopogon, despite its morphological variability, contrasts with some other, similarly widespread red algal taxa, in which molecular studies have revealed numerous discrete lineages (e.g. Lindstrom et al., Citation2011; Zuccarello et al., Citation2011; Clarkston & Saunders, Citation2012). One Compsopogon specimen (US-LA) stood out from the others, differing from them in cox1 (1.7–2.0%), though not in rbcL (≤0.7%). Apart from this single specimen, the lack of genetic variation in two species level markers among specimens from five continents and several oceanic islands is noteworthy. Compsopogon therefore appears to be somewhat unusual among red algae. However, there are other examples, such as Stylonema alsidii, another asexual red alga with a uniform morphology, which shows low genetic divergence worldwide (Zuccarello et al., Citation2008).
Previous culture-based studies, morphometric research and field observations have questioned the value of morphological characters used to define species of Compsopogon and distinguish the genus Compsopogonopsis (Nichols, 1964; Shyam & Sarma, 1980; Necchi et al., 1990; Necchi & Dip, 1992; Vis et al., 1992). Consequently, it has been suggested that the reported variation in morphology might reflect the existence of just one to three species. Other researchers have also concluded that most of the earlier described species are probably ecophenes of a single species (Shyam & Sarma, 1980). However, these previous studies on field and culture material, and also the present study, have not determined which conditions lead to the formation of one morphology or another. This is a matter that deserves attention in further studies. We sampled specimens with regular cortication (‘caeruleus’ morphology) and specimens with rhizoidal cortication (‘leptoclados’ morphology) from locations worldwide and observed little to no correlated genetic variation. Therefore, we support the conclusion of Rintoul et al. (1999) based on DNA data that there is a single monospecific genus in North America, but we extend this finding worldwide: there is only a single species of Compsopogon, for which the correct name is C. caeruleus.
Synonymy
Compsopogon caeruleus (Balbis ex C. Agardh) Montagne (1846), Flore d'Algérie Cryptogamie, première partie: 154.
HETEROTYPIC SYNONYMS (in addition to those reported in previous studies, particularly Necchi & Dip, 1992; Vis et al., 1992, Rintoul et al., 1999; Sheath & Sherwood, 2002):
C. aegyptiacus Aleem (1981), Bulletin of the Faculty of Science King Abdul Aziz University, 5: 60.
C. minutus C.C. Jao (1941), Sinensia, 12: 249, pl. 1,–8.
C. prolificus Yadava & Kumano, Japanese Journal of Phycology, 33: 19, –7 (1985)
C. sparsus S.L. Xie & Y.J. Ling (Citation1998), Acta Phytotaxonomica Sinica, 36: 81, pl. 2, –9.
C. tenellus Y.J. Ling & S.L. Xie (1998), Acta Phytotaxonomica Sinica, 36: 81, pl. 1, –7.
Acknowledgements
The collectors of specimens listed in are gratefully acknowledged. We acknowledge funding from the National Science Foundation (USA) grant number DEB0235676; FAPESP (Brazil) grants 2007/51270-7, 2008/00708-5; and CNPq grant 303952/2009-1.
References
- Ab'Saber , A.N . 2006 . Ecossistemas do Brasil , São Paulo : Editora Metalivros .
- Clarkston , B.E. and Saunders , G.W . 2012 . An examination of the red algal genus Pugetia (Kallymeniaceae, Gigartinales), with descriptions of Salishia firma gen. & comb. nov., Pugetia cryptica sp. nov. and Beringia wynnei sp . nov. Phycologia , 51 : 33 – 61 .
- Conklin , K.Y. , Kurihara , A. and Sherwood , A.R . 2009 . A molecular method for identification of the morphologically plastic invasive algal species Eucheuma denticulatum and Kappaphycus spp. (Rhodophyta, Gigartinales) in Hawaii . Journal of Applied Phycology , 21 : 691 – 699 .
- Drummond, A.J., Aston, B., Buxton, S., Cheung, M., Cooper, A. Heled, J., Kearse, M., Moir, R., Stones-Havas, S., Sturrock, S., Thierer, T. Wilson, A. (2012). Geneious v5.6. http://www.geneious.com (http://www.geneious.com)
- Entwisle , T.J. and Kraft , G.T . 1984 . Survey of red algae (Rhodophyta) of south-eastern Australia . Australian Journal of Marine and Freshwater Research , 35 : 213 – 259 .
- Freshwater , D.W. , Fredericq , S. , Butler , B.S. and Hommersand , M.H . 1994 . A gene phylogeny of the red algae (Rhodophyta) based on plastid rbcL . Proceedings of the National Academy of Sciences of the United States of America , 91 : 7281 – 7285 .
- Guindon , S. and Gascuel , O . 2003 . A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood . Systematic Biology , 52 : 696 – 704 .
- Hall, T.A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95–98. http://www.mbio.ncsu.edu/BioEdit/bioedit.html (http://www.mbio.ncsu.edu/BioEdit/bioedit.html)
- Hoef-Emden , K. , Shrestha , R.P. , Lapidot , M. , Weinstein , Y. , Melkonian , M. and Arad , S.M . 2005 . Actin phylogeny and intron distribution in bangiophyte red algae (Rhodoplantae) . Journal of Molecular Evolution , 61 : 360 – 371 .
- Huelsenbeck , J. P. and Ronquist , F . 2001 . MrBayes: Bayesian inference of phylogenetic trees . Bioinformatics , 17 : 754 – 755 .
- Kitayama , T . 2011 . First record of Compsopogon caeruleus (Balbis ex C. Agardh) Montagne (Compsopogonophyceae, Rhodophyta) from Ogasawara Islands, Japan . Bulletin of the National Museum of Natural Sciences, Series B , 37 : 169 – 174 .
- Krishnamurthy , V . 1962 . The morphology and taxonomy of the genus Compsopogon Montagne . Journal of the Linnean Society (Botany) , 58 : 207 – 222 .
- Kumano , S . 2002 . Freshwater Red Algae of the World , Bristol : Biopress .
- Lindstrom , S.C. , Hughey , J.R. and Martone , P.T . 2011 . New, resurrected and redefined species of Mastocarpus (Phyllophoraceae, Rhodophyta) from the northeast Pacific . Phycologia , 50 : 661 – 683 .
- Manny , B.A. , Edsall , T.A. and Wujek , D.E . 1991 . Compsopogon cf. coeruleus, a benthic red alga (Rhodophyta) new to the Laurentian Great Lakes . Canadian Journal of Botany , 69 : 1237 – 1240 .
- Milstein , D. and Oliveira , M.C . 2005 . Molecular phylogeny of Bangiales (Rhodophyta) based on small subunit rDNA sequencing: emphasis on Brazilian . Porphyra species. Phycologia , 44 : 212 – 221 .
- Montagne, J.F.M. C. (1846–1869). Algae. In Flore d'Algérie Cryptogamie, première partie. (By [M.C.] Durieu de Maisonneuve, [J.F.C.] Montagne, [J.B.G.M.] Bory de Saint-Vincent, L.-R. Tulasne, C. Tulasne & [J.H.] Lèveillè.) Imprimerie Impériale.
- Montagne , J.F.M.C . 1850 . Cryptogamia Guyanensis . Annales des Sciences Naturelles, Serie 3 , 14 : 284 – 309 . Algae
- Müller , K.M. , Lynch , M.D. and Sheath , R.G . 2010 . “ Bangiophytes: from one class to six; where do we go from here? ” . In Red Algae in the Genomic Age , Edited by: Seckbach , J. and Chapman , D.J. 243 – 259 . Dordrecht : Springer .
- Necchi , O., and Dip , M.R . 1992 . The family Compsopogonaceae (Rhodophyta) in Brazil . Algological Studies , 66 : 105 – 118 .
- Necchi , O., and Pascoaloto , D . 1995 . Morphometry of Compsopogon coeruleus (Compsopogonaceae, Rhodophyta) populations in a tropical river basin of southeastern Brazil . Algological Studies , 76 : 61 – 73 .
- Necchi , O., , Góes , R.M. and Dip , M.R . 1990 . Phenology of Compsopogon coeruleus (Balbis) Montagne (Compsopogonaceae, Rhodophyta) and evaluation of taxonomic characters of the genus . Japanese Journal of Phycology , 38 : 1 – 10 .
- Necchi , O., , Branco , C.C.Z. and Branco , L.H.Z . 1999a . Distribution of Rhodophyta in streams from São Paulo state, southeastern Brazil . Archiv für Hydrobiologie , 147 : 73 – 89 .
- O , Necchi , C.C.Z , Branco and R.R.V , Gomes . 1999 . Microhabitat and plant structure of Compsopogon coeruleus (Comsopogonaceae, Rhodophyta) populations in streams from São Paulo State, southeastern Brazil . Cryptogamie, Algologie , 20 : 75 – 87 . b
- Nichols , H.W . 1964 . Culture and developmental morphology of Compsopogon coeruleus . American Journal of Botany , 51 : 180 – 188 .
- Olson , D.M. , Dinerstein , E. , Wikramanayake , E.D. , Burgess , N.D. , Powell , G.V.N. , Underwood , E.C. , D'Amico , J.A. , Itoua , I. , Strand , H.E. , Morrison , J.C. , Lucks , C.J. , Allnutt , T.F. , Ricketts , T.H. , Kura , Y. , Lamoreux , J.F. , Wettengel , W.W. , Hedao , P. and Kassem , K.R . 2001 . Terrestrial ecoregions of the world: a new map of life on Earth . Bioscience , 51 : 934 – 938 .
- Omernik , J.M . 1987 . Ecoregions of the conterminous United States. Map (scale 1:7,500,000) . Annals of the Association of American Geographers , 77 : 118 – 125 .
- Patel , R.J. and Francis , M.A . 1969 . Some interesting observations on Compsopogon aeruginosus (J. Ag.) Kützing, a species new to India . Phykos , 8 : 46 – 51 .
- Posada , D. and Crandall , K.A . 1998 . MODELTEST: testing the model of DNA substitution . Bioinformatics , 14 : 817 – 818 .
- Rintoul , T.L. , Sheath , R.G. and Vis , M.L . 1999 . Systematics and biogeography of the Compsopogonales (Rhodophyta) with emphasis on the freshwater families in North America . Phycologia , 38 : 517 – 527 .
- Saunders , G.W . 2005 . Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future application . Philosophical Transactions of the Royal Society, , B 360 : 1879 – 1888 .
- Saunders , G.W. and McDevitt , D.C . 2012 . “ Methods for DNA barcoding photosynthetic protists emphasizing the macroalgae and diatoms ” . In DNA Barcodes: Methods and Protocols , Edited by: Kress , W.J. and Erickson , D.L. 207 – 222 . New York : Springer Science .
- Scott , J. , Thomas , J. and Saunders , B . 1988 . Primary pit connections in Compsopogon coeruleus (Balbis) Montagne (Compsopogonales, Rhodophyta) . Phycologia , 27 : 327 – 333 .
- Sheath , R.G. and Hambrook , J.A . 1990 . “ Freshwater ecology ” . In Biology of the Red Algae , Edited by: Cole , K.M. and Sheath , R.G. 423 – 453 . Cambridge : Cambridge University Press .
- Sheath , R.G. and Sherwood , A.R . 2002 . “ Phylum Rhodophyta (red algae) ” . In The Freshwater Algal Flora of the British Isles , Edited by: John , D.M. , Whitton , B.A. and Brook , A.J. 123 – 143 . Cambridge : Cambridge University Press .
- Sherwood , A.R. , Kurihara , A. , Conklin , K.Y. , Sauvage , T. and Presting , G.G . 2010 . The Hawaiian Rhodophyta Biodiversity Survey (2006–2010): a summary of principal findings . BMC Plant Biology , 10 : 258
- Shyam , R. and Sarma , Y.S.R.K . 1980 . Cultural observations on the morphology, reproduction and cytology of a freshwater red alga Compsopogon Mont. from India . Nova Hedwigia , 32 : 745 – 765 .
- Starmach , K . 1977 . Flora Słodkowodna Polski , vol. 14 , Warsaw and Krakow : Polska Akademia Nawuk Instytut Botaniki .
- Stewart , S.A. and Vis , M.L . 2007 . Investigation of two species complexes in Batrachospermum section Batrachospermum (Batrachospermales, Rhodophyta) . Phycologia , 46 : 380 – 385 .
- Thiers, B. (2012). Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York Botanical Garden's Virtual Herbarium. http://sweetgum.nybg.org/ih/ (http://sweetgum.nybg.org/ih/)
- Tomas , X. , Lopez , P. , Margalef-Mir , R. and Comin , F.A . 1980 . Distribution and ecology of Compsopogon coeruleus (Balbis) Montagne (Rhodophyta, Bangiophycideae) in eastern Spain . Cryptogamie, Algologie , 1 : 179 – 186 .
- Toro, M., Robles, S., Tejero, I., Prat, N., Solá, C. & Beltrán, D. (2009). 32. Aguas continentales corrientes. Ecosistemas lóticos. In VVAA. Bases Ecológicas preliminares para la conservación de los tipos de hábitat de interés comunitario en España, 1–134. Dirección General del Medio Natural, Política Forestal, Ministerio de Medio Ambiente y medio Rural y Marino, Madrid.
- Vis , M.L. , Sheath , R.G. and Cole , K.M . 1992 . Systematics of the freshwater red algal family Compsopogonaceae in North America . Phycologia , 31 : 564 – 575 .
- West , J.A. , Zuccarello , G.C. , Scott , J. , Pickett-Heaps , J. and Kim , G.H . 2005 . Observations on Purpureofilum apyrenoidigerum gen. et sp. nov. from Australia and Bangiopsis subsimplex from India (Stylonematales, Bangiophyceae, Rhodophyta) . Phycological Research , 53 : 49 – 66 .
- West , J.A. , Zuccarello , G.C. , Scott , J.A. , West , K.A. and Goer , S.L . 2007 . Pulvinus veneticus gen. et sp. nov. (Compsopogonales, Rhodophyta) from Vanuatu . Phycologia , 46 : 237 – 246 .
- Xie , S.-L. and Ling , Y.-J . 1998 . Two new species of Compsopogon (Rhodophyta) from Shanxi and Guangxi, China . Acta Phytotaxonomica Sinica , 36 : 81 – 83 .
- Yadava , R.N. and Kumano , S . 1985 . Compsopogon prolificus sp. nov. (Compsopogonaceae, Rhodophyta) from Allahabad, Uttar Pradesh in India . Japanese Journal of Phycology , 33 : 13 – 20 .
- Yoon , H.S. , Müller , K.M. , Sheath , R.G. , Ott , F.D. and Bhattacharya , D . 2006 . The major lineages of red algae (Rhodophyta) . Journal of Phycology , 42 : 482 – 492 .
- Ziegler , A.C. 2002 . University of Hawaii Press, Honolulu , Hawaiian Natural History, Ecology, and Evolution
- Zuccarello , G.C. , West , J.A. and Kikuchi , N . 2008 . Phylogenetic relationships within the Stylonematales (Stylonematophyceae, Rhodophyta): biogeographic patterns do not apply to Stylonema alsidii . Journal of Phycology , 44 : 384 – 393 .
- Zuccarello , G.C. , Yoon , H.S. , Kim , H.J. , Sun , L. , Loiseaux de Goër , S. and West , J.A . 2011 . Molecular phylogeny of the upright Erythropeltidales (Compsopogonophyceae, Rhodophyta): multiple cryptic lineages of Erythrotrichia carnea . Journal of Phycology , 47 : 627 – 637 .