Abstract
The Synura petersenii species complex represents a common, cosmopolitan and highly diverse taxon of autotrophic freshwater flagellates. In this paper, we describe and characterize four new species (S. borealis, S. heteropora, S. hibernica and S. laticarina) that have been identified during our extensive sampling of freshwater habitats in 15 European countries. Morphometric analyses of siliceous scales led to the significant phenotypic differentiation of all four newly described species, and their separation from other related species of the S. petersenii complex. Two of these newly described species (S. hibernica and S. borealis) can be clearly distinguished by characteristic large colonies consisting of elongated, lanceolate-shaped cells. Development of strongly elongated, narrow cells in S. hibernica could be explained by the adaptation of this species to oligotrophic conditions. Though morphologically distinct, S. borealis possesses an exceptionally high degree of genetic diversity, possibly indicating recent speciation and evolutionary diversification within this taxon. Three of the four newly described species exhibit restricted biogeographic distribution. The evolutionarily related S. borealis and S. laticarina occur only in Northern Europe, and seem to be adapted to colder areas. The most remarkable distribution pattern was observed for S. hibernica, which has a geographic distribution that is restricted to western Ireland.
Introduction
The order Synurales (Chrysophyceae, Stramenopiles) contains several genera of scale-bearing flagellates that are important components of phytoplankton communities of various freshwater bodies (Kristiansen & Preisig, Citation2007; Škaloud et al., Citation2013a). The most conspicuous genus of the order, Synura, is a colonial organism formed by a variable number of cells joined together at their posterior ends. Each cell is covered by imbricate silica scales (Wee, Citation1997), consisting of a perforated basal plate with various upturned or bent parts, and a secondary ornamentation, which is used to delimit the different species (Kristiansen, Citation1986). According to the recently published phylogeny of the Synurales, the genus Synura should be split into five sections: Echinulatae, Peterseniae, Spinosae, Splendidae and Synura (Škaloud et al., Citation2013a).Taxa belonging to the section Peterseniae are well characterized by body scales with a central keel that may end in a spine-like projection. The most widely recognized species of this section, Synura petersenii sensu lato, is considered to be one of the most widely distributed freshwater chrysophytes (Kristiansen, Citation1975), even causing taste and odour problems in water supplies (Nicholls & Gerrath, Citation1985). This species has been subjected to various ecophysiological experiments (e.g. Saxby-Rouen et al., Citation1997; Kim et al., Citation2008; Pichrtová & Němcová, Citation2011).
Due to the species-specific morphology of silica scales, Synura could be supposed to have one of the best morphological species concepts within protists. Synura scales are often well preserved in benthic sediments and used in palaeoecological studies. Because many taxa are distributed within a narrow range of ecological variables (e.g. pH, temperature and nutrient concentration), investigation of fossilized Synura scales is used as a tool to assess eutrophication, acidification and shifts in climate (Smol, Citation1995; Smol & Cumming, Citation2000). In addition, the records of fossilized Synura scales discovered in Eocene sediments are used to date the diversification and evolution of the genus (Boo et al., Citation2010; Siver, Citation2013).
Recent molecular phylogenetic investigations have revealed a conflict between the traditional morphological species concept based on ultrastructure of silica structures and the phylogenetic data. Synura petersenii sensu lato represents the most investigated species in this respect. First, Wee et al. (Citation2001) demonstrated the existence of two well-supported S. petersenii clades with different distribution patterns. Later, Kynčlová et al. (Citation2010) investigated a number of European isolates, and revealed the existence of six cryptic species. Almost concurrently, Boo et al. (Citation2010) published a multigene phylogeny of approximately 100 S. petersenii isolates, confirming the high degree of hidden, species-level diversity within this species. A taxonomic assessment of observed cryptic diversity redefined the species concept within the S. petersenii morphotype, and recognized six cryptic lineages as separate species (Škaloud et al., Citation2012).
Phylogenetic analyses performed by Boo et al. (Citation2010) and Škaloud et al. (Citation2012) clearly indicate that there are more than six hidden species within the S. petersenii complex. To further investigate the real level of species diversity within this complex, we performed extensive sampling in 15 European countries, including Greenland. According to the analyses of internal transcribed spacer (ITS) rDNA sequences, 58 of more than 800 investigated Synura isolates belonged to four novel, undescribed species. The principal aim of this study was to expand our knowledge and understanding of the diversity in the S. petersenii complex, and to describe and characterize all these new taxa. In addition, we performed a detailed morphological investigation of all cryptic species, and investigated whether they could be delineated by the morphology of silica scales.
Materials and methods
Origin and cultivation of the investigated strains
The strains used in this study were obtained during an investigation of silica-scaled chrysophytes in Europe and Greenland during the period 2008–2013 (). The material was usually sampled from plankton and metaphyton of various water bodies, using a plankton net with 20-μm mesh. To ensure the highest viability of Synura strains, individual colonies were isolated on the same day that they were sampled. The colonies were removed from the natural sample by micropipetting, and transferred into separate wells of a 96-well plate containing 400 μl of either MES- or HEPES-buffered DY IV liquid medium (Andersen et al., Citation1997). The wells were kept at 15°C (fridge bag TK 51, Ardes SpA, Ponte Nossa, Italy), under constant illumination of 50–200 μmol m−2 s−1 provided by 6 W LED diodes (LB115A-6W-X, Yuyao Lianliang Electric Appliance Co Ltd, Ningbo, China). After returning them to the laboratory, the strains were cultivated at 15°C (cooling box C5G, Helkama Oy, Helsinki, Finland), under illumination of 40 μmol m−2 s−1 and a 16-h light : 8-h dark cycle (TLD 18W/33 fluorescent lamps, Philips, Amsterdam, the Netherlands). In addition, isolated colonies were separately transferred to 200 μl PCR tubes and subsequently frozen at −24°C.
Table 1. Origin and sampling details of analysed strains.
DNA extraction, PCR and DNA sequencing
For DNA isolation, 100 µl of the growing culture was centrifuged in PCR tubes (6000 rpm for 3 min), and 30 µl of InstaGene matrix (Bio-Rad Laboratories, Hercules, CA, USA) was added to the pellet. The solution was vortexed for 10 s, incubated at 56°C for 30 min, and heated at 99°C for 8 min. After vortexing a second time, the tubes were centrifuged at 12000 rpm for 2 min, and the supernatant was directly used as a PCR template. Three molecular markers were amplified by PCR: nuclear ITS rDNA, chloroplast rbcL and mitochondrial coxI. The amplification of ITS rDNA was performed as described in Kynčlová et al. (Citation2010), using the primers Kn1.1 (5ʹ-CAA GGT TTC CGT AGG TGA ACC-3ʹ; Wee et al., Citation2001) and ITS4 (5ʹ-TCC TCC GCT TAT TGA TAT GC-3ʹ; White et al., Citation1990). Amplification of the rbcL marker was performed according to Jo et al. (Citation2011), using the primers rbcL_2F (5ʹ-AAA AGT GAC CGT TAT GAA TC-3ʹ; Daugbjerg & Andersen, Citation1997) and rbcL_R3 (5ʹ-GTA ATA TCT TTC TTC CAT AAA TCT AA-3ʹ; Jo et al., Citation2011). The cox1 marker was amplified according to Boo et al. (Citation2010), using the primers F692 (5ʹ-TTG TDT GGT CAG TTT TAA TTA C-3ʹ) and R1433 (5ʹ-GGC ATA CCT GCW ARA CCT AA-3ʹ; Boo et al., Citation2010). The PCR products were purified and sequenced at Macrogen Inc. in Seoul, Korea.
Phylogenetic analyses
A multiple alignment of the newly determined ITS rDNA, rbcL and coxI gene sequences and other sequences selected from the GenBank/EMBL/DDBJ databases was built using MAFFT, version 6, applying the Q-INS-i strategy (Katoh et al., Citation2002). The sequences were selected to encompass all known lineages within the Synura petersenii species complex (Table S1). ITS rDNA sequences were then aligned on the basis of their rRNA secondary structure information (Kynčlová et al., Citation2010) with MEGA 4 (Kumar et al., Citation2008). The three loci were concatenated, yielding an alignment of 2308 bases. The final matrix contained 51 ITS rDNA, 36 rbcL and 33 coxI sequences. A suitable partitioning strategy and partition-specific substitution models were selected in a multistep process (Verbruggen et al., Citation2010). Initially, a guide tree was obtained by carrying out a second-level maximum likelihood (ML) search on the unpartitioned dataset with a HKY + Γ8 model using TreeFinder (Jobb et al., Citation2004). Then, the dataset was divided by 17 different partitioning strategies (combining different levels of locus segmentation). Subsequently, Bayesian information criterion (BIC) calculations were performed for all 17 potential partitioning strategies, assuming the guide tree and HKY + Γ8 model for each partition. The five best-scoring partitioning strategies (lowest BIC scores) were retained for further analysis. In the next step, models of sequence evolution were selected for individual partitions using BIC. For each partition present in the five retained partitioning strategies, 12 different nucleotide substitution models were evaluated (F81, HKY, GTR and their combinations with Γ, I and Γ + I). Finally, the partitioning strategies were re-evaluated using the selected models for the particular partitions. This BIC-based model selection procedure selected the following model with eight partitions: (1) internal transcribed spacers ITS1 and ITS2 – GTR + Γ; (2) 5.8 S ribosomal locus – F81; (3), first codon position of the rbcL gene – GTR + Γ; (4) second codon position of the rbcL gene – F81; (5) third codon position of the rbcL gene – GTR + Γ; (6) first codon position of the coxI gene – GTR + Γ; (7) second codon position of the coxI gene – F81 + Γ; and (8) third codon position of the coxI gene – GTR + Γ.
The phylogenetic tree was inferred with Bayesian inference (BI) using MrBayes version 3.1.2 (Ronquist & Huelsenbeck, Citation2003). Two parallel MCMC runs were carried out for 10 million generations. The dataset was divided into eight partitions, for which different substitution models were selected according to the BIC-based model selection. Trees and parameters were sampled every 100 generations. Convergence of the two runs was assessed during the run by calculating the average standard deviation of split frequencies (SDSF). The SDSF value between simultaneous runs was 0.003908. The burn-in was determined using the ‘sump’ command. Bootstrap analyses were performed with ML and weighted parsimony (wMP) criteria using GARLI version 0.951 (Zwickl, Citation2006) and PAUP* version 4.0b10 (Swofford, Citation2002), respectively. ML analyses consisted of rapid heuristic searches (100 pseudoreplicates) using automatic termination (genthreshfortopoterm command set to 100 000). The dataset was divided into eight partitions with different substitution models. The wMP bootstrapping (1000 replications) was performed using heuristic searches with 100 random sequence addition replicates, tree bisection reconnection swapping, random addition of sequences (the number limited to 10 000 for each replicate), and gap characters treated as a fifth character state. Character weights were assigned using the rescaled consistency index on a scale of 0 to 1000. New weights were based on the mean fit values for each character over all trees in the memory.
Morphological investigations and statistical analyses
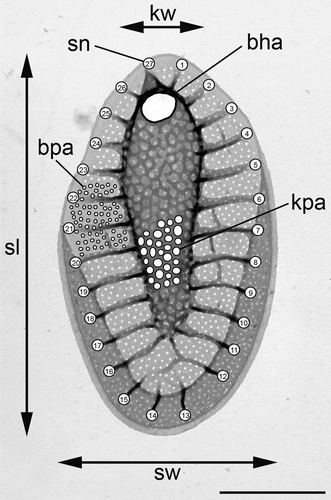
For morphological observations, the strains were cultivated in 50-ml Erlenmeyer flasks for 1–2 months. To avoid depletion of nutrients, the strains were grown under lower light and temperature regimes. Light microscopy (LM) observations were performed using an Olympus BX51 microscope. For scanning electron microscopy (SEM) observations, a drop of glutaraldehyde (GA)-fixed (2% GA overnight) cell suspension was sedimented for 60 min on polyL-lysine-coated glass coverslips to ensure appropriate cell adhesion. The coverslips were washed by repeated transfer into drops of deionized water dispensed onto the hydrophobic surface of a Parafilm strip, and subsequently dehydrated via acetone series. The cells were dried to a critical point with liquid carbon dioxide (Bal-Tec CPD 030), coated with platinum for 90 s with a Bal-Tec SCD 050 sputter coater, and observed with a JEOL JSM-740 1F FESEM scanning electron microscope. For transmission electron microscopy (TEM) investigations of silica scales, a drop from the living cultures was placed onto formvar-coated copper grids and dried. After washing in a series of water droplets, the grids were examined in a TEM JEOL 1011 electron microscope. At least three strains were randomly chosen from each of the clades identified by the molecular study, including the cultures deposited in the Culture Collection of Algae of Charles University in Prague (CAUP) and the National Center for Marine Algae and Microbiota in Maine (designated as CCMP). For each clade, the following characters were measured in 30 randomly selected scales: (1) scale length; (2) scale width; (3) area of a base hole; (4) average area of a keel pore; (5) average area of a base-plate pore; (6) keel width; and (7) number of struts (). The measurements were performed using the program ImageJ 1.45 s (Schneider et al., Citation2012). Average values of keel and base-plate pores were obtained by measuring areas of approximately 30 and 60 pores per scale, respectively. Statistical analyses of measured data (principal component and canonical discriminant analyses) were performed using Statistica 8.0 (StatSoft, Inc., Tulsa, Oklahoma, USA).
Results
Phylogenetic analyses
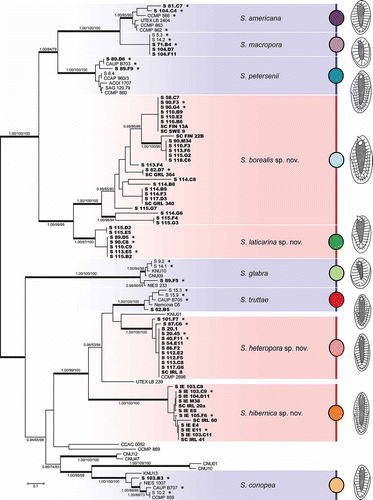
The Bayesian analysis based on the concatenated dataset (ITS rDNA, rbcL and cox1 sequences) revealed the existence of at least 16 lineages within the Synura petersenii species complex (). In addition to the six described species (S. americana, S. conopea, S. glabra, S. macropora, S. petersenii and S. truttae) and lineages resolved in previous studies, the phylogenetic analysis detected the existence of four distinct, novel lineages. The first two lineages (here referred to as S. borealis sp. nov. and S. laticarina sp. nov.) formed a well-supported clade together with S. americana, S. macropora and S. petersenii. The latter two newly recognized lineages (here referred to as S. heteropora sp. nov. and S. hibernica sp. nov.) were inferred to be members of another well-supported clade including S. truttae and the strains UTEX LB 239 and KNU01. Synura borealis represented the most diverse novel lineage, containing a number of isolates clustering into several distinct lineages. The most common genotype was represented by eight isolates originating from Estonia, Finland, Norway and Sweden. By contrast, S. laticarina was represented by seven sequences clustered into two related sub-clades. The third novel lineage, S. heteropora, consisted of 13 isolates originating from Austria, Czech Republic, Estonia, Great Britain, Ireland, Sweden and Norway. The strain CCMP 2898, which was isolated from an Austrian lake, was inferred as a member of this lineage. The last newly recognized lineage, S. hibernica, consisted of 12 isolates that all originated from western Ireland.
Fig. 2. Phylogeny of the genus Synura, section Peterseniae, obtained by Bayesian inference of the concatenated ITS rDNA, rbcL and cox1 dataset. The analysis was performed under a partitioned model, using different substitution models for each partition. Values at the nodes indicate statistical support estimated by three methods; MrBayes posterior node probability (left), maximum likelihood bootstrap (middle), and maximum parsimony bootstrap (right). Only statistical supports higher than 0.90/50/50 are shown. Thick branches highlight nodes receiving the highest posterior probability (PP) support (1.00). Newly sequenced strains are marked in bold. Strains used for the statistical analyses of morphological features are marked by asterisks. Scale bar represents the expected number of substitutions per site.
Morphological observations and taxa descriptions
Molecular phylogenetic analyses show the existence of four novel lineages in the genus Synura, section Peterseniae. The detailed TEM investigation of siliceous scales demonstrated their clear distinctness from all species without molecular characterization (S. obesa, S. australiensis, ‘S. petersenii’ f. columnata, ‘S. petersenii’ f. praefracta, and ‘S. petersenii’ f. taymyrensis). Therefore, these novel lineages represent four new species, which we describe and illustrate below.
Synura borealis Škaloud & Škaloudová, sp. nov.
()
Figs. 3–10. Scale morphology of Synura borealis sp. nov. (Fig. 3: LM; Figs 4, 5: SEM; Figs 6–10: TEM). Scale bars represent 10 µm (Fig. 3) and 1 μm (Figs 4–10). Fig. 3. Colony consisting of elongated, lanceolate-shaped cells (strain S 90.G4). Fig. 4. Single cell surrounded by a layer of siliceous scales (S 90.G4). Fig. 5. Body scale (S 90.G4). Fig. 6. Body scale with transverse folds interconnecting the struts. Note the over-layered pore pattern at the scale keel (S 62.D7). Fig. 7. Body scale with obviously anteriorly widened keel (S 62.D7). Fig. 8. Apical scale (S 62.D7). Fig. 9. Apical scales with prominently protruding keel tips (S 58.C7). Fig. 10. Rear scale (S 62.D7).
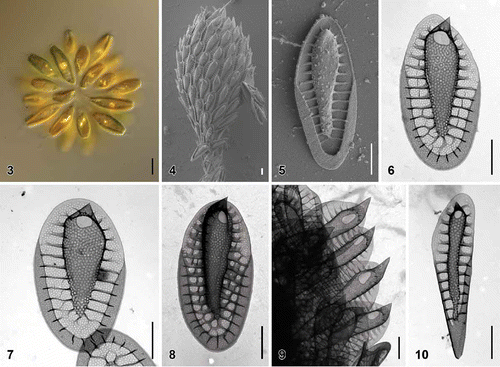
| Description: | = | Colonies are spherical, up to 86 µm in diameter, consisting of approximately 16–38 cells associated by their posterior ends. Cells are significantly elongated, lanceolate-shaped, posteriorly elongated into the tail, 31–42 µm long and 7–12 µm wide (). Each cell is surrounded by a layer of imbricate siliceous scales (). Body scales are 4.0–5.8 µm long and 1.6–2.6 µm wide, consisting of a basal plate with a centrally raised keel protruding into an acute tip ( and ). The keel is often anteriorly widened, ornamented by medium-sized pores (diameter, 54–88 nm). Anteriorly, keel pores are produced on both sides, so that the keel pore pattern is notably over-layered (). The ratio between scale and keel width varies from 1.7 to 2.9. The basal plate is ornamented by numerous small pores (diameter, 17–26 nm), and anteriorly perforated by a large, rounded or elongated base hole (diameter, 0.27–0.55 µm). Numerous struts (28–38), often interconnected by transverse folds, extend regularly from the keel to the scale perimeter. Apical scales are 3.2–4.2 µm long and 1.7–2.5 µm wide (). The keel of the apical scales ends in a prominent, acute tip (). Rear scales are long and narrow, 4.3–5.7 µm long and 1.1–1.6 µm wide (). |
| Etymology: | = | The specific epithet ‘borealis’ refers to the northern (boreal) occurrence of the species, which is found in northern Europe and Greenland. |
| Type locality: | = | Unnamed lake, Disko Island, Greenland (69.28913°N, –53.49322°W). |
| Holotype: | = | Synura borealis strain S 62.D7, frozen material deposited in the Culture Collection of Algae of Charles University in Prague (CAUP). presents an illustration of the holotype. |
| Holotype DNA barcode: | = | GenBank Accession no. HG514176. |
| Distribution (): | = | Estonia, Finland, Greenland, Norway and Sweden. |
Synura heteropora Škaloud, Škaloudová & Procházková, sp. nov.
()
Figs. 11–19. Scale morphology of Synura heteropora sp. nov. (Fig. 11: LM; Figs 12, 13, 16: SEM; Figs 14, 15, 17–19: TEM). Scale bars represent 10 μm (Fig. 11) and 1 μm (Figs 12–19). Fig. 11. Colony consisting of densely grouped, pyriform cells (strain S 20.45). Fig. 12. Single cell surrounded by a layer of siliceous scales (S 54.E11). Fig. 13. Body scale (S 54.E11). Fig. 14. Body scale (S 20.45). Fig. 15. Body scale with transverse folds interconnecting the struts (S 87.C6). Fig. 16. Apical scale (S 54.E11). Fig. 17. Apical scale with a pronounced, rounded keel tip (S 101.F7). Figs 18, 19. Rear scales (S 101.F7).
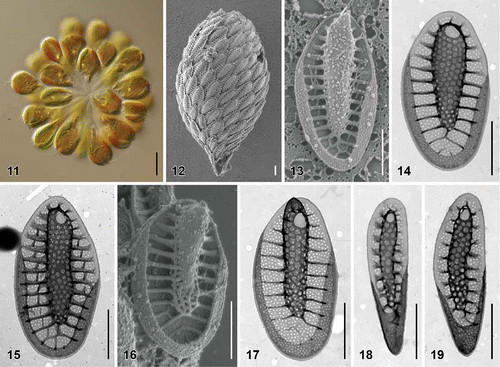
| Description: | = | Colonies are spherical, up to 50 µm in diameter, consisting of approximately 16–36 cells associated by their posterior ends (). Cells are pyriform, posteriorly elongated into the tail, 20–25 µm long and 7–11 µm wide. Each cell is surrounded by a layer of imbricate siliceous scales (). Body scales are 2.5–3.8 µm long and 1.1–1.9 µm wide, consisting of a basal plate with a centrally raised keel protruding into an acute tip (). The keel is cylindrical, occasionally slightly widened anteriorly, and ornamented by larger pores (diameter, 49–100 nm) (). The ratio between scale and keel width varies from 1.7 to 3.9. The basal plate is ornamented by numerous small pores (diameter, 20–31 nm), and anteriorly perforated by a rounded base hole (diameter, 0.19–0.42 µm). Numerous struts (22–28), often interconnected by transverse folds, extend regularly from the keel to the scale perimeter (). Apical scales are 2.6–3.0 µm long and 1.3–1.6 µm wide (). The keel of the apical scales ends in a rounded tip (). Rear scales are 1.8–3.2 µm long and 0.6–1.0 µm wide (, ). |
| Etymology: | = | Named for the different sizes of the keel- and base-plate pores. |
| Type locality: | = | Crinan Canal, Scotland, UK (56.060461° N, 5.481813° W). |
| Holotype: | = | Synura heteropora strain S 20.45, frozen material deposited in the Culture Collection of Algae of Charles University in Prague (CAUP). presents an illustration of the holotype. |
| Holotype DNA barcode: | = | GenBank Accession no. HG514198. |
| Distribution (): | = | Austria, Czech Republic, Estonia, Ireland, Norway, Sweden and the UK. |
Synura hibernica Škaloud & Škaloudová, sp. nov.
()
Figs. 20–28. Scale morphology of Synura hibernica sp. nov. (Fig. 20: LM; Figs 21, 22: SEM; Figs 23–28: TEM). Scale bars represent 10 μm (Fig. 20) and 1 μm (Figs 21–28). Fig. 20. Colony consisting of significantly elongated cells (strain S IE 104.D11). Fig. 21. Single cell surrounded by a layer of siliceous scales (S IE 104.D11). Fig. 22. Body scale (S IE 104.D11). Fig. 23. Body scale (S IE 103.C9). Fig. 24. Body scale (S IE E11). Fig. 25. Apical scale with prominently protruding keel tip (S IE E11). Fig. 26. Lateral view on apical scales with protruding keel tips (environmental sample, Lough Anillaun, Co. Galway, Ireland). Fig. 27. Rear scale (S IE M38). Fig. 28. Rear scale (S IE E11).
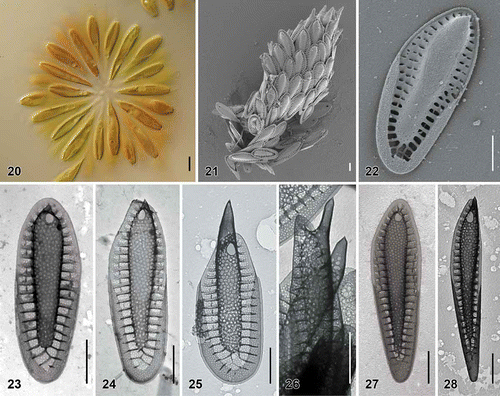
| Description: | = | Colonies are spherical, up to 94 µm in diameter, consisting of approximately 12–52 cells associated by their posterior ends (). Cells are significantly elongated, anteriorly cylindrical, posteriorly tapering into the tail, 26–47 µm long and 6–12 µm wide. Each cell is surrounded by a layer of imbricate siliceous scales (). Body scales are 3.4–5.6 µm long and 1.2–2.0 µm wide, consisting of a basal plate with a centrally raised keel protruding into an acute tip (). The keel is cylindrical, ornamented by medium-sized pores (diameter, 49–89 nm) (). The ratio between scale and keel width varies from 1.9 to 3.9. The basal plate is ornamented by numerous small pores (diameter, 18–27 nm), and anteriorly perforated by a rounded base hole (diameter, 0.14–0.38 µm). A large number of struts (30–47), often interconnected by transverse folds, extend regularly from the keel to the scale perimeter (). Apical scales are 2.9–3.3 µm long and 1.6–1.9 µm wide (). The keel of the apical scales ends in a prominent, acute tip (), sometimes terminated by two short teeth. Rear scales are long and narrow, 3.0–4.9 µm long and 0.9–1.0 µm wide (). |
| Etymology: | = | The specific epithet ‘hibernica’ refers to the common occurrence of the species in Ireland. |
| Type locality: | = | Glanmore Lake, Ireland (51.732724° N, 9.767121° W). |
| Holotype: | = | Synura hibernica strain S IE 104.D11, frozen material deposited in the Culture Collection of Algae of Charles University in Prague (CAUP). presents an illustration of the holotype. |
| Holotype DNA barcode: | = | GenBank Accession no. HG514216. |
| Distribution (): | = | Currently known only in Ireland. |
Synura laticarina Škaloud & Škaloudová, sp. nov.
()
Figs. 29–36. Scale morphology of Synura laticarina sp. nov. (Fig. 29: LM; Figs 30 and 31: SEM; Figs 32–36: TEM). Scale bars represent 10 μm (Fig. 29) and 1 μm (Figs 30–36). Fig. 29. Colony consisting of pyriform cells (strain S 90.C8). Fig. 30. Single cell surrounded by a layer of siliceous scales (S 90.C8). Fig. 31. Body scale (S 90.C8). Fig. 32. Body scale with anteriorly widened keel (S 89.D5). Fig. 33. Body scale. Note over-layered pore pattern at the scale keel (S 89.D5). Fig. 34. Body scale with transverse folds interconnecting the struts (S 90.C8). Fig. 35. Apical scale (S 89.D5). Fig. 36. Rear scale (S 89.D5).
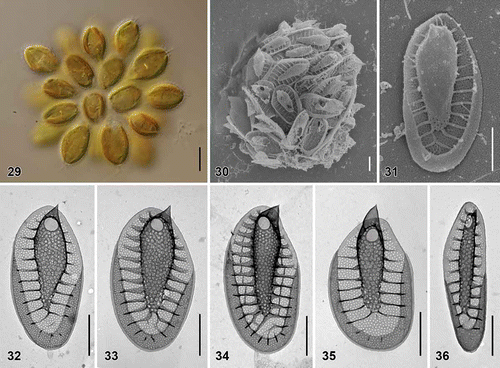
| Description: | = | Colonies are spherical, up to 64 µm in diameter, consisting of approximately 12–28 cells associated by their posterior ends (). Cells are pyriform, posteriorly elongated into the tail, 21–32 µm long and 7–13 µm wide. Each cell is surrounded by a layer of imbricate siliceous scales (). Body scales are 3.1–4.3 µm long and 1.6–2.1 µm wide, consisting of a basal plate with a centrally raised keel protruding into an acute tip (). The keel is often anteriorly widened, ornamented by medium-sized pores (diameter, 52–76 nm) (). Anteriorly, keel pores are produced on both sides, so that the keel pore pattern is notably over-layered (). The ratio between scale and keel width varies from 1.9 to 2.9. The basal plate is ornamented by numerous small pores (diameter, 19–25 nm), and anteriorly perforated by a rounded base hole (diameter, 0.20–0.33 µm). Numerous struts (24–32), often interconnected by transverse folds, extend regularly from the keel to the scale perimeter (). Apical scales are 2.5–3.0 µm long and 1.7–2.0 µm wide (). The keel of the apical scales ends in a prominent, sometimes rounded tip. Rear scales are 2.7–4.3 µm long and 0.7–1.3 µm wide (). |
| Etymology: | = | The specific epithet is derived from the Latin ‘latus’ (broad) and ‘carina’ (keel), referring to the remarkable anterior widening of the keel. |
| Type locality: | = | Ojala Lake, Finland (61.847778° N, 26.291447° E). |
| Holotype: | = | Synura laticarina strain S 89.D5, frozen material deposited in the Culture Collection of Algae of Charles University in Prague (CAUP). presents an illustration of the holotype. |
| Holotype DNA barcode: | = | GenBank Accession no. HG514221. |
| Distribution (): | = | Estonia, Finland, Norway and Sweden. |
Morphological analyses
To investigate morphological differences among the 10 recognized, closely related Synura species in detail, we morphologically characterized each species by measuring 30 randomly chosen body scales. The species were obviously heterogeneous in the size of body scales (). Synura borealis, S. hibernica and S. petersenii had significantly longer scales than those of the remaining species. In addition, two groups of species could be recognized by the scale width: S. conopea, S. heteropora, S. hibernica and S. truttae had much narrower scales than those of the other species. Obvious morphological differences also were observed in the number of struts (), which represented the least variable morphological character. Therefore, some species could be clearly recognized only by counting the number of struts in a few scales. Heterogeneity also was observed in the size of all three pores measured. The base-plate pore area was the most discriminating character among the species (), with exceptionally large pores presented in S. macropora. All four newly recognized species were characterized by rather small base-plate pores. Both the keel pore and the base hole area showed relatively high size variability, which restricted their use as good discriminant features (). Synura borealis and S. truttae were well characterized by a notably large base hole. Strong heterogeneity was observed in the base-plate area to keel area ratio (). The keel pores of S. macropora and S. glabra were approximately comparable in size to the base-plate pores; however, those of S. borealis, S. heteropora, S. hibernica and S. laticarina were usually distinctively larger. Synura americana, S. hibernica and S. laticarina were well characterized by an anteriorly widened keel (). The central morphological characteristics of these 10 lineages, and other members of the genus Synura, section Peterseniae, are presented in . A key to these species is given in Table S2.
Figs. 37–42. Morphological comparisons and statistical analyses of siliceous scales. Figs 37–40. Scatterplots of morphological features measured in the 10 investigated species; average values and standard deviations are given. Fig. 37. Scatterplot of scale length versus scale width. Fig. 38. Scatterplot of base-plate pore area versus number of struts. Fig. 39. Scatterplot of base hole area versus keel pore area. Fig. 40. Scatterplot of keel pore to base-plate pore area ratio versus keel width. Fig. 41. Principal component analysis (PCA) of the entire measured morphological features dataset. Fig. 42. Canonical discriminant analysis (CDA) of the same dataset.
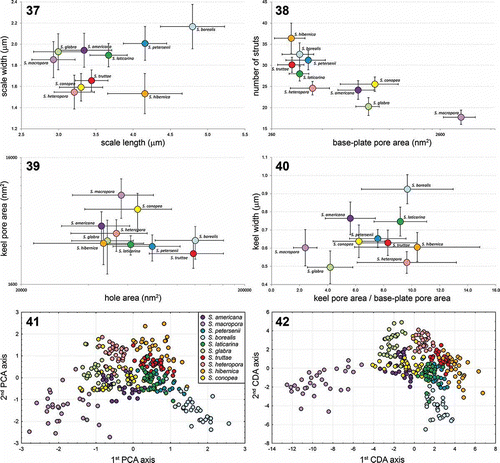
Table 2. Scale characteristics of selected Synura species (section Peterseniae) with similar scale morphologies. Newly described species are given in bold. For terminology of measured characteristics, see .
The principal component analysis (PCA) of the entire dataset resulted in a relatively well-defined grouping of scales belonging to the particular species (). Scales of S. macropora and S. borealis formed two distinct clusters with negative values on the first PCR axis. By contrast, scales of S. conopea were mostly intermixed with other species. The canonical discriminant analysis (CDA) yielded much better grouping of scales based on their morphological data (). With the exception of S. conopea and S. americana, all species formed distinct, separate clusters. The discriminant analysis (DA) indicated strongly significant differentiation among the 10 species (Wilk’s λ = 0.0008; P < 0.00001). The forward stepwise analysis indicated that all tested morphological characters were significant for species recognition (P < 0.00001), and selected the base-plate pore area (Partial Wilk’s λ = 0.20), the keel width (Partial Wilk’s λ = 0.41) and the number of struts (Partial Wilk’s λ = 0.46) as the three best discriminating characters. The discrimination of species was highly significant even when only these three characters were analysed (Wilk’s λ = 0.0058; P < 0.00001). Congruently, whereas the first CDA axis was principally correlated with the base-plate pore area and the number of struts (correlation coefficients −0.82 and 0.63, respectively), the second CDA axis correlated with keel and scale widths (correlation coefficients 0.68 and 0.64, respectively). The average correct discrimination of individual scales on the basis of their morphology reached 92% (). The lowest correct discrimination levels were recovered in S. conopea (70.0%) and S. laticarina (80.0%).
Table 3. Classification matrix of the canonical discriminant analyses of all seven morphological characters/three best discriminating characters (base-plate pore area, keel width, number of struts). Rows: observed classification. Columns: predicted classification.
Biogeography
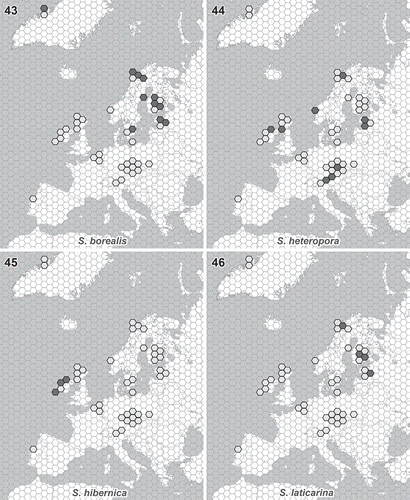
A 6-year sampling of Synura isolates performed in 15 different countries enabled us to determine their biogeographic distribution across the European continent. Different distribution patterns can be recognized in four newly described Synura species (). Synura heteropora is distributed across much of Europe, whereas the remaining three species are more restricted in their occurrence. Evolutionarily related S. borealis and S. laticarina exhibit similar biogeographic patterns, occurring in the northern regions (). Synura laticarina is regionally more restricted, currently reported only from Estonia, Finland, Norway and Sweden (). The most restricted distribution pattern was observed in S. hibernica. This species was found only in the blanket peat bogs located along the western coast of Ireland ().
Figs. 43–46. Distribution of newly described Synura species. Light grey hexagons show all studied regions where the occurrence of any Synura species has been recorded. Filled hexagons show the known distribution pattern of the new species. Fig. 43. S. borealis. Fig. 44. S. heteropora. Fig. 45. S. hibernica. Fig. 46. S. laticarina. The hexagon edge length corresponds to 80 km (hexagon area ≈ 16 600 km2).
Discussion
Synura petersenii sensu lato is one of the most widely distributed and common groups of freshwater microorganisms. It is relatively easily cultivated and investigated with molecular methods (Wee et al., Citation2001; Boo et al., Citation2010; Kynčlová et al., Citation2010; Škaloud et al., Citation2012, Citation2013a), so it represents an ideal model taxon for investigating evolutionary patterns in protists. Particular species are relatively young in evolutionary terms, diverging on the order of several million years ago (Jo et al., Citation2013). Therefore, it allows us to investigate speciation processes, rates of morphological and ecological differentiation of species, and overall species diversity within this recently diverging group of microorganisms. This study was the next step necessary for uncovering the real species diversity and to delimit and characterize particular species within this complex.
Overall species diversity
Biological variation within Synura petersenii is very complex. Before information was available about genetic differentiation, S. petersenii was generally considered as a single species, with recognition of a few forms or varieties (Kristiansen & Preisig, Citation2007). In fact, this taxon contains a number of morphologically similar, yet genetically distinct species. In addition to the four newly proposed species, S. petersenii s.l. now includes 10 well-defined, genetically characterized species. Phylogenetic analysis clearly recognizes six lineages comprised of several strains deposited in various culture collections, which very probably represent new, yet undescribed species (). Several taxa belonging to the section Peterseniae are currently uncharacterized with molecular markers, and their phylogenetic position is still unknown. All these taxa, including S. australiensis (Playfair, Citation1915), ‘S. petersenii’ f. columnata (Siver, Citation1988), ‘S. petersenii’ f. praefracta (Asmund, Citation1968), ‘S. petersenii’ f. taymyrensis (Kristiansen et al., Citation1997), and S. obesa (Němcová et al., Citation2008), could form additional lineages within S. petersenii s.l. Therefore, it is very difficult to estimate the total number of species in this taxon. In an extensive molecular investigation of more than 100 S. petersenii s.l. strains, Boo et al. (Citation2010) identified a large number of genotypes clustered into seven groups. The strains originated from different areas located in four continents. Of the four new species proposed in the present study, only S. heteropora was identified with a previously recognized genotype, the strain CCMP 2898 belonging to clade G I Boo et al. (Citation2010). Obviously, there is still a large degree of hidden diversity that cannot be fully resolved without additional molecular and morphological data. However, even if we can predict that several tens of currently undiscovered taxa exist, it is probable that the most frequently occurring species are already known and characterized.
Species delineation and morphological evolution
The six species delineated in previous studies (Kynčlová et al., Citation2010; Škaloud et al., Citation2012) were shown to be distinguishable by the siliceous scale morphology. However, the present study includes descriptions of four additional, morphologically highly similar taxa. Morphometric analyses of siliceous scales enabled the significant phenotypic differentiation of all species of the S. petersenii complex, including the newly described species. The CDA analysis significantly recognized all 10 investigated species (), and identified S. conopea as the species with the lowest correct discrimination level (70%). When analysing only the three best discriminating characters (base-plate pore area, keel width and number of struts), the correct discrimination level decreased to 57% in S. conopea and S. petersenii, but remained quite high in the majority of species (). Because the presented percentages represent post-hoc classifications, we can expect a lower accuracy when new scales will be classified. However, the CDA analyses clearly indicated that simply measuring three morphological features in four body scales should be sufficient to correctly classify each of the species.
We detected a new lineage sister to S. americana, S. glabra and S. petersenii that we interpreted as two novel species, the genetically diverse S. borealis and the more molecularly homogeneous S. laticarina (). Synura borealis possessed an exceptionally high degree of genetic diversity, particularly in the cox1 gene, which could be interpreted as resulting from several independent species. However, the recognition of just these two species was strongly supported by our morphological data. The silica scales of all S. borealis strains shared characteristic morphological features, including the anteriorly widened keel with notably over-layered pore pattern, and large scale dimensions. Further, S. borealis and S. laticarina species could be undoubtedly recognized under the light microscope. All investigated S. borealis strains were characterized by large colonies consisting of significantly elongated, lanceolate-shaped cells, whereas S. laticarina produced notably smaller colonies containing rounded, pyriform cells. Therefore, we interpret the high genetic diversity in S. borealis as an indication of recent speciation and ongoing evolutionary differentiation. Further investigation of S. borealis genotypes, including their ecophysiological differentiation, ecological preferences and distributional patterns could provide important insights into the drivers of species diversification and mechanisms of population-level structuring in Synura species.
Particular species of the S. petersenii group obviously underwent some degree of morphological differentiation, although they are relatively young in evolutionary terms. Some species were more significantly differentiated with respect to siliceous scales than others, and can be recognized at first glance. For example, S. macropora can be unambiguously identified by its extremely large base-plate pores (). We can only speculate about the causes of this rapid morphological evolution. Large pores could significantly decrease the density of the cell, and hence reduce the sinking rate. However, cells with larger pores could be more susceptible to viral infection (Losic et al., Citation2006). Synura macropora seems to occur only in eutrophic habitats characterized by high conductivity values (; Škaloud et al., Citation2013b). It was shown that eutrophic conditions decreased the availability of silica, which often caused a shift from heavily silicified to less silicified diatoms in freshwater biotopes (Rabalais et al., Citation1996). Similarly, adaptation of S. macropora to eutrophic conditions could lead to the formation of less silicified scales characterized by large base-plate pores.
Synura hibernica represents another morphologically well-defined species characterized by very narrow, long siliceous scales bearing a large number of struts (). This species can even be distinguished under the light microscope by the characteristically large colonies consisting of strongly elongated, narrow cells. The significantly different morphology of this Irish taxon has been discussed previously by Řezáčová & Škaloud (Citation2005), who emphasized its striking similarity with the tropical species S. australiensis. Obviously, the characteristically elongated shape of the siliceous scales is related to the elongated cell shape in both species. Synura australiensis and S. hibernica represent another example of rapid morphological evolution within the S. petersenii group, leading to the speciation of organisms producing big colonies consisting of long and narrow cells. Unfortunately, in the absence of molecular data for S. australiensis, it remains unclear whether these two taxa are evolutionarily related to each other. Significant morphological transformation may have been caused by an ecological niche-shift during the speciation of both taxa, similarly to that of S. macropora. In protists, a negative correlation between cell size and temperature has been proposed by Atkinson et al. (Citation2003). However, S. hibernica is currently known only from Ireland, whereas the distribution of S. australiensis is restricted to tropical and subtropical regions (Kristiansen & Preisig, Citation2007). Therefore, we can rule out temperature-related morphological speciation of these two taxa.
The development of elongated cells in S. australiensis and S. hibernica could be explained by adaptation to oligotrophic conditions. It is known that most oligotrophs achieve a high surface-to-volume ratio, which increases the organisms’ capacity to scavenge available nutrients (Reynolds, Citation2006). Elongation of Synura cells would be the most effective way to significantly increase this ratio. All 12 isolated strains of S. hibernica originated from nutrient-poor localities, with conductivity values ranging between 23–87 µS cm−1 (). To our knowledge, all records of S. australiensis have been associated with clear, oligotrophic water bodies (Croome & Tyler, Citation1985; Cronberg, Citation1989, Citation1996; Saha & Wujek, Citation1990; Wee et al., Citation1993; Hansen, Citation1996). The measured conductivity of these water samples was 10–123 µS cm−1.
Biogeographic patterns
The biogeography of protists has become a highly controversial topic over the last 10 years (Martiny et al., Citation2006; Caron, Citation2009). It has been postulated that the small size, extremely large populations and high dispersal potential of protists result in the cosmopolitan distribution of the vast majority of species (Finlay, Citation2002; Finlay & Fenchel, Citation2004). Conversely, limited geographical distributions have been implied by Foissner (Citation1999), primarily based on the observed restricted distribution of flagship species with easily recognizable morphologies and easily demonstrable presence/absence (Foissner, Citation2006, Citation2008). Synura petersenii s.l. represents a common group of freshwater microorganisms, known to occur in all continents except Antarctica (Kristiansen, Citation2008). Because its molecular diversity has been investigated in three continents (Wee et al., Citation2001; Boo et al., Citation2010; Škaloud et al., Citation2012), it represents an ideal taxon to study the distribution patterns in protists.
The first evaluation of biogeographic patterns in the S. petersenii group was published by Boo et al. (Citation2010), who described the presence of restricted biogeographic distributions for many species. However, Škaloud et al. (Citation2012) compared the genetic data with published morphological observations, and reported much broader distributions for some of the investigated taxa. For example, putative North American endemic S. americana, although dominantly occurring in the USA and Canada, was also reported in South America and Europe (Cronberg, Citation1989; Škaloud et al., Citation2012). However, the restricted distribution of several lineages is evident (e.g. clades D and F sensu Boo et al. Citation2010). Our results support the restricted distribution of Synura species, including the strict regional endemism observed in S. hibernica. To the best of our knowledge, all four newly proposed species have been reported only in Europe and Greenland (Figs 43–46). A comparison of the morphological characteristics with previously published reports reveals that S. borealis and S. hibernica have been observed previously in Greenland and Ireland, respectively (Kristiansen, Citation1992; Řezáčová & Škaloud, Citation2005); however, we found no other reports outside Europe.
Three of four newly described species exhibit restricted biogeographic distribution. Evolutionarily related S. borealis and S. laticarina seem to be adapted to colder areas, and their distributions appear to be limited by high summer temperatures (). The occurrence of S. borealis in Greenland indicates no obvious dispersal limitations, so we expect its circumboreal distribution. The most remarkable distribution pattern, however, is that of S. hibernica. This species is restricted in its geographic distribution to western Ireland (). Although we found S. hibernica in nine localities located along the western coast of Ireland (), it was never detected in any of the other sampled localities. To check this highly unusual distribution pattern, we undertook two expeditions in 2008 and 2012 to investigate the diversity of Synura species in neighbouring Scotland. Ecological and limnological conditions are very similar in western Ireland and Scotland: both areas are characterized by the occurrence of extensive peat deposits restricted to a cool oceanic climate, known as blanket bogs (Moore, Citation1982). Moreover, the British Isles lie at the intersection of major bird migration corridors, and several bird species frequently pass through one part of the Isles to reach another (Newton, Citation2010). Therefore, the algal flora of the western British lakelands, including western Ireland and northwestern Scotland, are highly similar. The significant similarity of western British desmid flora was reported previously (West & West, Citation1909). Although we sampled a total of 71 localities in northwestern Scotland, we did not discover a single colony of S. hibernica. This distribution pattern, which is restricted to an extremely small biogeographic area, is remarkable and entirely outstanding. Future work should study the mechanisms underlying the obvious dispersal limitation of S. hibernica, including ecological and habitat requirements, desiccation tolerance and mechanisms of cyst formation.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at http://dx.doi.org/10.1080/09670262.2014.905710
Table S1. Strain names, geographic origins, and the corresponding ITS rDNA, rbcL, and cox1 GenBank accession numbers for the taxa used in the phylogenetic analyses. Newly obtained sequences are given in bold.
Table S2. Key to species of the genus Synura, section Peterseniae.
Alignment S1. The final, partitioned alignment of concatenated ITS rDNA, rbcL, and cox1 sequences used for phylogenetic reconstructions.
Supplementary material
Download Zip (37.2 KB)Acknowledgements
The authors would like to thank Fabio Rindi and Michael D. Guiry (Ryan Institute, National University of Ireland, Galway); Thomas Pröschold and John G. Day (Culture Collection of Algae and Protozoa, Scottish Marine Institute, Oban, UK); David John (Natural History Museum, London, UK); Ole Stecher and Frantz Nielsen (Arctic Station, University of Copenhagen, Greenland); Eugen Rott (Institute of Botany, University of Innsbruck, Austria); Helena Bestová, Lucie Jelínková, Katarína Nemjová, Pavla Slámová, Pavel Svoboda and Vojtěch Scharfen (Department of Botany, Charles University in Prague, Czech Republic) for their kind support during sampling. This work was supported by the Czech Science Foundation [grant number P506/11/P056].
References
- Andersen, R.A., Morton, S.L. & Sexton, J.P. (1997). Provasoli-Guillard National Center for Culture of Marine Phytoplankton 1997 – list of strains. Journal of Phycology, 33 (suppl.): 1–75.
- Asmund, B. (1968). Studies on Chrysophyceae from some ponds and lakes in Alaska. VI. Occurrence of Synura species. Hydrobiologia, 31: 497–515.
- Atkinson, D., Ciotti, B.J. & Montagnes, D.J.S. (2003). Protists decrease in size linearly with temperature: ca. 2.5% °C-l. Proceedings of the Royal Society of London Series B – Biological Sciences, 270: 2605–2611.
- Boo, S.M., Kim, H.S., Shin, W., Boo, G.H., Cho, S.M., Jo, B.Y., Kim, J.-H., Kim, J.H., Yang, E.C., Siver, P.A., Wolfe, A.P., Bhattacharya, D., Andersen, R.A. & Yoon, H.S. (2010). Complex phylogeographic patterns in the freshwater alga Synura provide new insights into ubiquity vs. endemism in microbial eukaryotes. Molecular Ecology, 19: 4328–4338.
- Caron, D.A. (2009). Past president’s address: protistan biogeography: why all the fuss? Journal of Eukaryotic Microbiology, 56: 105–112.
- Cronberg, G. (1989). Scaled chrysophytes from the tropics. Nova Hedwigia, Beiheft, 95: 191–232.
- Cronberg, G. (1996). Scaled chrysophytes from the Okavango Delta, Botswana, Africa. Nova Hedwigia, Beiheft, 114: 91–108.
- Croome, R.L. & Tyler, P.A. (1985). Synura australiensis (Mallomonadaceae, Chrysophyceae), a light and electron microscopical investigation. Nordic Journal of Botany, 5: 399–401.
- Daugbjerg, N. & Andersen, R.A. (1997). Phylogenetic analysis of the rbcL sequences from haptophytes and heterokont algae suggest their chloroplasts are unrelated. Molecular Biology and Evolution, 14:1242–1251.
- Finlay, B.J. (2002). Global dispersal of free-living microbial eukaryote species. Science, 296: 1061–1063.
- Finlay, B.J. & Fenchel, T. (2004). The ubiquity of small species: patterns of local and global diversity. Bioscience, 54: 777–784.
- Foissner, W. (1999). Protist diversity: estimates of the near-imponderable. Protist, 150: 363–368.
- Foissner, W. (2006). Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozoologica, 45: 111–136.
- Foissner, W. (2008). Protist diversity and distribution: some basic considerations. Biodiversity and Conservation, 17: 235–242.
- Hansen, P. (1996). Silica-scaled Chrysophyceae and Synurophyceae from Madagascar. Archiv für Protistenkunde, 147: 145–172.
- Jo, B.Y., Shin, W., Boo, S.M., Kim, H.S. & Siver, P.A. (2011). Studies on ultrastructure and three-gene phylogeny of the genus Mallomonas (Synurophyceae). Journal of Phycology, 47: 415–425.
- Jo, B.Y., Shin, W., Kim, H.S., Siver, P.A. & Andersen, R.A. (2013). Phylogeny of the genus Mallomonas (Synurophyceae) and descriptions of five new species on the basis of morphological evidence. Phycologia, 52: 266–278.
- Jobb, G., von Haeseler, A. & Strimmer, K. (2004). TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evolutionary Biology, 4: 18.
- Katoh, K., Misawa, K., Kuma, K. & Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acid Research, 30: 3059–3066.
- Kim, J.H., Shin, M.O., Lee, K.L. & Kim, H.S. (2008). Effect of environmental conditions on the growth of Synura petersenii (Synurophyceae) in vitro and two eutrophic water bodies in Korea. Nova Hedwigia, 86: 529–544.
- Kristiansen, J. (1975). On the occurrence of the species of Synura. Verhandlungen der Internationalen Vereinigung for Theoretische und Angewandte Limnologie, 19: 2709–2715.
- Kristiansen, J. (1986). The ultrastructural bases of Chrysophyte systematics and phylogeny. CRC Critical Reviews in Plant Science, 4: 149–211.
- Kristiansen, J. (1992). Silica-scaled chrysophytes from West Greenland: Disko Island and the Søndre Strømfjord region. Nordic Journal of Botany, 12: 525–536.
- Kristiansen, J. (2008). Dispersal and biogeography of silica-scaled chrysophytes. Biodiversity and Conservation, 17: 419–426.
- Kristiansen, J. & Preisig, H.R. (2007). Chrysophyte and haptophyte algae, part 2: Synurophyceae. In Süsswasserflora von Mitteleuropa. Vol. 1 –2 (Büdel, B. et al., editors), 1–252. Spektrum Akademischer Verlag, Springer, Berlin.
- Kristiansen, J., Düvel, L. & Wegeberg, S. (1997). Silica-scaled chrysophytes from the Taymyr Peninsula, Northern Siberia. Nova Hedwigia, 65: 337–351.
- Kumar, S., Dudley, J., Nei, M. & Tamura, K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics, 9: 299–306.
- Kynčlová, A., Škaloud, P. & Škaloudová, M. (2010). Unveiling hidden diversity in the Synura petersenii species complex (Synurophyceae, Heterokontophyta). Nova Hedwigia, Beiheft, 136: 283–298.
- Losic, D., Rosengarten, G., Mitchell, J.G. & Voelcker, N.H. (2006). Pore architecture of diatom frustules: potential nanostructured membranes for molecular and particle separations. Journal of Nanoscience and Nanotechnology, 6: 1–8.
- Martiny, J.B., Bohannan, B.J., Brown, J.H., Colwell, R.K., Fuhrman, J.A., Green, J.L., Horner-Devine, M.C., Kane, M., Krumins, J.A., Kuske, C.R., Morin, P.J., Naeem, S., Ovreås, L., Reysenbach, A.L., Smith, V.H. & Staley, J.T. (2006). Microbial biogeography: putting microorganisms on the map. Nature Reviews, Microbiology, 4: 102–112.
- Moore, P.D. (1982). Beneath the blanket bogs of Britain. Natural History, 91: 48–55.
- Němcová, Y., Nováková, S. & Řezáčová-Škaloudová, M. (2008). Synura obesa sp. nov. (Synurophyceae) and other silica-scaled chrysophytes from Abisko (Swedish Lapland). Nova Hedwigia, 86: 243–254.
- Newton, I. (2010). Bird Migration. Harper Collins, London.
- Nicholls, K.H. & Gerrath, J.F. (1985). The taxonomy of Synura (Chrysophyceae) in Ontario with special reference to taste and odour in water supplies. Canadian Journal of Botany, 63: 1482–1493.
- Pichrtová, M. & Němcová, Y. (2011). Effect of temperature on size and shape of silica scales in Synura petersenii and Mallomonas tonsurata (Stramenopiles). Hydrobiologia, 673: 1–11.
- Playfair, G.I. (1915). Freshwater algae of the Lismore District: with an appendix on the algal fungi and Schizomycetes. Proceedings of the Linnean Society of New South Wales, 40: 310–362.
- Rabalais, N.N., Turner, R.E., Justic, D., Dortch, Q., Wiseman Jr., W.J. & Sen Gupta, B.K. (1996). Nutrient changes in the Mississippi River and system responses on the adjacent continental shelf. Estuaries, 19: 386–407.
- Reynolds, C.S. (2006). The Ecology of Phytoplankton. Cambridge University Press, Cambridge.
- Řezáčová, M. & Škaloud, P. (2005). Silica-scaled chrysophytes of Ireland. With an appendix: geographic variation of scale shape of Mallomonas caudata. Nova Hedwigia, Beiheft, 128: 101–124.
- Ronquist, F. & Huelsenbeck, J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572–1574.
- Saha, L.C. & Wujek, D.E. (1990). Scale-bearing chrysophytes from tropical Northeast India. Nordic Journal of Botany, 10: 343–354.
- Saxby-Rouen, K.J., Leadbeater, B.S.C. & Reynolds, C.S. (1997). The growth response of Synura petersenii (Synurophyceae) to photon flux density, temperature, and pH. Phycologia, 36: 233–243.
- Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9: 671–675.
- Siver, P.A. (1988). A new form of the common chrysophycean alga Synura petersenii. Transactions of the American Microscopical Society, 107: 380–385.
- Siver, P.A. (2013): Synura cronbergiae sp. nov., a new species described from two Paleogene maar lakes in northern Canada. Nova Hedwigia, 97: 179–187.
- Škaloud, P., Kynčlová, A., Benada, O., Kofroňová, O. & Škaloudová, M. (2012). Toward a revision of the genus Synura, section Petersenianae (Synurophyceae, Heterokontophyta): morphological characterization of six pseudo-cryptic species. Phycologia, 51: 303–329.
- Škaloud, P., Kristiansen, J. & Škaloudová, M. (2013a). Developments in the taxonomy of silica-scaled chrysophytes – from morphological and ultrastructural to molecular approaches. Nordic Journal of Botany, 31: 385–402.
- Škaloud, P., Škaloudová, M., Pichrtová, M., Němcová, Y., Kreidlová, J. & Pusztai, M. (2013b). www. chrysophytes.eu – a database on distribution and ecology of silica-scaled chrysophytes in Europe. Nova Hedwigia, Beiheft, 142: 141–147.
- Smol, J.P. (1995). Application of chrysophytes to problems in paleoecology. In Chrysophyte Algae. Ecology, Phylogeny and Development (Sandgren, C.D., Smol, J.P. & Kristiansen, J., editors), 303–330. Cambridge University Press, Cambridge.
- Smol, J.P. & Cumming, B.F. (2000). Tracking longterm changes in climate using algal indicators in lake sediments. Journal of Phycology, 36: 986–1011.
- Swofford, D.L. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA.
- Verbruggen, H., Maggs, C.A., Saunders, G.W., Le Gall, L., Yoon, H.S. & De Clerck, O. (2010). Data mining approach identifies research priorities and data requirements for resolving the red algal tree of life. BMC Evolutionary Biology, 10: 16.
- Wee, J. L. (1997). Scale biogenesis in Synurophycean protists: phylogenetic implications. CRC Critical Reviews in Plant Sciences, 16: 497–534.
- Wee, J.L., Booth, D.J. & Bossier, M.A. (1993). Synurophyceae from the southern Atlantic Coastal Plain of North America: a preliminary survey in Louisiana, USA. Nordic Journal of Botany, 13: 95–106.
- Wee, J.L., Fasone, L.D., Sattler, A., Starks, W.W. & Hurley, D.L. (2001). ITS/5.8S DNA sequence variation in 15 isolates of Synura petersenii Korshikov (Synurophyceae). Nova Hedwigia, Beiheft, 122: 245–258.
- West, W. & West, G.S. (1909). The British freshwater phytoplankton, with special reference to desmid-plankton and the distribution of British desmids. Proceedings of the Royal Society, London, 81B: 165–206.
- White, T.J., Bruns, T., Lee, S. & Taylor, J.W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications (Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J., editors), 315–322. Academic Press, New York.
- Zwickl, D.J. (2006). Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation, University of Texas at Austin, Austin.