Abstract
Two new Ulvella species, U. elegans R. Nielsen & K. Gunnarsson and U. islandica R. Nielsen & K. Gunnarsson are described. These microfilamentous marine green algae were found in the sublittoral zone in northern Iceland, epiphytic on Euthora cristata and associated with a calcareous polychaete tube, respectively. Unialgal cultures were established from field-collected material for morphological observations. In culture, Ulvella elegans was characterized by rosettes of monostromatic pseudoparenchyma consisting of radiating filaments with a margin of mutually free filaments. Each cell had one pyrenoid. Hairs were not observed. Ulvella islandica had a heterotrichous morphology, consisting of dense tufts of upright broad branches and much narrower, rhizoid-like branches. Acrochaete-type hairs occurred; these are hyaline non-septate merocytic extensions from a more or less bulbous base, which may be separated from the vegetative cell below. Most cells had one pyrenoid except for a few broad cells which had two or three. In a phylogenetic reconstruction based on the chloroplast-encoded tufA gene, the sequences for the two species were clearly distinct from any other Ulvella sequence available for this gene. Ulvella islandica was placed in a clade together with U. lens, U. wittrockii, U. reticulata and U. pseudorepens. Ulvella elegans occupied a branch deep in the phylogeny but the position was poorly supported.
Introduction
Modern taxonomy of microfilamentous green algae is based on observations in nature and in culture, complemented with molecular investigations (Nielsen et al., Citation2013). Several species often grow mixed together on the same substratum in nature, making it difficult to distinguish individual plants. Species-specific characters such as hairs and the size and morphology of sporangia are often missing at the time of collection, making identification impossible. Culture studies enable separation of species and permit additional characters to develop (Kornmann, Citation1959, Citation1960; Nielsen & Pedersen, Citation1977). Molecular methods have supplied data to reveal phylogenetic relationship between genera and species (e.g. O’Kelly et al., Citation2004). In addition, molecular data supply important information for assessing species boundaries and for testing morphology-based species circumscriptions (Rinkel et al., Citation2012; Nielsen et al., Citation2013).
A recent revision of Ulvella based on culture studies, supported by molecular analyses of the chloroplast-encoded tufA gene, documented that species previously referred to Acrochaete, Endophyton, Entocladia, Pringsheimiella, Pseudodictyon and Ulvella belong to a single genus for which Ulvella has priority (Nielsen et al., Citation2013). The concept of Ulvella was thus extended and several new species described. Species were distinguished by their growth form in culture, cell shape and size, the number of pyrenoids per cell, the positions of Acrochaete-type hairs (hyaline non-septate merocytic extensions from a more or less bulbous base, which may be separated from the vegetative cell below: Nielsen, Citation1979) and sporangia, the shape and size of sporangia, and whether germlings are with or without an empty spore-wall. The phylogenetic relationships of species were investigated by analyses of chloroplast tufA sequences (Nielsen et al., Citation2013).
In a detailed study of the microfilamentous marine green algae of Iceland, based on morphological structures of naturally occurring plants and of numerous unialgal isolates (Nielsen & Gunnarsson, in prep), two new Ulvella species were detected. The aim of the work reported here was to describe these new species using a combination of phenotypic and genotypic characters.
Materials and methods
Cultures were established from field-collected material. Isolate IS06013-1 was initiated from plants on calcareous polychaete tubes sampled at 30 m depth at Laugakambur, Hrísey, Iceland (66° 01.95′ N, 18° 24.47′ W) on 13 June 2006. Isolate IS06015 was initiated from epiphytes on Euthora cristata sampled between 1–5 m depth at Laugakambur, Hrísey (66° 01.75′ N, 18° 24.47′ W), also on 13 June 2006. Crude cultures were established from fragments of host material placed in test tubes with growth medium. Unialgal cultures were obtained for each species by isolating and serial washing single plants of the target species into clean plastic Petri dishes with 15 ml of growth medium, using a fine glass pipette. The strains are available from the Scandinavian Culture Collection of Algae & Protozoa, Copenhagen (SCCAP). The growth medium was a modified Provasoli medium (MV30: Christensen, Citation1982) with GeO2 added to prevent diatom growth (Lewin, Citation1966). The cultures were maintained at 4 ºC with a 12 : 12 h light : dark cycle at an irradiance of c. 16 µmol photons m–2 s–1 provided by fluorescence lamps (Philips TL 40W, Philips, Amsterdam, the Netherlands). Observations were made once or twice per week of plants that were kept growing actively by transfer to fresh medium every other week or monthly. Photos were taken with an Olympus digital camera DP-70 mounted on an Olympus AX70 microscope (Olympus, Tokyo, Japan). A saltwater immersion 20× objective was used to study the morphology of prostrate plants growing in plastic Petri dishes.
Following the procedure of Doyle & Doyle (Citation1987), total genomic DNA was extracted from two cultures kept frozen at −18 °C. PCR amplifications and sequencing were performed using primers tufAF and tufAR (Famà et al., Citation2002) following the procedure described in Nielsen et al. (Citation2013). Sequence editing was done using Sequencher version 4.10 (Gene Codes Corporation, Ann Arbor, Michigan, USA). Sequences were deposited in GenBank under accession numbers KF444923 for isolate IS06013-1 (U. elegans) and KF444924 for IS06015 (U. islandica). The two new tufA sequences were manually inserted into the matrix previously used by Nielsen et al. (Citation2013). A list of specimens included in the matrix is provided in .
Table 1. Species included in the phylogenetic analysis with associated data.
Phylogenetic analyses were based on Bayesian inference, maximum likelihood and parsimony. MrBayes (v. 3.1.2, Ronquist & Huelsenbeck, Citation2003) was used to perform Bayesian analysis with a general time reversible (GTR) substitution model. The choice of model was based on jmodeltest ver. 2.1.3 (Darriba et al., Citation2012) which selected the best fit for our data matrix of tufA sequences from 56 different models of DNA evolution. We used partitioning of the codon positions for the protein-encoded chloroplast gene and ran two independent Markov Chain Monte Carlo (MCMC) (each comprising one cold and three heated chains) with 4 × 106 generations. Parameter values and trees were sampled and saved every 1000th generation. The numbers of substitution types allowed were two for first and second codon positions (lset = 2) and six types for third codon positions (lset = 6). We assumed that all the model parameters were unlinked and rate multipliers were variable across partitions. We plotted the log likelihood values as a function of generations, which showed that the lnL values converged at c. 5530 after 20 000 generations leaving 3981 trees. These were imported into PAUP* vers. 4.0b8 (Swofford, Citation2001) to produce a 50% majority rule consensus tree. Posterior probabilities were also obtained from the 3981 trees.
Maximum likelihood analysis was performed using PhyML (Guindon et al., Citation2010) via the online version available on the Montpellier bioinformatics platform at http://www.atgc-montpellier.fr/phyml. For this we used a GTR model with the parameter settings suggested by Modeltest (v. 3.7, Posada & Crandall, Citation1998). We used 1000 bootstrap replications in maximum likelihood to evaluate the robustness of the tree topology.
Parsimony analyses were performed using PAUP* and the following options: heuristic search, 100 random addition sequences, five trees held each step and TBR swapping. PAUP* v. 4.0b8 was preferred to the most recent version 10 (Swofford, Citation2002), as the latter outputs erroneous tree lengths and an excessive number of tree islands. Uninformative characters were excluded and all characters were treated as equally weighted and non-additive. Bootstrap support was calculated using PAUP* v. 4.0b8 running 1000 replicates. In each replicate we ran 10 random addition sequences saving no more than 500 trees per replicate.
Results
The tufA matrix included 50 sequences (43 Ulvella spp. and seven outgroup) and 894 characters. The 50% majority rule consensus tree derived from Bayesian inference is shown in , which shows the posterior probabilities and also bootstrap support values from the maximum likelihood and parsimony analyses. The parsimony analysis included 227 informative characters and resulted in 1154 equally parsimonious trees of length 756 with a consistency index of 0.491, and a retention index of 0.692.
Fig. 1. 50% majority rule consensus tree derived from Bayesian inference. Support values on branches are posterior probabilities, bootstrap support from maximum likelihood, and bootstrap values from parsimony. Bootstrap values below 50 are indicated with a dash. The two new species are shown in bold. For details of other taxa included in the analyses see .
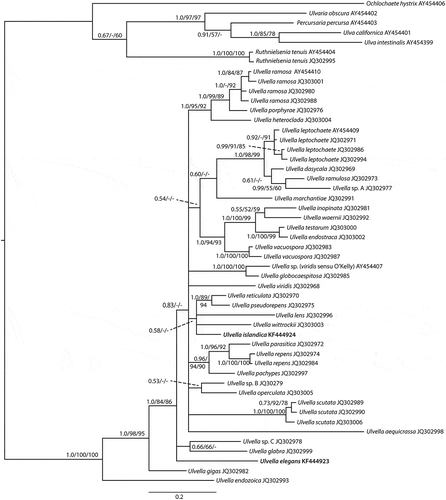
Isolate IS06015 (KF444924) was placed in a clade together with U. lens, U. wittrockii, U. reticulata and U. pseudorepens. This clade had a very low posterior probability and no bootstrap support from either maximum likelihood or parsimony (). The position of isolate IS06015 within the clade was also unresolved. However, based on the level of tufA sequence divergence between IS06015 and the other species of the clade we recognize it as a distinct species, described below as Ulvella islandica.
Isolate IS06013-1 (KF444923) was placed at a relatively basal position within Ulvella, but its position was unresolved with respect to a poorly supported clade including U. glabra and the unnamed Ulvella sp. C., and the large clade including all other Ulvella species except U. gigas and U. endozoica. Based on the pronounced level of sequence divergence from all other species, we recognize IS06013-1 as a distinct species, which we name U. elegans.
Ulvella elegans R. Nielsen & K. Gunnarsson, sp. nov.
Figs 2–6
Figs 2–6. Ulvella elegans. 2. Mature plant with a pseudoparenchymatous central area and mutually free distal filaments. 3. Young plant of radiating filaments of cylindrical vegetative cells with a single pyrenoid. 4. Same plant as in shown in different focus. 5. Barrel-shaped mature and empty sporangia. 6. Germlings. Scale bars = 20 µm (–) and 10 µm (, ).
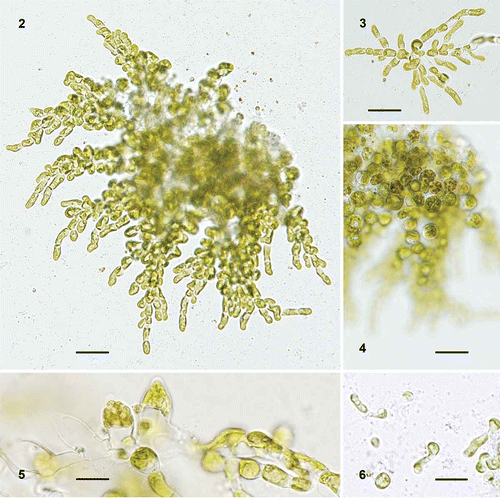
Description: Monostromatic rosettes or mutually free filaments with alternate or opposite branches. The distal, cylindrical cells are 3.5–6.5 µm wide and 2–4 times as long; the rounded middle cells 10.5–12.5 µm across. Vegetative cells contain a parietal chloroplast with one pyrenoid. Hairs have not been observed. Sporangia are 11–13.8 µm tall, with a conical apex. On release, spores germinate unilaterally and remain a part of the developing plants.
Holotype: Dried sample of isolate IS06013-1 maintained in the Botanical Museum, Copenhagen (C) as no. CAT 2481.
Type locality: Laugakambur, Hrísey, Iceland, 30 m depth, on a polychaete tube.
Etymology: The name refers to the elegant look of the monostromatic rosettes with free marginal filaments of plants kept in culture.
Individuals formed a rosette in contact with a solid substratum in culture. Cells in the middle part were confluent and formed a monostromatic, pseudoparenchymatous layer, while the margin consisted of mutually free filaments (, ). Plants growing freely formed bushes of free filaments developed around a dense middle part. Cells in the middle were rounded to almost globular and 10.5–12.5 µm in diameter, whereas distal cells were 3.5–6.5 µm wide and 2–4 times as long. Vegetative cells contained a parietal chloroplast with a single pyrenoid. Hairs were not observed. Sporangia developed from the middle cells of substrate-attached plants or from intercalary cells in freely growing plants (, ). They became almost barrel-shaped with a conical apex and were 11–13.8 µm tall. On release, spores germinated unilaterally and remained part of the developing plants ().
Ulvella islandica R. Nielsen & K. Gunnarsson, sp. nov.
Figs 7–12
Figs 7–12. Ulvella islandica, morphology of heterotrichous plants in culture. 7. Tuft of broad cylindrical filaments with rounded cells at the base. 8. Basal part of tuft with rhizoid-like filaments. 9. Vegetative cell with a parietal chloroplast and one pyrenoid. 10. Acrochaete-type hair. 11. Cylindrical, apical sporangium. 12. Germling with evacuated spore-wall attached. Scale bars = 20 µm (, ) and 10 µm (–).
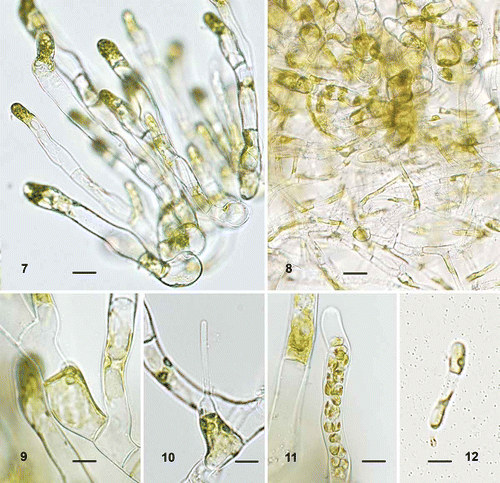
Description: Tufts of upright branches of cylindrical cells, 10–15 µm wide, with rounded cells at base, 20–32 µm across. Narrow, alternately branched filaments resembling rhizoids develop from both kinds of cells at the base of the tuft; they have cylindrical cells, 4.5–6 µm wide. Vegetative cells contain a parietal, slightly lobed chloroplast with one (to three) pyrenoid(s). Acrochaete-type hairs occur apically on cells of broad filaments. Sporangia develop from similar cells and become elongate, linear-cylindrical.
Holotype: Dried sample of isolate IS06015 maintained at the Botanical Museum, Copenhagen (C) as no. CAT 2480.
Type locality: Laugakambur, Hrísey, Iceland, 1–5 m depth, on Euthora cristata.
Etymology: The name refers to the origin of the type material.
Mature plants in culture had a heterotrichous morphology, consisting of a dense tuft of upright broad branches with rounded cells at the base (). The distal cylindrical cells of the broad branches were 10–15 µm in width, whereas the rounded basal cells were 20–32 µm across. Irregularly branched narrow filaments, 4.5–6 µm wide, developed from the rounded cells and the lower cylindrical cells, and formed a matted cell mass (). Most cells contained a single pyrenoid in a parietal, slightly lobed chloroplast (); however, two or three pyrenoids were observed in a few broad cells. Acrochaete-type hairs occurred apically on cells of broad filaments (). Sporangia developed from similar cells and became linear-cylindrical (). Settled spores germinated unilaterally; an evacuated, often brown-pigmented, spore-wall and a germ-tube were visible in young, few-celled plants ().
Discussion
The discovery of two new species of microfilamentous marine green algae from Iceland confirms the prediction of new species of Ulvella by Nielsen et al. (Citation2013). Species of Ulvella are generally separated by growth form, the shape and size of the cells, the number of pyrenoids per cell, the position of Acrochaete-type hairs, and the shape and size of sporangia. These characters are usually better developed in plants in culture than in samples collected in nature, where some characters may be missing at the time of collecting. Therefore, the morphology of the new species reported here was described from actively growing cultured algae. The rosettes of U. elegans make this species most similar to U. marchantiae, but the two differ in the morphology of the sporangia, which are bottle-shaped with a short neck in U. marchantiae and barrel-shaped with a conical apex in U. elegans. The presence of Acrochaete-type hairs was considered an important diagnostic character by Nielsen Citation(1979) and Acrochaete-type hairs were observed only in U. marchantiae (Nielsen & McLachlan, Citation1986, as Acrochaete marchantiae), not in U. elegans. The lack of Acrochaete-type hairs is shared by U. glabra, U. ramosa and U. testarum but none of these species seems to be closely related to U. elegans (). Based on the phylogenetic distribution of these characteristic hairs in species of Ulvella, they appear to have been repeatedly lost during evolution; their absence is probably not a consequence of culture conditions as isolates lacking Acrochaete-type hairs have been subject to the same culture methods as hair-producing Ulvella species.
The heterotrichous morphology of U. islandica is shared by several Ulvella species, among them U. pseudorepens, which belongs to the same clade as U. islandica. The two can be easily distinguished by the number of pyrenoids, with several per cell in U. pseudorepens and only one in the majority of the cells in U. islandica. Ulvella repens is heterotrichous with relatively broad upright filaments (Nielsen et al., Citation2013) like U. islandica. A matted mass of rhizoidal-like branches only occurs in U. islandica.
Though phylogenetic analyses have clarified some relationships among the species of Ulvella, others remain unresolved including the exact phylogenetic position and sister group of each of the two new species described here. In order to gain a better understanding of their phylogeny, future analyses of additional genes will most likely be helpful.
Acknowledgements
We are grateful to Svanhildur Egilsdóttir, Tryggvi Sveinsson, Erlendur Bogason, Juliet Brodie and Barbara Rinkel for their company and help during the sampling expedition. We thank Charlotte Hansen and Hannah Blossom for assistance with the molecular work. A Carlsberg Foundation grant supported the molecular work and equipment. Financial and logistic support was provided by the EU-program ‘Synthesys’ during analyses of the culture collections at Botanical Garden and Museum, Natural History Museum of Denmark.
References
- Christensen, T. (1982). Alger i naturen og i laboratoriet. Nucleus, Copenhagen.
- Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9: 772.
- Doyle, J.J. & Doyle, J.F. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19: 11–15.
- Famà, P., Wysor, B., Kooistra, W.H.C.F. & Zuccarello, G.C. (2002). Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene. Journal of Phycology, 38: 1040–1050.
- Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59: 307–321.
- Kornmann, P. (1959). Die heterogene Gattung Gomontia I. Der sporangiale Anteil, Codiolum polyrhizum. Helgoländer Wissenschaftliche Meeresuntersuchungen, 6: 229–238.
- Kornmann, P. (1960). Die heterogene Gattung Gomontia II. Der fädige Anteil, Eugomontia sacculata nov. gen. nov. spec. Helgoländer Wissenschaftliche Meeresuntersuchungen, 7: 59–71.
- Lewin, J. (1966). Silicon metabolism in diatoms. V. Germanium dioxide, a specific inhibitor of diatom growth. Phycologia, 6: 1–12.
- Nielsen, R. (1979). Culture studies on the type species of Acrochaete, Bolbocoleon and Entocladia (Chaetophoraceae, Chlorophyceae). Botaniska Notiser, 132: 441–449.
- Nielsen, R. & McLachlan, J. (1986). Acrochaete marchantiae comb. nov. and Trichothyra irregularis gen. et sp. nov. with notes on other species of small filamentous green algae from St. Lucia (West Indies). Nordic Journal of Botany, 6: 515–524.
- Nielsen, R. & Pedersen, P.M. (1977). Separation of Syncoryne reinkei nov. gen., nov. sp. from Pringsheimiella scutata (Chlorophyceae, Chaetophoraceae). Phycologia, 16: 411–416.
- Nielsen, R., Petersen, G., Seberg, O., Daugbjerg, N., O’Kelly, C.J. & Wysor, B. (2013). Revision of the genus Ulvella (Ulvellaceae, Ulvophyceae) based on morphology and tufA gene sequences of species in culture, with Acrochaete and Pringsheimiella placed in synonymy. Phycologia, 52: 37–56.
- O’Kelly, C.J., Wysor, B. & Bellows, W.K. (2004). Gene sequence diversity and the phylogenetic position of algae assigned to the genera Phaeophila and Ochlochaete (Ulvophyceae, Chlorophyta). Journal of Phycology, 40: 789–799.
- Posada, D. & Crandall, K.A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics, 14: 817–818.
- Rinkel, B.E., Hayes, P., Gueidan, C. & Brodie, J. (2012). A molecular phylogeny of Acrochaete and other endophytic green algae (Ulvales, Chlorophyta). Journal of Phycology, 48: 1020–1027.
- Ronquist, F. & Huelsenbeck, J.P. (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572–1574.
- Swofford, D.L. (2001). PAUP* phylogenetic analysis using parsimony (*and other methods), version 4.0b8. Sinauer Associates, Sunderland, MA.
- Swofford, D.L. (2002). PAUP* phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland, MA.
