Abstract
In past decades, environmental nitrogen fixation has been attributed almost exclusively to the action of enzymes in the well-studied molybdenum-dependent nitrogen fixation system. However, recent evidence has shown that nitrogen fixation by alternative pathways may be more frequent than previously suspected. In this study, the nitrogen fixation systems employed by lichen-symbiotic cyanobacteria were examined to determine whether their diazotrophy can be attributed, in part, to an alternative pathway. The mining of metagenomic data (generated through pyrosequencing) and PCR assays were used to determine which nitrogen-fixation systems are present in cyanobacteria from the genus Nostoc associated with four samples from different geographical regions, representing different lichen-forming fungal species in the genus Peltigera. A metatranscriptomic sequence library from an additional specimen was examined to determine which genes associated with N2 fixation are transcriptionally expressed. Results indicated that both the standard molybdenum-dependent system and an alternative vanadium-dependent system are present and actively transcribed in the lichen symbiosis. This study shows for the first time that an alternative system is utilized by cyanobacteria associated with fungi. The ability of lichen-associated cyanobacteria to switch between pathways could allow them to colonize a wider array of environments, including habitats characterized by low temperature and trace metal (e.g. molybdenum) availability. We discuss the implications of these findings for environmental studies that incorporate acetylene-reduction assay data.
Introduction
A lichen has traditionally been defined as a specific structure formed by a fungus (mycobiont) that develops a symbiotic relationship with a green alga and/or a cyanobacterium (photobiont). More recently, lichens have come to be viewed as complex ecosystems, hosting diverse microbial lineages from across the tree of life (Arnold et al., Citation2009; Hodkinson & Lutzoni, Citation2009; Bates et al., Citation2011; Cardinale et al., Citation2012; Hodkinson et al., Citation2012b). While evidence suggests that certain bacterial and fungal lineages may have strong preferences for living in association with lichens, as members of the lichen microbiome (Arnold et al., Citation2009; Hodkinson & Lutzoni, Citation2009; U’Ren et al., Citation2010, Citation2012; Bates et al., Citation2011; Schneider et al., Citation2011; Hodkinson et al., Citation2012b), the relationship between the two main associates (mycobiont and photobiont) is usually considered to be unique because the partners appear to be adapted to the symbiotic lifestyle (Ahmadjian, Citation1993) and comprise the bulk of the lichen biomass. Cyanolichens – those lichens with heterocystous cyanobacterial photobionts – are special among lichen symbioses because the photobiont fixes nitrogen (N2). Cyanolichens contribute significant levels of fixed nitrogen to ecosystems worldwide (Crittenden & Kershaw Citation1978; Gunther, Citation1989; Nash, Citation2008a), making them key for understanding global nutrient cycling and terrestrial ecology.
The most common type of cyanobacterial lichen photobiont is Nostoc (Nostocaceae) (Antoine, Citation2004; Nash, Citation2008a), which is the symbiont associated with members of the lichen-forming fungal genus Peltigera (Tschermak-Woess, Citation1988; Miadlikowska & Lutzoni, Citation2004; O’Brien et al., Citation2005; Hodkinson et al., Citation2012a), the group that is the focus of our research (). Nitrogen fixation by lichen-associated Nostoc has been attributed solely to the enzyme complex encoded by the nifHDK operon (Brigle et al., Citation1985; Thiel et al., Citation2002). The two components of this enzyme complex are an iron (Fe) protein (dinitrogenase reductase 1, encoded by nifH) and a molybdenum–iron (Mo–Fe) protein (dinitrogenase 1, for which nifD and nifK encode the α- and β-subunits, respectively) (Gillum et al., Citation1977; Hausinger & Howard, Citation1983; Brigle et al., Citation1985; Georgiadis et al., Citation1992).
Fig. 1. A cyanolichen thallus formed by the lichenized fungus (mycobiont) Peltigera rufescens with a cyanobacterial photobiont from the genus Nostoc.

Studies of the cyanobacterial genus Anabaena (a member of the same family as Nostoc, Nostocaceae) revealed that certain strains, all of which appear to be either obligate or facultative symbionts with plants, have an additional nitrogenase complex that is encoded by the vnf gene cluster (Thiel, Citation1993; Zehr et al., Citation2003; Raymond et al., Citation2004; Boison et al., Citation2006). The presence of the latter has also been inferred in a single strain of the cyanobacterial genus Nostoc (PCC 7422, isolated from plant tissue of Cycas sp.; Nostocaceae) through Southern hybridization, growth on Mo-free media, and ethane formation from acetylene (Masukawa et al., Citation2009); however, neither the presence of vnf genes in the strain nor the phylogenetic placement of the strain within Nostoc has been confirmed using sequence data. One of the two components of the vnf-encoded complex is an iron (Fe) protein (dinitrogenase reductase 2, encoded by vnfH) that is essentially identical in sequence to the product of nifH, and is cross-functional with it as an electron donor in Anabaena variabilis (Pratte et al., Citation2006). The other component is a vanadium–iron (V–Fe) protein (dinitrogenase 2, for which vnfDG encodes the fused α-/δ-subunit and vnfK encodes the β-subunit) (Robson et al., Citation1986, Citation1989; Joerger et al., Citation1990; Zehr et al., Citation2003; Raymond et al., Citation2004; Boison et al., Citation2006). In A. variabilis, the vnfEN genes are also essential for N2 fixation (functioning as a scaffold for catalytic cluster formation), although these genes are apparently absent in many strains of bacteria that utilize this system (e.g. Azotobacter spp.) (Thiel, Citation1996). A third system encoded by the anfHDGK operon (which is dependent upon Fe only and expressed under both Mo- and V-limited conditions) is known from lineages outside of the phylum Cyanobacteria (Bishop & Joerger, Citation1990).
The three major enzyme complexes for N2 fixation can be viewed in terms of a cascade of efficiency, with the Mo-dependent (nif-encoded) complex being most efficient, followed by the V-dependent (vnf-encoded) complex, and finally the Fe-dependent (anf-encoded) complex (Robson et al., Citation1986, Citation1989; Hales et al., Citation1986a, Citation1986b; Chisnell et al., Citation1988; Eady, Citation1989, Citation2003; Eady et al., Citation1988; Joerger et al., Citation1990; Walmsley & Kennedy, Citation1991; Raina et al., Citation1993; Bellenger et al., Citation2011). Paradoxically, Mo is the least abundant of the three crucial biometals in the continent crust, whereas V is approximately two orders of magnitude more abundant, and Fe is by far the most abundant of the three (Erickson, Citation1973; Wedepohl, Citation1995). Under special conditions, such as when Mo is limiting in particular microenvironments, N2 fixation via the alternative pathways may be favoured (Silvester, Citation1989; Barron et al., Citation2009; Boyd et al. Citation2011; Wurzburger et al., Citation2012). It is also known that the V-dependent nitrogenase is more efficient than the Mo-dependent nitrogenase at low temperatures (Miller & Eady, Citation1988), and that, while the V-dependent nitrogenase is typically considered to be repressed by Mo (based primarily on studies performed at room temperature), this enzyme is not documented to be regulated by Mo at any temperature under 14°C (Walmsley & Kennedy, Citation1991).
While alternative N2 fixation has been considered rare, Betancourt et al. (Citation2008) demonstrated that diazotrophs using these systems can be isolated from diverse natural environments. The purpose of this study was to determine whether the V-dependent N2 fixation pathway is used by strains of Nostoc associated with cyanolichens from the genus Peltigera. To accomplish this, we used a sequence-based approach: (1) probing metagenomic libraries from three Peltigera samples for sequences representing vnf genes, (2) using PCR and Sanger sequencing both to supplement the lowest coverage metagenomic library and to determine the presence/absence of vnf genes in an additional sample, and (3) probing a metatranscriptomic sequence library from an additional sample to examine whether RNA is transcribed from the vnf operon.
Materials and methods
Metagenomic analyses
To determine the presence or absence of cyanobacterial vnf genes in Peltigera lichen thalli, metagenomes from three different samples were examined (). A metagenomic sequence library was constructed from a P. dolichorhiza thallus (extract: DNA-2289) using Roche 454 sequencing as described by Magain et al. (Citation2010). Peltigera malacea and P. membranacea metagenomic assemblies were constructed from Roche 454 sequence reads as described by Xavier et al. (Citation2012). The nucleotide sequence of a full-length cyanobacterial vnf operon was retrieved from the NCBI nr/nt collection (A. variabilis ATCC 29413; GenBank accession: CP000117, GI: 75699950; bases 5009717 to 5019000 were extracted in FASTA format) and a BLASTn search was conducted using BLAST+ 2.2.24 against the metagenomic sequence libraries (Altschul et al., Citation1997), which had been formatted as BLAST databases using the mpiformatdb command in MPIBLAST 1.6.0. Sequences in the metagenomes that returned results with bitscores above 65 and E-values below 1.00E-07 were identified as putative vnf sequences (see sequences archived in GenBank for final bitscores and E-values through online BLAST).
Table 1. Nostoc-containing cyanolichen samples analysed as part of this study. ‘Data-type’ gives the type of sequence data generated: MG = Roche 454-based shotgun metagenome; MT = Roche 454-based metatranscriptome; PCR = PCR-amplified, Sanger-sequenced set of vnf gene fragments.
PCR for vnf genes
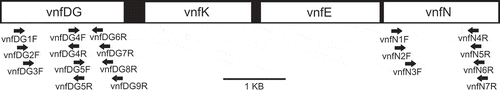
PCR and Sanger sequencing were performed on two Peltigera samples (1) to obtain longer sequences from the vnf operon in the sample of a P. dolichorhiza thallus discussed above, and (2) to determine whether sequences encoding vnf genes were present in one additional Peltigera sample representing P. neopolydactyla (and, if so, to obtain representative sequences of vnfDG and vnfN from this sample). To design primers for the amplification of cyanobacterial vnf genes, Anabaena vnf operon sequences from GenBank (GI: 75699950, 37694441, 83034994 and 37694450) were aligned with fragments that showed high sequence similarity (determined using the BLASTn criteria defined above under ‘Metagenomic analyses’) from both the P. dolichorhiza metagenome discussed above and a P. praetextata metatranscriptome (Hodkinson, Citation2011; ; see below, ‘Metatranscriptomic analyses’). Regions in which sequences were conserved between the GenBank reference sequences and at least one metagenomic or metatranscriptomic sequence fragment were targeted for primer design.
For PCR reactions to amplify fragments of vnfDG, primers vnfDG1F, 2F and 3F were combined with all six reverse vnfDG primers (; ); additionally, vnfDG4F and 5F were combined with vnfDG9R (giving a total of 20 primer combinations for the vnfDG region). For vnfN amplification, all forward primers were combined with all reverse primers (a total of 12 possible combinations for the vnfN region). PCR conditions and thermocycler settings followed Hodkinson & Lutzoni (Citation2009). Two Peltigera DNA extracts were analysed: DNA-2289, also used for generating the P. dolichorhiza metagenomic dataset described in detail by Magain et al. (Citation2010), and DNA-2308, extracted from a sample of P. neopolydactyla using a modified CTAB extraction protocol (). Sequencing reactions were carried out with ABI Big Dye terminator v1.1 cycle sequencing kit (Applied Biosystems, Foster City, California, USA) diluted to 1/64th reactions using BDX64 BigDye enhancing buffer (MCLAB, South San Francisco, California, USA) according to standard BDX64 protocols with the same primers used for amplification; electrophoresis was performed using an ABI 3130xl Genetic Analyzer (Applied Biosystems) and sequence reads were assembled using Sequencher 4.9 (Gene Codes Corporation, Ann Arbor, Michigan, USA).
Table 2. Cyanobacterial vnf primers designed for this study.
Metatranscriptomic analyses
To determine whether transcription of genes from the vnf cluster could be detected in a sample of Peltigera, we probed a metatranscriptomic sequence library of a single sample from a lichen thallus dominated by the fungus P. praetextata and the cyanobacterium Nostoc sp. (). Sample collection, permanent voucher storage, nucleic acid extraction, rRNA subtraction, cDNA preparation and Roche 454 GS FLX sequencing were performed as outlined by Hodkinson (Citation2011). Sequence data were uploaded for overall assessment of metatranscriptome content to the MG-RAST server (http://metagenomics.nmpdr.org/; Meyer et al., Citation2008) and the dataset was made public under project 3216, metagenome 4453629.3. Transcripts identified by MG-RAST as being derived from genes involved in N2 fixation were downloaded and MegaBLAST searches were performed against the NCBI nr/nt database to confirm their taxonomic affinities (Zhang et al., Citation2000). Additionally, BLASTn searches for N-fixation genes were conducted on the entire metatranscriptomic dataset as previously outlined above (in ‘Metagenomic analyses’) using the metatranscriptomic sequence file as a BLAST database to identify transcripts potentially derived from N2 fixation genes that were not detected by MG-RAST.
Phylogenetic inference
To determine the affinities of the vnf sequences found in lichen thalli, a phylogeny was inferred using a vnfD sequence dataset assembled from the following: (1) composite sequences made of sequence fragments from PCR and metagenomic libraries that showed high sequence similarity to the vnfD-like portion of the vnfDG-encoding sequence in A. variabilis (determined using BLASTn criteria defined above); (2) the top BLASTn hits for the P. membranacea vnfD-like portion of the vnfDG-encoding sequence in the NCBI nr/nt database (‘maximum bitscore’ of 100+); and (3) the list of sequences found by searching for ‘vnfD’ in the NCBI nucleotide database in September 2012. Sequences from the second and third categories were downloaded from GenBank in FASTA format and were processed using a PERL script written to rename the sequences for easy downstream processing (‘genbank_fasta_renamer.pl,’ available here as Supplementary file 3). When multiple GenBank records existed for the same strain, only the most complete sequence was retained; sequences with significant unalignable portions within the vnfD region were likewise removed. Sequences were aligned by hand and ambiguously aligned regions were designated as exclusion sets using Mesquite 2.74 (Maddison & Maddison, Citation2010). The full sequence vnfD alignment file is available in NEXUS format in Supplementary File 1. To prepare the alignment for analyses with RAxML 7.2.6 (Stamatakis, Citation2006), characters in exclusion sets were manually deleted and the alignment was exported in extended PHYLIP format. Maximum likelihood phylogenetic analyses were performed with three partitions for the three different codon positions. A set of 500 bootstrap replicates was run (Pattengale et al., Citation2009), followed by a thorough topology search with the ‘-f a’ function in RAxML, using a GTRGAMMA model. A general-time-reversible (GTR) model was selected because its use was not prohibited by time or computing resources. Simplified models (that can be viewed as special cases of GTR) have often been favoured in cases where it can be demonstrated that inferences would not suffer significantly from their use (based on LRT or AIC) (Posada & Crandall, Citation1998); however, given the speed provided by RAxML, there was no need in this case to simplify the model. Since the use of a gamma distribution is the best way to account for among-site rate heterogeneity (Mayrose et al., Citation2005), GTRGAMMA was the logical choice for a model. While some researchers have attempted to take into account the proportion of invariant sites while using gamma, we did not consider it appropriate, as it creates a strong correlation created between I and α, which could bias results (Yang, Citation1993; Gu et al., Citation1995; Sullivan et al., Citation1999; Minin et al., Citation2003; Mayrose et al., Citation2005; Ren et al., Citation2005; Yang, Citation2006). The procedures outlined above for vnfD were used to analyse sequences from the vnfN region (the alignment is given in Supplementary File 2), with the following differences in dataset assembly: (1) in addition to sequence data from PCR and metagenomic libraries, data from the P. praetextata metatranscriptomic sequence library were also included (this consisted of a single vnfN sequence fragment); (2) the P. membranacea vnfN sequence fragment was used for obtaining top BLASTn hits from the NCBI nr/nt database (as above, a ‘maximum bitscore’ of 100+ was used); and (3) the term ‘vnfN’ was used to query the NCBI nucleotide database in September 2012.
Sequences from the vnf gene cluster in Nostoc were deposited in GenBank under accession numbers KF662359–KF662370 (see Table S1).
Results
For the Peltigera dolichorhiza sample, DNA-2289, PCR with Sanger sequencing and Roche 454-based shotgun metagenomic sequencing produced vnf sequences showing high similarity to Anabaena vnf sequences as determined by BLAST searches (GenBank accessions KF662362 and KF662369). Sequences for the cyanobacterial vnf gene cluster were found by probing the metagenomes of P. malacea (GenBank accessions KF662359 and KF662365) and P. membranacea (GenBank accessions KF662360, KF662364, KF662366 and KF662370). Portions of cyanobacterial vnfDG and vnfN genes were successfully amplified through PCR and sequenced from Peltigera neopolydactyla (DNA-2308; GenBank accessions KF662361 and KF662368). Cyanobacterial genes from the nif and vnf clusters were found through MG-RAST analyses of the metatranscriptomic dataset generated from the single sample of P. praetextata (MG-RAST project 3216, metagenome 4453629.3). All nif and vnf fragments from the metatranscriptome were identified as belonging to the Cyanobacteria. For the single P. praetextata sample, two transcripts were found from the vnf cluster, one representing the vnfN gene (GenBank accession KF662367), and one representing a part of the vnfDG gene that is partially at the interface between the vnfD-like and the vnfG-like portions and partially within the vnfG-like portion (GenBank accession KF662363). The results from MG-RAST were corroborated by BLAST analyses of the entire metatranscriptomic dataset against the Anabaena vnf cluster.
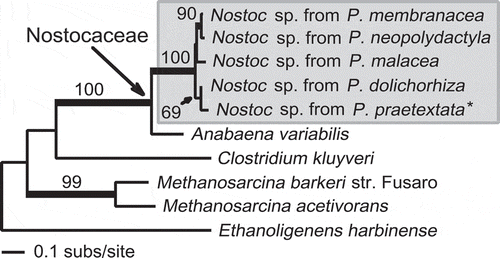
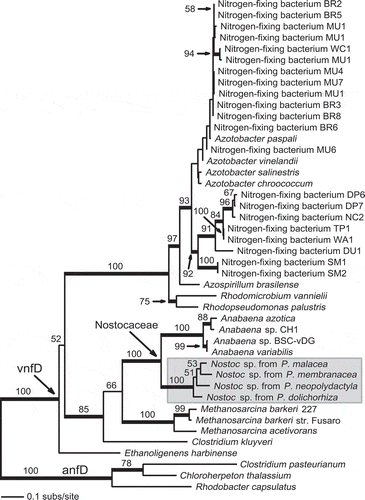
Alignment and phylogenetic analyses of vnfD sequences (and sequences representing the vnfD-like portion of vnfDG) demonstrated the relationship of sequences derived from the Peltigera thalli to Anabaena sequence fragments, which is best characterized as close but distinct, since all Nostoc-derived (lichen-associated) sequences formed a clade separate from the Anabaena clade (). The vnfN-encoding sequence detected in the metatranscriptome (generated from a single P. praetextata thallus) was inferred to be part of the Nostoc clade, which contained all other vnfN-encoding sequences derived from lichen thalli ().
Fig. 3. Maximum likelihood (ML) phylogeny of vnfD (including the vnfD-like portion of vnfDG for strains in which a fusion of adjacent genes has occurred) demonstrating the placement of lichen-associated sequences as a clade sister to Anabaena (Nostocaceae). Values above branches represent ML bootstrap proportions (BP) ≥ 50%; branches in bold indicate ML-BP support ≥ 70%.
Discussion
The evidence presented here demonstrates the presence and transcription of genes encoding enzymes for two N2 fixation pathways in lichen-associated Nostoc. The alternative (V-dependent) system, which has not previously been detected in this type of cyanobacterium, is shown to be present in all five Peltigera-dominated cyanolichens examined, which represent different fungal species from different geographical regions, and is demonstrated to be transcriptionally expressed in the one sample examined in this manner ().
The presence of two N2 fixation pathways presumably gives the cyanobacteria in this association the ability to use either one pathway or another when different trace elements become scarce or unavailable (Kutsche et al., Citation1996; Premakumar et al., Citation1998). For this reason, they will have a greater ability to colonize a wide array of environments, particularly those with habitats characterized by low temperature and metal availability. The V-dependent pathway is known to be less efficient for N2 fixation under typical circumstances (Eady, Citation1996, Citation2003; McKinlay & Harwood, Citation2011). However, if Mo is not present in a high enough concentration, or temperatures are too low for the Mo-dependent pathway to operate effectively, the V-dependent pathway can allow N2 fixation to continue (Miller & Eady, Citation1988; Young, Citation2005). Given that vanadium is more abundant on the Earth’s continental surface than molybdenum (Erickson, Citation1973; Wedepohl Citation1995), and that Mo has been shown to limit N2 fixation in certain ecosystems, this flexibility could be quite significant for lichens, which often live in nutrient-poor habitats and acquire much of their mineral nutritional requirements simply through aerial deposition (Nash, Citation2008b).
It is noteworthy that all transcripts representing N2 fixation genes were identified as belonging to the Cyanobacteria in the single sample for which transcripts were examined. These results perhaps suggest that the various non-cyanobacterial inhabitants of cyanolichen thalli that are potential nitrogen fixers (e.g. diverse members of Rhizobiales from families or genera containing known nitrogen fixers: Hodkinson et al., Citation2012b) do not fix a significant amount of nitrogen on the ecosystem level. However, one cannot discount the possibility that these results could stem from a more punctuated, less continuous strategy of N2 fixation in the majority of bacteria, compared to heterocystous cyanobacteria. Additional transcriptional observations are required at various time-points (e.g. under different conditions or at different developmental stages) to understand the relative fixed nitrogen contributions of cyanobacteria and the myriad non-photoautotrophic potential nitrogen fixers found in association with cyanolichen thalli.
Our findings demonstrate that N2 fixation by lichen-symbiotic cyanobacteria is more complex than previously suspected, and that the possession of different nitrogenases could potentially ‘buffer’ cyanolichens against environmental stresses, such as Mo availability and variation in abiotic factors (e.g. temperature). The fact that the V-dependent nitrogenase is found in a number of symbiotic strains of heterocystous cyanobacteria, while it is not known from any exclusively free-living cultured strains of the same genera, suggests a possible link with symbiosis. However, further speculation regarding the possibility of a greater evolutionary advantage of such a dual system of N2 fixation in symbiotic vs. free-living cyanobacteria will require more research into the fine-scale movements of micronutrients in various symbiotic and non-symbiotic microcosms.
The new findings presented here can also help scientists to gain a better understanding of N2 fixation in lichen-rich environments because they inform the interpretation of results from the acetylene reduction assay (ARA), the most common method used for examining amounts of environmental N2 fixation (Koch & Evans, Citation1966; Schollhorn & Burris, Citation1966; Stewart et al., Citation1967; Hardy et al., Citation1968; Silvester, Citation1989; Matzek & Vitousek, Citation2003; Pinto-Tomás et al., Citation2009; Davis et al., Citation2010; Cassar et al., Citation2012). ARA results can be misleading when N2 fixation is accomplished through alternative means because alternative pathways do not produce the same amounts of ethylene (Dilworth et al., Citation1988), which is the final product measured to estimate amounts of N2 fixation (note that alternative pathways produce ethane in addition to ethylene). In all cases, an ARA/N2 conversion factor, which can be determined experimentally using 15N enrichment experiments, must be applied to properly estimate dinitrogen fixation (Kurina & Vitousek, Citation2001). However, due mostly to the destructive, time-intensive nature of the enrichment, it is often assumed that N2 fixation through alternative means is negligible and the ARA/N2 conversion factor for the Mo-dependent nitrogenase is applied (Hällbom & Bergman, Citation1979; Blacklock et al., Citation1980; Cusack et al., Citation2009), despite the current controversy regarding the conversion of ARA values to N2 fixation rates (Belnap, Citation2001). Since cyanobacteria from lichens represent a major source of fixed nitrogen in a number of ecosystems, our findings could be quite significant for a proper understanding of global nutrient cycling and require scientists to re-evaluate many ecosystem-wide studies of nitrogen cycling. As highlighted in a recent review by Reed et al. (Citation2011), determining which nitrogenases are responsible for N2 fixation in the environment is a critical matter; demonstrating the importance of alternative means of N2 fixation would significantly impact current conceptual models relating N2 fixation to trace element control. Most studies on alternative N2 fixation are the result of lab research on very specific culturable N2 fixers. With the exception of the work by Betancourt et al. (Citation2008), the presence of alternative nitrogenases has been poorly studied in the natural environment and has never been demonstrated in cyanolichens. Thus, our results invite reconsideration of current conceptual models for N2 fixation in cyanolichens and open new avenues of research such as vanadium dynamics and homeostasis in cyanolichens and surrounding environments.
Supplemental data
Supplemental data for this article can be accessed here http://dx.doi.org/10.1080/09670262.2013.873143
Supplementary Table S1. Sequences used in phylogenetic analyses.
Supplementary File 1. VnfD alignment (nexus).
Supplementary File 2. VnfN alignment (nexus).
Supplementary File 3. PERL script for renaming the GenBank sequences.
Supplementary material
Download Zip (24.5 KB)Acknowledgements
Paul Bishop and Telisa Loveless are thanked for sparking B.P.H.’s interest in alternative nitrogen fixation pathways. We would like to thank Lisa Bukovnik, Bernie Ball, Jolanta Miadlikowska, Kathryn Picard, Tami McDonald, Daniele Armaleo, Terri Porter, Greg Bonito, Jason Jackson, Neil Gottel, Chris Schadt, Sarah Hodkinson and Molly McMullen, all of whom have provided important research assistance. This work was supported in part by Duke Biology Grants-in-Aid to B.P.H. from the Keever Endowment, a Mycological Society of America Graduate Fellowship to B.P.H., a grant from the Icelandic Research Fund, and a subcontract (112442) to Daniele Armaleo, Fred Dietrich and F.L. as part of the Pacific Northwest National Laboratory (PNNL) foundational scientific focus area (FSFA) under DOE-BER's genomic sciences programme in collaboration with Scott Baker and Jon Magnuson. Analyses were conducted using the Duke Shared Cluster Resource (DSCR), with outstanding services provided by John Pormann and Tom Milledge, and the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by a grant from the National Science Foundation (OCI-1053575) and was made available to B.P.H. through XSEDE Allocation Awards (DEB110024 & DEB130002). Contributions by L.F. and B.G. were made possible through NSF award DEB-0919284.
References
- Ahmadjian, V. (1993). The lichen symbiosis. John Wiley & Sons, New York.
- Altschul, S.F., Madden, T.L., Schaeffer, A.A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25: 3389–3402.
- Antoine, M.E. (2004). An ecophysiological approach to quantifying nitrogen fixation by Lobaria oregana. Bryologist, 107: 82–87.
- Arnold, A.E., Miadlikowska, J., Higgins, K.L., Sarvate, S.D., Gugger, P., Way, A., Hofstetter, V., Kauff, F. & Lutzoni, F. (2009). A phylogenetic estimation of trophic transition networks for ascomycetous fungi: are lichens cradles of symbiotrophic fungal diversification? Systematic Biology, 58: 283–297.
- Barron, A.R., Wurzburger, N., Bellenger, J.P., Wright, S.J., Kraepiel, A.M.L. & Hedin, L.O. (2009). Molybdenum limitation of asymbiotic nitrogen fixation in tropical forest soils. Nature Geoscience, 2: 42–45.
- Bates, S.T., Cropsey, G.W.G., Caporaso, J.G., Knight, R. & Fierer, N. (2011). Bacterial communities associated with the lichen symbiosis. Applied and Environmental Microbiology, 77: 1309–1314.
- Bellenger, J.P., Wichard, T., Xu, Y. & Kraepiel, A.M.L. (2011). Essential metals for nitrogen fixation in a free-living N2-fixing bacterium: chelation, homeostasis and high use efficiency. Environmental Microbiology, 13: 1395–1411.
- Belnap, J. (2001). Factors influencing nitrogen fixation and nitrogen release in biological soil crusts. In Biological soil crusts: structure, function, and management, ecological studies series 150 (Belnap, J. & Lange, O.L., editors), 241–261. Springer, Berlin.
- Betancourt, D.A., Loveless, T.M., Brown, J.W. & Bishop, P.E. (2008). Characterization of diazotrophs containing Mo-independent nitrogenases, isolated from diverse natural environments. Applied and Environmental Microbiology, 74: 3471–3480.
- Bishop, P.E. & Joerger, R.D. (1990). Genetics and molecular biology of alternative nitrogen fixation systems. Annual Review of Plant Physiology and Plant Molecular Biology, 41: 109–125.
- Blacklock, N.E., Goldsmith, F.B. & Patriquin, D.G. (1980). Nitrogen fixation (C2H2 reduction) by lungwort lichen (Lobaria pulmonaria (L.) Hoffm.) on red maple (Acer rubrum L.). Proceedings of the Nova Scotian Institute of Science, 30: 89–107.
- Boison, G., Steingen, C., Stal, L.J. & Bothe, H. (2006). The rice field cyanobacteria Anabaena azotica and Anabaena sp. CH1 express vanadium-dependent nitrogenase. Archives of Microbiology, 186: 367–376.
- Boyd, E.S., Hamilton, T.L. & Peters, J.W. (2011). An alternative path for the evolution of biological nitrogen fixation. Frontiers in Microbiology, 2(205): 1–11.
- Brigle, K.E., Newton, W.E. & Dean, D.R. (1985). Complete nucleotide sequence of the Azotobacter vinelandii nitrogenase structural gene cluster. Gene, 37: 37–44.
- Cardinale, M., Steinová, J., Rabensteiner, J., Berg, G. & Grube, M. (2012). Age, sun and substrate: triggers of bacterial communities in lichens. Environmental Microbiology Reports, 4: 23–28.
- Cassar, N., Bellenger, J.P., Karr, J., Jackson, R.B., Karr, J. & Barnett, B.A. (2012). N2 fixation estimates in real-time by cavity ring-down laser absorption spectroscopy. Oecologia, 168: 335–342.
- Chisnell, J.R., Premakumar, R. & Bishop, P.E. (1988). Purification of a second alternative nitrogenase from a nifHDK deletion strain of Azotobacter vinelandii. Journal of Bacteriology, 170: 27–33.
- Crittenden, P.D. & Kershaw, K.A. (1978). Discovering the role of lichens in the nitrogen cycle in boreal-arctic ecosystems. Bryologist, 81: 258–267.
- Cusack, D.F., Silver, W. & McDowell, W.H. (2009). Biological nitrogen fixation in two tropical forests: ecosystem-level patterns and effects of nitrogen fertilization. Ecosystems, 12: 1299–1315.
- Davis, S.C., Parton, W.J., Dohleman, F.G., Smith, C.M., Del Grosso, S., Kent, A.D. & DeLucia, E.H. (2010). Comparative biogeochemical cycles of bioenergy crops reveal nitrogen-fixation and low greenhouse gas emissions in a Miscanthus × giganteus agro-ecosystem. Ecosystems, 13: 144–156.
- Dilworth, M.J., Eady, R.R. & Eldridge, M.E. (1988). The vanadium nitrogenase of Azotobacter chroococcum: reduction of acetylene and ethylene to ethane. Biochemical Journal, 249: 745–751.
- Eady, R.R. (1989). The vanadium nitrogenase of Azotobacter. Polyhedron, 8: 1695–1700.
- Eady, R.R. (1996). Structure–function relationships of alternative nitrogenases. Chemical Reviews, 96: 3013–3030.
- Eady, R.R. (2003). Current status of structure function relationships of vanadium nitrogenase. Coordination Chemistry Reviews, 237: 23–30.
- Eady, R.R., Richardson, T.H., Miller, R.W., Hawkins, M. & Lowe, D.J. (1988). The vanadium nitrogenase of Azotobacter chroococcum. Purification and properties of the Fe protein. Biochemical Journal, 256: 189–196.
- Erickson, R.L. (1973). Crustal abundance of elements, and mineral reserves and resources. In United States Mineral Resources, Geological Survey Professional Paper 820 (Brobts, D.A. & Pratt, W.P., editors), 21–25. United States Government Printing Office, Washington, D.C.
- Georgiadis, M.M., Komiya, H., Chakrabarti, P., Woo, D., Kornuc, J.J. & Rees, D.C. (1992). Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science, 257: 1653–1659.
- Gillum, W.O., Mortenson, L.E., Chen, J.S. & Holm, R.H. (1977). Quantitative extrusions of the Fe4S4 cores of the active sites of ferredoxins and the hydrogenase of Clostridium pasteurianum. Journal of the American Chemical Society, 99: 584–595.
- Gu, X., Fu, Y.X. & Li, W.H. (1995). Maximum likelihood estimation of the heterogeneity of substitution rate among nucleotide sites. Molecular Biology and Evolution, 12: 546–557.
- Gunther, A.J. (1989). Nitrogen fixation by lichens in a subarctic Alaskan watershed. Bryologist, 92: 202–208.
- Hales, B.J., Langosch, D.J. & Case, E.E. (1986a). Isolation and characterization of a second nitrogenase Fe-protein from Azotobacter vinelandii. Journal of Biological Chemistry, 261: 15301–15306.
- Hales, B.J., Case, E.E., Morningstar, J.E., Dzeda, M.F. & Mauterer, L.A. (1986b). Isolation of a new vanadium-containing nitrogenase from Azotobacter vinelandii. Biochemistry, 25: 7251–7255.
- Hällbom, L. & Bergman, B. (1979). Influence of certain herbicides and a forest fertilizer on the nitrogen fixation by the lichen Peltigera praetextata. Oecologia, 40: 19–27.
- Hardy, R.W.F., Holsten, R.D., Jackson, E.K. & Burns, R.C. (1968). Acetylene ethylene assay for N fixation – laboratory and field evaluation. Plant Physiology, 43: 1185–1207.
- Hausinger, R.P. & Howard, J.B. (1983). Thiol reactivity of the nitrogenase Fe-protein from Azotobacter vinelandii. Journal of Biological Chemistry, 258: 13486–13492.
- Hodkinson, B.P. (2011). A phylogenetic, ecological, and functional characterization of the lichen microbiome. PhD thesis, Duke University, Durham, North Carolina, U.S.A.
- Hodkinson, B.P. & Lutzoni, F. (2009). A microbiotic survey of lichen-associated bacteria reveals a new lineage from the Rhizobiales. Symbiosis, 49: 163–180.
- Hodkinson, B.P., Gottel, N.R., Schadt, C.W. & Lutzoni, F. (2012a). Data from: Photoautotrophic symbiont and geography are major factors affecting highly structured and diverse bacterial communities in the lichen microbiome. Dryad Digital Repository, DOI:10.5061/dryad.t99b1.
- Hodkinson, B.P., Gottel, N.R., Schadt, C.W. & Lutzoni, F. (2012b). Photoautotrophic symbiont and geography are major factors affecting highly structured and diverse bacterial communities in the lichen microbiome. Environmental Microbiology, 14: 147–161.
- Joerger, R.D., Loveless, T.M., Pau, R.N., Mitchenall, L.A., Simon, B.H. & Bishop, P.E. (1990). Nucleotide sequences and mutational analysis of the structural genes for nitrogenase 2 of Azotobacter vinelandii. Journal of Bacteriology, 172: 3400–3408.
- Koch, B. & Evans, H.J. (1990). Reduction of acetylene to ethylene by soybean root nodules. Plant Physiology, 41: 1748–1750.
- Kurina, L.M. & Vitousek, P.M. (2001). Nitrogen fixation rates of Stereocaulon vulcani on young Hawaiian lava flows. Biogeochemistry, 55: 179–194.
- Kutsche, M., Leimkühler, S., Angermüller, S. & Klipp, W. (1996). Promoters controlling expression of the alternative nitrogenase and the molybdenum uptake system in Rhodobacter capsulatus are activated by NtrC, independent of sigma54, and repressed by molybdenum. Journal of Bacteriology, 178: 2010–2017.
- Maddison, W.P. & Maddison, D.R. (2010). Mesquite: a modular system for evolutionary analysis version 2.74. http://mesquiteproject.org
- Magain, N., Forrest, L.L., Sérusiaux, E. & Goffinet, B. (2010). Microsatellite primers in the Peltigera dolichorhiza complex (lichenized ascomycete, Peltigerales). American Journal of Botany, 97: e102–e104.
- Masukawa, H., Zhang, X., Yamazaki, E., Iwata, S., Nakamura, K., Mochimaru, M., Inoue, K. & Sakurai, H. (2009). Survey of the distribution of different types of nitrogenases and hydrogenases in heterocyst-forming cyanobacteria. Marine Biotechnology, 11: 397–409.
- Matzek, V. & Vitousek, P. (2003). Nitrogen fixation in bryophytes, lichens, and decaying wood along a soil–age gradient in Hawaiian montane rain forest. Biotropica, 35: 12–19.
- Mayrose, I., Friedman, N. & Pupko, T. (2005). A Gamma mixture model better accounts for among site rate heterogeneity. Bioinformatics, 21: ii151–ii158.
- McKinlay, J.B. & Harwood, C.S. (2011). Harnessing bacteria that use light to produce hydrogen. Microbe, 6: 345–351.
- Meyer, F., Paarmann, D., D’souza, M., Olson, R., Glass, E.M., Kubal, M., Paczian, T., Rodriguez, A., Stevens, R., Wilke, A., Wilkening, J. & Edwards, R.A. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics, 9: 386.
- Miadlikowska, J. & Lutzoni, F. (2004). Phylogenetic classification of peltigeralean fungi (Peltigerales, Ascomycota) based on ribosomal RNA small and large subunits. American Journal of Botany, 91: 449–464.
- Miller, R.W. & Eady, R.R. (1988). Molybdenum and vanadium nitrogenases of Azotobacter chroococcum. Low temperature favours N2 reduction by vanadium nitrogenase. Biochemistry Journal, 256: 429–432.
- Minin, V., Abdo, Z., Joyce, P. & Sullivan, J. (2003). Performance-based selection of likelihood models for phylogeny estimation. Systematic Biology, 52: 674–683.
- Nash, T.H., III. (2008a). Nitrogen, its metabolism and potential contribution to ecosystems. In Lichen biology, 2nd edition (Nash, T.H., III, editor), 216–233. Cambridge University Press, Cambridge.
- Nash, T.H., III. (2008b). Nutrients, elemental accumulation and mineral cycling. In Lichen biology, 2nd edition (Nash, T.H., III, editor), 234–251. Cambridge University Press, Cambridge.
- O’Brien, H.E., Miadlikowska, J. & Lutzoni, F. (2005). Assessing host specialization in symbiotic cyanobacteria associated with four closely related species of the lichen fungus Peltigera. European Journal of Phycology, 40: 363–378.
- Pattengale, N., Alipour, M., Bininda-Emonds, O., Moret, B. & Stamatakis, A. (2009). How many bootstrap replicates are necessary? In Lecture Notes in Computer Science: Research in Computational Molecular Biology, Volume 5541 (Batzoglou, S., editor), 184–200. Springer, Berlin.
- Pinto-Tomás, A.A., Anderson, M.A., Suen, G., Stevenson, D.M., Chu, F.S.T., Cleland, W.W., Weimer, P.J. & Currie, C.R. (2009). Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science, 326: 1120–1123.
- Posada, D. & Crandall, K.A. (1998). MODELTEST: Testing the model of DNA substitution. Bioinformatics, 14: 817–818.
- Pratte, B.S., Eplin, K. & Thiel, T. (2006). Cross-functionality of nitrogenase components NifH1 and VnfH in Anabaena variabilis. Journal of Bacteriology, 188: 5806–5811.
- Premakumar, R., Pau, R.N., Mitchenall, L.A., Easo, M. & Bishop, P.E. (1998). Regulation of the transcriptional activators AnfA and VnfA by metals and ammonium in Azotobacter vinelandii. FEMS Microbiology Letters, 164: 63–68.
- Raina, R., Bageshwar, U.K. & Das, H.K. (1993). The ORF encoding a putative ferredoxin-like protein downstream of the vnfH gene in Azotobacter vinelandii is involved in the vanadium-dependent alternative pathway of nitrogen fixation. Molecular and General Genetics, 236: 459–462.
- Raymond, J., Siefert, J.L., Staples, C.R. & Blankenship, R.E. (2004). The natural history of nitrogen fixation. Molecular Biology and Evolution, 21: 541–554.
- Reed, S.C., Cleveland, C.C. & Townsend, A.R. (2011). Functional ecology of free-living nitrogen fixation: a contemporary perspective. Annual Review of Ecology, Evolution and Systematics, 42: 489–512.
- Ren, F., Tanaka, H. &, Yang, Z. (2005). An empirical examination of the utility of codon-substitution models in phylogeny reconstruction. Systematic Biology, 54: 808–818.
- Robson, R.L., Eady, R.R., Richardson, T.H., Miller, R.W., Hawkins, M. & Postgate, J.R. (1986). The alternative nitrogenase of Azotobacter chroococcum is a vanadium enzyme. Nature, 322: 388–390.
- Robson, R.L., Woodley, P.R., Pau, R.N. & Eady, R.R. (1989). Structural genes for the vanadium nitrogenase from Azotobacter chroococcum. EMBO Journal, 8: 1217–1224.
- Schneider, T., Schmid, E., de Castro, J.V., Cardinale, M., Eberl, L., Grube, M., Berg, G. & Riedel, K. (2011). Structure and function of the symbiosis partners of the lung lichen (Lobaria pulmonaria L. Hoffm.) analyzed by metaproteomics. Proteomics, 11: 2752–2756.
- Schollhorn, R. & Burris, R.H. (1966). Study of intermediates in nitrogen fixation. Federation Proceedings, 25: 710.
- Silvester, W.B. (1989). Molybdenum limitation of asymbiotic nitrogen-fixation in forests of Pacific Northwest America. Soil Biology and Biochemistry, 21: 283–289.
- Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22: 2688–2690.
- Stewart, W.D.P., Fitzgerald, G.P. & Burris, R.H. (1967). In situ studies on N fixation using acetylene reduction technique. Proceedings of the National Academy of Sciences of the United States of America, 58: 2071–2078.
- Sullivan, J., Swofford, D. & Naylor, G. (1999). The effect of taxon sampling on estimating rate heterogeneity parameters of maximum-likelihood models. Molecular Biology and Evolution, 16: 1347–1356.
- Thiel , T. (1993). Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. Journal of Bacteriology, 175: 6276–6286.
- Thiel, T. (1996). Isolation and characterization of the VnfEN genes of the cyanobacterium Anabaena variabilis. Journal of Bacteriology, 178: 4493–4499.
- Thiel, T., Meeks, J.C., Elhai, J., Potts, M., Larimer, F., Lamerdin, J., Predki, P. & Atlas, R. (2002). Nitrogen fixation: analysis of the genome of the cyanobacterium Nostoc punctiforme. In Nitrogen fixation: global perspectives (Finan, T.M., O’Brian, M.R., Layzell, D.B., Vessey, J.K. & Newton, W., editors), 88–92. CABI Publishing, New York.
- Tschermak-Woess, E. (1988). The algal partner. In CRC Handbook of Lichenology (Galun, M., editor), 39–92. CRC Press, Boca Raton, FL.
- U’Ren, J., Lutzoni, F., Miadlikowska, J. & Arnold, A.E. (2010). Community analysis reveals close affinities between endophytic and endolichenic fungi in mosses and lichens. Microbial Ecology, 60: 340–353.
- U’Ren, J.M., Lutzoni, F., Miadlikowska, J., Laetsch, A.D. & Arnold, A.E. (2012). Host and geographic structure of endophytic and endolichenic fungi at a continental scale. American Journal of Botany, 99: 898–914.
- Walmsley, J. & Kennedy, C. (1991). Temperature-dependent regulation by molybdenum and vanadium of expression of the structural genes encoding three nitrogenases in Azotobacter vinelandii. Applied and Environmental Microbiology, 57: 622–624.
- Wedepohl, K.H. (1995). The composition of the continental crust. Geochimica et Cosmochimica Acta, 59: 1217–1232.
- Wurzburger, N., Bellenger, J.P., Kraepiel, A.M.L. & Hedin, L.O. (2012). Molybdenum and phosphorus interact to constrain asymbiotic nitrogen fixation in tropical forests. PLoS ONE, 7: e33710.
- Yang, Z. (1993). Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Molecular Biology and Evolution, 10: 1396–1401.
- Yang, Z. (2006). Computational Molecular Evolution. Oxford University Press, Oxford.
- Young, J. (2005). The phylogeny and evolution of nitrogenases. In Genomes and genomics of nitrogen-fixing organisms [nitrogen fixation: origins, applications, and research progress, Vol. 3] (Palacios, R. & Newton, W.E., editors), 221–241. Springer, Dordrecht.
- Xavier, B.B., Miao, V.P.W., Jónsson, Z.O. & Andrésson, Ó.S. (2012). Mitochondrial genomes from the lichenized fungi Peltigera membranacea and Peltigera malacea: features and phylogeny. Fungal Biology, 116: 802–814.
- Zehr, J.P., Jenkins, B.D., Short, S.M. & Steward, G.F. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environmental Microbiology, 5: 539–554.
- Zhang, Z., Schwartz, S., Wagner, L. & Miller, W. (2000). A greedy algorithm for aligning DNA sequences. Journal of Computational Biology, 7: 203–214.