The Mediterranean red algal flora is diverse but current knowledge of its diversity is at best fragmentary. Here, a new species of Kallymenia from Croatia is described based on morphological and molecular data. Members of the genus Kallymenia share similar morphology making their generic identification relatively easy, whereas species level identification is notoriously difficult. In this paper an integrative systematics study using three gene markers, cox1 (COI), rbcL and nuclear LSU, allowed us to (i) confirm the identity of four existing members of this genus inhabiting the Mediterranean Sea: K. feldmannii, K. lacerata, K. patens and K. requienii; (ii) detect the presence of K. reniformis only in the Atlantic, and (iii) reveal the presence of a new member of Kallymenia from the Mediterranean Sea, K. ercegovicii Vergés & Le Gall, sp. nov., which is described based on distinctive morphological and molecular characters. Kallymenia ercegovicii is distinguished, using three molecular markers, from all Kallymenia species for which these sequences are available. Morphologically, the new species can be distinguished from other polycarpogonial Kallymenia by a set of characters including a deeply lobed thallus, large inner cortical cells, stellate but non-glanglionic medullary cells and non-ostiolate cystocarps surrounded by a filamentous net composed of elongated cells forming fascicles. An initial phylogeny of the genus Kallymenia was inferred from cox1 (COI), rbcL and nuclear LSU sequences. Kallymenia ercegovicii was resolved with full support within the same lineage as K. reniformis (the generitype), K. feldmannii and K. patens, whereas K. lacerata and K. requienii were only distantly related.
Introduction
The macroalgal flora of the Mediterranean Sea has been extensively studied and most of our current knowledge of ‘morphological’ species diversity has accrued since the second half of the 18th century (Thuret & Bornet, Citation1878; De Toni, Citation1897). In a recent review of the biodiversity of the Mediterranean Sea by Coll et al. (Citation2010), Enric Ballesteros listed c. 650 species of red algae of which 150 were recorded as endemic and 73 as introduced species (see also Boudouresque & Verlaque, Citation2002; Boudouresque, Citation2004), reflecting the Mediterranean Sea as a hotspot of biodiversity (Tortonese, Citation1985; Myers et al., Citation2000; Boudouresque & Verlaque, Citation2002; Boudouresque, Citation2004; Klein et al., Citation2005; Coll et al., Citation2010). Since the mid-1990s, phycologists have increasingly relied on molecular data to assess phylogenetic relationships among various taxa and the advent of DNA barcoding revealed that red algal alpha diversity was underestimated due partly to cryptic diversity. Despite considerable knowledge of the Mediterranean red algal flora, only a few studies have assessed alpha diversity using molecular tools (e.g. Rodriguez Prieto & De Clerck, Citation2009; Le Gall & Saunders, Citation2010a, Citation2010b; Manghisi et al., Citation2010; Wolf et al., Citation2011; Vergés et al., Citation2013a, Citation2013b). These studies have revealed at least six new taxa for the region, which, in most cases, were unexpected (either cryptic or exogenous). In light of these observations, we initiated an assessment of the alpha diversity of the red algal flora of the Mediterranean Sea and we present herein our results on the kallymeniacean genus Kallymenia.
The genus Kallymenia (Kallymeniaceae, Rhodophyta), described by J. Agardh (Citation1842), currently encompasses 43 taxonomically accepted species and has a worldwide distribution (Guiry & Guiry, Citation2013). Six species have been recorded in the Mediterranean Sea: K. feldmannii Codomier, K. lacerata Feldmann, K. requienii (J. Agardh) J. Agardh, K. patens (J. Agardh) Codomier ex Parkinson, K. reniformis (Turner) J. Agardh and K. spathulata (J. Agardh) Codomier ex Parkinson (Codomier, Citation1971; Vergés & Rodríguez-Prieto, Citation2006), as well as two unidentified taxa called Kallymenia sp. 1 and Kallymenia sp. 2 collected in Italy (Abdelahad & D’Archino, Citation2011). Until now only K. reniformis and two forms of K. spathulata, f. pennata and f. luxurians, have been reported, as ‘Halarachnion spathulatum’ (Ercegovic, Citation1949, Citation1957; Giaccone, Citation1978), from the Croatian coast of the Adriatic Sea.
In recent years, molecular systematic tools have been used to clarify boundaries within the family Kallymeniaceae and allowed the description of new genera and species (Hommersand et al., Citation2009; Clarkston & Saunders, Citation2010; D’Archino et al., Citation2010, Citation2011, Citation2012; Arakaki et al., Citation2011) using either rbcL (D’Archino et al., Citation2010; Arakaki et al., Citation2011), rbcL and LSU (D’Archino et al., Citation2011; Clarkston & Saunders, Citation2012), LSU and COI-5′ (Clarkston & Saunders, Citation2010, Citation2012) or a combination of several gene markers, LSU, COI-5′, ITS, UPA and EF2 (Clarkston & Saunders, Citation2013).
In the current paper, we report on an integrative approach coupling comprehensive sampling with the acquisition of both molecular and morphological data, which led us to uncover and describe a new member of the genus Kallymenia for the Mediterranean Sea. Phylogenetic analysis inferred from mitochondrial (cox1, here termed COI), chloroplast (rbcL) and nuclear (LSU) genes among Mediterranean species of Kallymenia is discussed, and we emphasize the phylogenetic significance within the genus of the number of carpogonial branches per carpogonial branch system.
Materials and methods
Sample collection
Specimens of Kallymenia were collected by SCUBA between 2 and 50 m depth at various sites along the French coast of the Mediterranean and between 3 and 16 m depth along the Atlantic coast of Europe between 2004 and 2008 (–) in the course of floristic surveys. Croatia was sampled only once, in June 2003. Samples were dried as herbarium specimens and housed at the Herbarium of the Natural History Museum of Paris [PC, abbreviations follow Thiers (Citation2013)], and for molecular analyses they were cleaned, dried and preserved in silica gel.
Table 1. Samples of Kallymenia spp. and Meredithia microphylla used in the phylogenetic analysis, voucher or isolate number, locality, GenBank accession number and reference are indicated.
Table 2. Collection data for Kallymenia spp. specimens examined.
Molecular sequencing and analysis
Material used for molecular analysis is listed in , along with valid names, details of collection data, vouchers and BOLD and GenBank accession numbers for COI-5′, rbcL and LSU sequences. Total DNA was extracted using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions except for the extraction buffer which was prepared in the lab as follows: 1M Tris-base, 1M Tris-HCl, 0.05M Na2EDTA, 0.2M NaCl, 2.5M potassium acetate, 10% Tween 20 and 0.2 mg ml−1 Pro K. A total of 42 COI sequences were generated for this study. For each sequence a total of 670 base pairs of the COI-5′ region was amplified using the forward primers GazF1 (Saunders, Citation2005) and GWSFn (Le Gall & Saunders, Citation2010a) variously combined with the reverse primers GazR1 (Saunders, Citation2005) and GWSRx (Saunders, Citation2009).
Both rbcL (including the rbcL-rbcS spacer) and LSU (28S) were amplified for each of the seven species included in this study. RbcL was amplified in three fragments covering a total of 1647 bp using the following combinations of primers: F-rbcLstart/R-753, F-577/R1381, and F-993/R-rbcS start (Freshwater & Rueness, Citation1994). LSU (28S) was amplified as three overlapping fragments using primers T01N/T20, T04/T08 and T05/T15, and using the PCR primers and the internal primers T10, T16N, T19N, T22, T24, T25, T30, T33, following protocols of Harper & Saunders (Citation2001) and Le Gall & Saunders (Citation2010a). Purification and sequencing reactions were performed by Genoscope (www.genoscope.fr, Evry, France). Forward and reverse electropherograms were edited and assembled with Codoncode (Dedham, Massachusetts, USA) and multiple sequence alignments were constructed for each marker using SeaView (Gouy et al., Citation2010). A combined gene dataset was constructed by merging CO1 (663 bp), rbcL (878 bp) and LSU (2764 bp) data for the seven species included in this study using the webtool FastaBox 1.41 (Villesen, Citation2007). For the COI datasets, genetic species groups were determined by distance analyses using the neighbour-joining algorithm in SeaView. Phylogenetic analyses were conducted on the combined dataset and the LSU-only alignment by Bayesian inference using MrBayes version 3.2.1 (Ronquist et al., Citation2012); ML and distance bootstrap values were also calculated using SeaView.
For the combined dataset, sequence data were partitioned by genes. Analyses were run with four heated Monte Carlo Markov Chains for 2 000 000 generations. Output trees and data were sampled every 100 generations. Appropriate burn-in for each run was determined by plotting the overall likelihood against generations prior to estimating the posterior probability distribution. In all analyses, likelihood values were stable after the first 200 000 generations. Final results were based on the pooled samples from the stationary phase of the two independent runs.
Anatomy and morphology
Anatomical and reproductive features were observed in squash preparations, using 10% HCl mixed with acidified 1% aniline blue, and by sections made with a razor blade and subsequently stained in an acidified 1% aniline blue/distilled water solution and mounted permanently in 50% Karo® corn syrup (Bestfoods, Englewood Cliffs, California, USA). Habit views were reproduced with a HP Officejet 6500A scanner (Hewlett-Packard, Palo Alto, California, USA); photomicrographs were taken with a MRc5 camera (Zeiss, Berlin, Germany) attached to an Axio Imager A2 microscope (Zeiss, Berlin). Slides have been deposited in PC.
Results and discussion
Integrative systematics approach reveals a new species of Kallymenia
DNA barcode for species assignment
COI sequences were obtained for 41 specimens of Kallymenia from Atlantic and Mediterranean coasts of Europe and a specimen of Meredithia microphylla (J. Agardh) J. Agardh was included as an outgroup. Analysis of this barcode dataset resolved six species among the Kallymenia specimens, diverging from 5.0 to 14.5% among species groups (uncorrected p-distance, ). All the specimens from the Atlantic clustered together and were assigned to the species Kallymenia reniformis based on their anatomical and reproductive features. Observation of morphological and anatomical characters led us to identify four of the five clusters of Mediterranean specimens which were assigned to Kallymenia lacerata (8 specimens), Kallymenia requienii (3 specimens), Kallymenia feldmannii (6 specimens) and Kallymenia patens (5 specimens). Kallymenia lacerata and K. requienii were only distantly related (p-distance of 14.5%) and these two taxa displayed a greater divergence (p-distance >10%) from all of the remaining species of Kallymenia, among which there was less than 6.4% divergence. Infraspecific variation was only observed within Kallymenia patens (up to 0.5% divergence) and K. requienii (up to 0.16% divergence). The specimen LLG0376 collected at Vis Island, Croatia was resolved as a sister taxon to Kallymenia feldmannii with a p-distance of 5.2%, a value which reflects interspecific variation, as the threshold between infraspecific and interspecific variation in the CO1 for red algae ranges from 0.5 to 2% according to previous studies (e.g. Saunders, Citation2005; Robba et al., Citation2006; Clarkston & Saunders, Citation2010; Le Gall & Saunders, Citation2010a, Citation2010b; Clarkston & Saunders, Citation2013). Moreover, analysis resolved K. patens as the sister taxon to the Atlantic species K. reniformis with a similar p-distance of 5%. We therefore considered this Croatian specimen to be a distinct species, here named Kallymenia ercegovicii Vergés & Le Gall, sp. nov., and we conducted a thorough morphological and anatomical study to complement molecular data with morpho-anatomical characters in order to provide a suite of diagnostic features.
Fig. 1. Genetic species groups displayed as a tree inferred from DNA barcode sequences (COI) with interspecific divergences among Kallymenia species and the outgroup Meredithia microphylla (uncorrected p-distances). Kallymenia reniformis specimens were collected in the Atlantic Ocean, and the rest of the samples used in this analysis were collected in the Mediterranean Sea.
Taxonomic treatments
Kallymenia ercegovicii Vergés & Le Gall, sp. nov. (–)
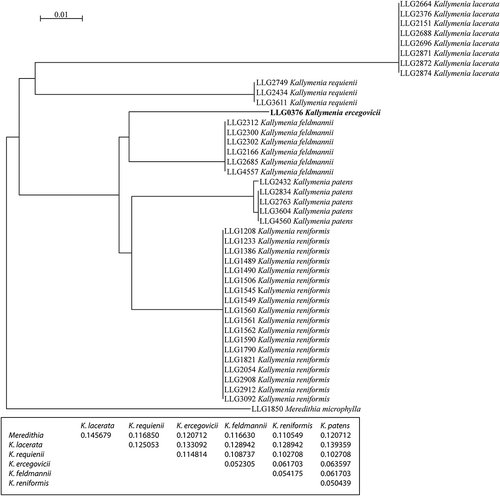
Figs 2–8. Kallymenia ercegovicii Vergés & Le Gall, sp. nov. (PC0152698): habit, morphology, anatomical and male reproductive structures. Scale bar = 1 cm (), 1000 µm (), 40 µm (, , ), 100 µm (, ). . Holotype with the perennial part of the thallus indicated by the arrow. . Enlargement of the perennial part (arrow). . Cross-section of thallus with the cortical layers indicated (1st = first cortical layer; 2nd = second cortical layer; 3rd = third cortical layer; 4th = fourth cortical layer; 5th = fifth cortical layer. . Squash preparation showing mid-cortical cells (mcc) with no, or only very short, arms and a large cell volume; inner cortical cell (icc) with arms developing in all directions and a medullary stellate cell (msc). . Outer cortical cells arranged in rosettes around cells of the next cortical layer. . Medullary stellate cell (mc). . Male gametangia (arrows) developing from spermatangial mother cells among outer cortical cells.
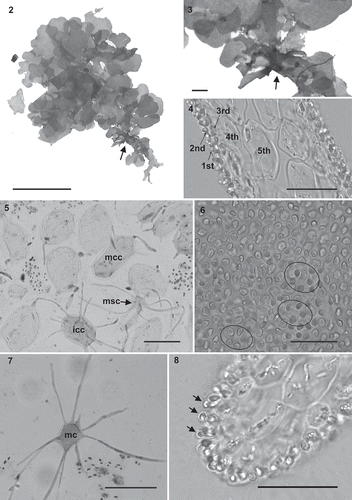
Figs 9–11. Kallymenia ercegovicii Vergés & Le Gall, sp. nov. (PC0152698): Reproductive structures. Scale bar = 40 µm. –. Polycarpogonial female reproductive structures (1st: first carpogonial branch cell, cp: carpogonium, hg: hypogynous cell, sc: supporting cell, t: trichogyne). . Apical part of a connecting filament (arrow).
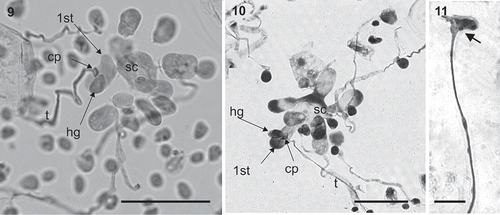
Figs 12–14. Kallymenia ercegovicii Vergés & Le Gall, sp. nov. (PC0152698): Cystocarps. Scale bar = 200 µm (, ), 100 µm (). –. Carposporophytes in surface view with fragmented involucre (arrows). . Cross-section of a carposporophyte showing the fascicles of the involucre (arrows).
| Diagnosis: | = | Thallus erect, foliose, up to 3.3 cm high, 2.8 cm broad and 140 μm thick. Frond very lobed, with an entire margin. Inner cortex of hyaline stellate cells, up to 160 μm in diameter. Medullary cells non-refractive, with the cell body up to 50 μm diameter and arms up to 60 μm long. Polycarpogonial, with a rounded supporting cell, up to 5 (–8) rounded subsidiary cells, and up to 7 3-celled carpogonial branches. Auxiliary cell system not observed. Connecting filaments present. Mature carposporophyte up to 950 μm in diameter surrounded by a well-developed involucre of cells arranged in fascicles. Spermatangia (2–4 µm in diameter) rounded, covering the surface, cut off from the outer cortical cells. Tetrasporangia unknown. |
| Holotype: | = | PC0152698. |
| Type locality: | = | Vis Island, Croatia (43º01.53′ N, 16º 22.57′ E), growing on rocks in the entrance of a cave at 10 m depth, 20 June 2007; leg. L. Le Gall & J.M. Utgé. |
| Etymology: | = | Named in honour of A. Ercegovic for his significant contributions to the knowledge of the Adriatic marine flora. |
| Distribution: | = | Known only from the type locality. |
| Habit: | = | Plants are erect, perennial, sessile, up to 3.3 cm long, 2.8 cm broad and 140 µm thick. Fronds are deeply lobed, rose-red in colour and membranous (). |
| Vegetative structure: | = | The thallus structure is multiaxial (), with a compact cortex and a lax medulla of slender filaments intermixed with a network of stellate cells. All the cortical cells except the outer ones have secondary pit-connections to the cells of the same layer, forming a network parallel to the frond surface, and also to the cells of the neighbouring layers, or in the case of the inner cortical cells, with the medullary stellate cells or filaments. Inner cortical cells stellate, hyaline, with a cell body up to 70 µm in diameter (); the next layers of the cortical cells are composed of enlarged hyaline cells, slightly stellate with very short arms, with a cell body up to 160 µm diameter (); cells of the deeper cortical layers are ovoid or irregular in shape, hyaline, measuring up to 25 µm in diameter in the inner layer and up to 10 µm in the outer layer; outer cortical cells rounded, rose-red, up to 5 µm in diameter and arranged forming a rosette (). Medulla composed of stellate cells (, ), hyaline, with a cell body up to 50 µm in diameter and arms up to 60 µm long that link the cells to each other, to the medullary filaments or to the inner cortical cells. Medullary filaments are hyaline, composed of one or more cells, branched or not. |
| Reproduction: | = | Thallus monoecious. Spermatangia formed at the surface of the thallus, 2–4 µm in diameter, rounded, cut off singly by the outer cortical cells (). Non-procarpic, with numerous carpogonial branch systems located in the middle and apical parts of the thallus, polycarpogonial, arising on cells of the inner cortex and developing through the outer cortex. Carpogonial branch systems consisting of a rounded supporting cell (–) that supports up to 5 (–8) ovoid subsidiary cells and up to 7 3-celled carpogonial branches which are borne on the supporting cell; carpogonium prolonged by a trichogyne up to 120 µm long (–). Only one fusion cell was observed that developed initial connecting filaments; mature connecting filaments were very scarce and were unbranched and swollen at their apices (). Neither auxiliary cell systems nor the place where the gonimoblast filaments starts to develop were observed. |
Cystocarps, growing towards the medulla, are protruding and 300–950 µm in diameter (). An ostiole is absent; carpospores are liberated through the broken filamentous involucre that surrounds the carposporangia. The involucre arises from vegetative cells and consists of fascicles of cells diminishing in size outwards. Inner fascicle cells are elongated, up to 25 µm long, and the outer cells are rounded or ovoid, up to 5 µm in diameter (). When cystocarps break these fascicles remain visible in surface view (). Carpospores, 13–17 µm in diameter, form dense masses and are intermixed with filaments ().
Remarks
The distinguishing characteristics of Kallymenia ercegovicii are its small, deeply lobed thallus, the large size of the inner cortical layers (cells up to 160 µm), the polycarpogonial cell system, and the net formed by the arrangement of the fascicles of cells around the cystocarp. These features are unique and this species differs from the other polycarpogonial species of Kallymenia described to date (). Gross morphology and vegetative traits in K. ercegovicii are similar to the rest of the polycarpogonial species except Kallymenia lacerata which has atypical medullary cells differing in shape and larger in size. Regarding reproductive structures, post-fertilization stages are similar in most of the polycarpogonial species but remain undescribed for K. ercegovicii. Kallymena rosea Womersley & R.E.Norris and K. lacerata appear to differ from the other Kallymenia species. Kallymenia rosea is the only member in which carpogonial branches and subsidiary cells of the auxiliary cell system have an additional cell; in K. lacerata only aborted fusion cells have been detected and neither mature connecting filaments nor auxiliary cell systems have been observed, suggesting that it may be an apomictic species (Vergés & Rodríguez-Prieto Citation2006).
Table 3. Comparison of the polycarpogonial species of Kallymenia.
Reproductive stages observed in Kallymenia ercegovicii followed the same pattern as in the type species of the genus, Kallymenia reniformis (Norris, Citation1957; Hommersand & Ott, Citation1970; Vergés, Citation2001). We could see numerous carpogonial cell systems, but only in a few examples of mature carpogonial cell systems was the subsidiary cell fusing with the supporting cell, which corresponds to the initial step in the formation of the fusion cells. Moreover, long connecting filaments were observed through the thallus, confirming the non-procarpic nature of this species (). Finally, fusion cells, auxiliary cell systems and initial stages of cystocarp development were never detected despite thorough observations around the cystocarps, suggesting that this specimen was immature, or apomictic. The lack of more fertile specimens prevented finding early post-fertilization stages, which remain undescribed.
Common traits in Kallymeniaceae are the presence of filaments around and intermixed with carposporangia, and cystocarps generally protruding, with an ostiole where carpospores are liberated towards the surface or, in non-ostiolate species, the cortex breaks and carpospores pass through. The new species has a much-developed involucre around the mass of carpo-sporangia and gonimoblast filaments which breaks up when carpospores are mature, so the spores reach the surface. This kind of structure is unique and does not match any description found in the literature for Kallymenia (Feldmann, Citation1942; Norris, Citation1957, Citation1964; Abbot, Citation1968; Hommersand & Ott, Citation1970; Codomier Citation1971; Womersley & Norris, Citation1971; Irvine, Citation1983; Womersley, Citation1994; Vergés, Citation2001).
Phylogenetic affinities of Kallymenia ercegovicii
Phylogenetic analyses were undertaken to resolve interspecific relationships among European Kallymenia, and to determine their relationship with Kallymenia ercegovicii, based on a combined dataset including LSU, rbcL as well as the COI generated in this study () and an LSU only alignment including data from GenBank (). Phylograms inferred from Bayesian analyses are presented for the combined alignments (–LnL = 10 508), and LSU alignment (–LnL = 12 128) and include posterior probabilities and bootstrap results for ML and distance analyses. The combined analysis resolved Kallymenia feldmannii, K. patens, K. reniformis and the herein described K. ercegovicii within a fully supported lineage in all analyses. Kallymenia ercegovicii was resolved with strong to full support as a sister species of the lineage encompassing K. patens and K. reniformis. Our analysis revealed that typical polycarpogonial species of the genus Kallymenia from the North Atlantic and the Mediterranean Sea (K. feldmannii, K. patens, K. reniformis and K. ercegovicii) are closely related, which suggests they share a recent common ancestor. Nevertheless, K. ercegovicii can be distinguished from the other polycarpogonial Kallymenia using any of the molecular markers included in this study or a combination of morphological characters including the thallus length and shape, the large dimensions of the cortical cells (up to 160 µm in K. ercegovicii, and up to 100 µm in the most similar species), the stellate (but not ganglionic) shape of medullary cells and the absence of a prominent cystocarp involucre with fascicles of cells (). Kallymenia requienii and K. lacerata were only distantly related to the other European species of Kallymenia and our phylogenetic analysis failed to resolve their relationships.
Fig. 15. Phylogenetic tree inferred from Bayesian analysis of a concatenated alignment of CO1, rbcL and LSU. Thick branches lead to nodes strongly supported in all analyses (Bayesian posterior probabilities of 1, ML and NJ bootstrap support above 90%).
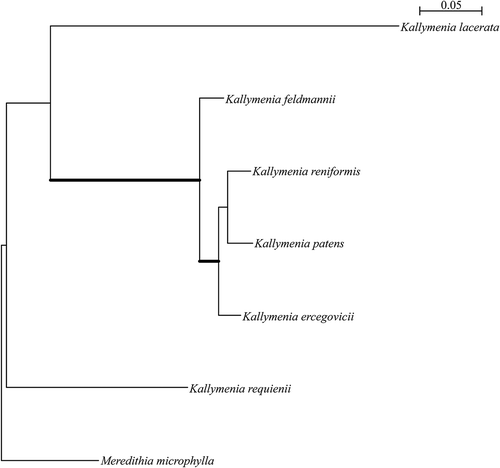
Fig. 16. Phylogenetic tree inferred from Bayesian analysis of the LSU. Thick branches lead to nodes strongly supported in all analyses (Bayesian posterior probabilities of 1, ML and NJ bootstrap support above 90%). Voucher specimens for sequences generated in this study are indicated in bold.
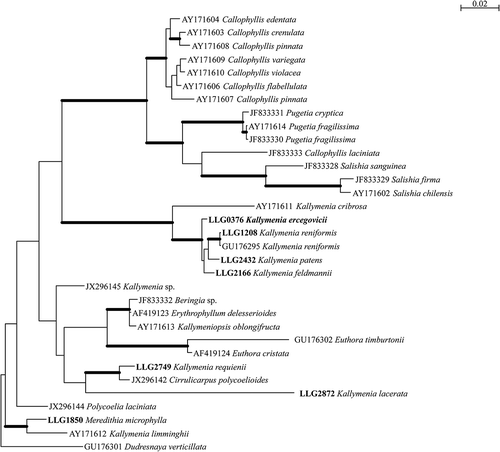
Similarly, phylogenetic analysis of the LSU alignment resolved Kallymenia feldmannii, K. patens, K. reniformis and K. ercegovicii within a fully supported lineage, allied to Kallymenia cribrosa Harvey, an Australian perforated monocarpogonial species (Womersley, Citation1994). Relationships among the existing European Kallymenia were not resolved. Kallymenia requienii, the only Mediterranean monocarpogonial species of the genus Kallymenia, allies with Cirrulicarpus polycoelioides (J. Agardh) Womersley with strong support in all analyses. Both taxa are monocarpogonial and share anatomical and reproductive characters including several cortical layers of cells diminishing in size outwards, stellate medullary cells present, non-procarpic, a single carpogonial branch per system, and supporting, subsidiary and fusion cells lobed. Therefore the main morphological feature which differentiates the genus Cirrulicarpus from Kallymenia is the branching pattern of the thallus (Womersley, Citation1994). An extended analysis with more Cirrulicarpus samples including the type species, Cirrulicarpus gmelinii (Grunow) Tokida & Masaki, should be undertaken to resolve the phylogenetic relationships between these taxa.
Finally, the phylogenetic affinities of Kallymenia lacerata were unresolved in all analyses, this species joining either, with weak support, the genus Euthora J. Agardh in distance analysis or the lineage encompassing Cirrulicarpus polycoelioides and Kallymenia requienii in Bayesian and ML analyses, challenging the affiliation of this taxon with the genus Kallymenia for which the generitype is Kallymenia reniformis, an Atlantic species included in our analysis, whose lectotype is an original illustration based on material collected from Salterton, Devon, UK (Turner Citation1808–1809). It is noteworthy that Kallymenia lacerata, which is a polycarpogonial species, exhibits a distinctive pattern in post-fertilization stages (Vergés & Rodríguez-Prieto, Citation2006). Our phylogenetic analyses join many published studies in challenging the current generic boundaries in the Kallymeniaceae (e.g. D’Archino et al., Citation2010). Clearly, considerable systematic research remains for this intriguing group of algae with their multitude of patterns of pre- and post-fertilization development.
Conclusions
The discovery of this new species of the genus Kallymenia confirms the high diversity of this genus in the Mediterranean, and is a new contribution to the knowledge of biodiversity in the Mediterranean Sea. The location of the Mediterranean Sea at the margins of Africa, Europe and West Asia and its history of geological upheaval (Maldonado, Citation1985; Garcia-Castellanos et al., Citation2009) have led to extremely diverse spatial and temporal diversity of habitats which has shaped the extraordinary species diversity. The present study emphasizes the value of an integrative systematic approach using molecular data in addition to morphological characters in order to elucidate the biodiversity that remains undescribed in the Mediterranean algal flora.
Acknowledgements
The molecular data were mostly generated at the Service de Systématique Moléculaire du Muséum National d’Histoire Naturelle (CNRS – UMS 2700) thanks to funds provided by the ATM ‘Taxonomie moléculaire: DNA Barcode et gestion durable des collections’. Sequencing was performed by the Genoscope through the projects entitled ‘Speed ID’ and ‘Bibliotheque du Vivant’. Some CO1 sequence data for this study were generated in the Saunders laboratory at the University of New Brunswick (UNB), Canada, through financial support of the Canadian Barcode of Life Network from Genome Canada through the Ontario Genomics Institute, the Natural Sciences and Engineering Research Council of Canada, as well as other sponsors listed at www.BOLNET.ca, with infrastructure support from the Canada Foundation for Innovation and New Brunswick Innovation Foundation. Special thanks are extended to T. Moore, D. McDevit and K. Dixon for their technical support at UNB. A. Vergés is grateful for the Synthesys award received in 2009 that supported the study of Kallymenia specimens located at the BM herbarium in the Natural History Museum, London. We are grateful to two anonymous reviewers and to the editors for their valuable comments which have substantially improved the manuscript.
References
- Abbott, I.A. (1968). Studies in some foliose red algae of the Pacific coast. III. Dumontiaceae, Weeksiaceae, Kallymeniaceae. Journal of Phycology, 4: 180–198.
- Abdelahad, N. & D’Archino, R. (2011). Formation of secondary connecting filaments, a new post-fertilisation stage in Mediterranean species of Kallymenia (Kallymeniaceae, Rhodophyta). Cryptogamie, Algologie, 32: 351–362.
- Agardh, J.G. (1842). Algae maris Mediterranei et Adriatici, observationes in diagnosin specierum et dispositionem generum. pp. [i]–x, 1–164. Fortin, Masson et Cie, Paris.
- Arakaki, N., Alveal, K., Ramírez, M.E. & Fredericq S. (2011). The genus Callophyllis (Kallymeniaceae, Rhodophyta) from the central-south Chilean coast (33° to 41° S), with the description of two new species. Revista Chilena de Historia Natural, 84: 481–499.
- Boudouresque, C.F. (2004). Marine biodiversity in the Mediterranean: status of species, populations and communities. Scientific Reports of Port-Cros National Park, France, 20: 97–146.
- Boudouresque, C.F. & Verlaque, M. (2002). Biological pollution in the Mediterranean Sea: invasive versus introduced macrophytes. Marine Pollution Bulletin, 44: 32–38.
- Clarkston, B.E. & Saunders, G.W. (2010). A comparison of two DNA barcode markers for species discrimination in the red algal family Kallymeniaceae (Gigartinales, Florideophyceae), with a description of Euthora timburtonii sp. nov. Botany, 88: 119–131.
- Clarkston, B.E. & Saunders, G.W. (2012). An examination of the red algal genus Pugetia (Kallymeniaceae, Gigartinales), with descriptions of Salishia firma gen. & comb. nov., Pugetia cryptica sp. nov. and Beringia wynnei sp. nov. Phycologia, 51: 33–61.
- Clarkston, B.E. & Saunders, G.W. (2013). Resolving species diversity in the red algal genus Callophyllis (Kallymeniaceae, Gigartinales) in Canada using molecular assisted alpha taxonomy. European Journal of Phycology, 48: 27–46.
- Codomier, L. (1971). Recherches sur les Kallymenia (Cryptonémiales, Kallymeniacées). I. Les espèces Méditerranéennes. Vie et Milieu, 22: 1–54.
- Coll, M., Piroddi, C., Steenbeek, J., Kaschner, K., Ben Rais Lasram, F., Aguzzi, J. et al. (2010). The biodiversity of the Mediterranean Sea: estimates, patterns, and threats. PLOS ONE, 5: e11842.
- D’Archino, R., Nelson, W.A. & Zuccarello, G.C. (2010). Psaromenia (Kallymeniaceae, Rhodophyta): a new genus for Kallymenia berggrenii. Phycologia, 49: 73–85.
- D’Archino, R., Nelson, W.A. & Zuccarello, G.C. (2011). Diversity and complexity in New Zealand Kallymeniaceae (Rhodophyta): recognition of the genus Ectophora and description of E. marginata sp. nov. Phycologia, 50: 241–255.
- D’Archino, R., Nelson, W.A. & Zuccarello, G.C. (2012). Stauromenia australis, a new genus and species in the family Kallymeniaceae (Rhodophyta) from southern New Zealand. Phycologia, 51: 451–460.
- De Toni, G.B. (1897). Sylloge algarum omnium hucusque cognitarum. Vol. IV. Florideae. Sectio I. pp. [i]–xx, [i]–lxi + [1]–388. Patavii [Padua].
- Ercegovic, A. (1949). Sur quelques algues rouges, rares ou nouvelles, de l’Adriatique. Acta Adriatica, 4: 91–121 [ reprint 1–81].
- Ercegovic, A. (1957). La flore sous-marine de l’Ilot de Jabuka. Acta Adriatica, 8: 1–130.
- Feldmann, J. (1942). Les Kallymenia (Rhodophycées, Cryptonemiales) des côtes d’Algérie. Bulletin de la Société d’Histoire Naturelle de l’Afrique du Nord, 33: 7–14.
- Freshwater, D.W. & Rueness, J. (1994). Phylogenetic relationships of some European Gelidium (Gelidiales, Rhodophyta) species, based on rbcL nucleotide sequence analysis. Phycologia, 33: 187–194.
- Ganesan, E.K. (1976). On Kallymenia westii sp. nov. (Rhodophyta, Cryptonemiales) from the Caribbean Sea. Boletín del Instituto Oceanográfico, Universidad de Oriente, 15: 169–175.
- Garcia-Castellanos, D., Estrada, F., Jimenez-Munt, I., Gorini, C., Fernandez, M., Verges, J. & De Vicente, R. (2009). Catastrophic flood of the Mediterranean after the Messinian salinity crisis. Nature, 462: 778–781.
- Giaccone, G. (1978). Revisione della flora marina de Mare Adriatico. Annuario Parco Marino Miramare, 6: 1–118.
- Gouy, M., Guindon, S. & Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution, 27: 221–224.
- Guiry, M.D. & Guiry, G.M. (2013). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 12 July 2013.
- Harper, J.T. & Saunders, G.W. (2001). Molecular systematics of the Florideophyceae (Rhodophyta) using nuclear large and small subunit rDNA sequence data. Journal of Phycology, 37: 1073–1082.
- Hommersand, M.H. & Ott, D.W. (1970). Development of the carposporophyte of Kallymenia reniformis (Turner) J. Agardh. Journal of Phycology, 6: 322–331.
- Hommersand, M.H., Moe, R.L., Amsler, C.D. & Fredericq, S. (2009). Notes on the systematics of Antarctic and subantarctic Rhodophyta with descriptions of four new genera and five new species. Botanica Marina, 52: 509–534.
- Huvé, H. & Passelaigue, F. (1970). A propos de quelques Rhodophycées foliacées de la région de Marseille. Bulletin de la Société Phycologique de France, 15: 43–48.
- Irvine, L.M. (1983). Seaweeds of the British Isles. Volume 1. Rhodophyta. Part 2A. Cryptonemiales (sensu stricto), Palmariales, Rhodymeniales. British Museum (Natural History), London.
- Klein, J., Ruitton, S., Verlaque, M. & Boudouresque, C.F. (2005). Species introductions, diversity and disturbances in marine macrophyte assemblages of the northwestern Mediterranean Sea. Marine Ecology Progress Series, 290: 79–88.
- Le Gall, L. & Saunders, G.W. (2010a). DNA barcoding is a powerful tool to uncover algal diversity: a case study of the Phyllophoraceae (Gigartinales, Rhodophyta) in the Canadian flora. Journal of Phycology, 46: 374–389.
- Le Gall, L. & Saunders, G.W. (2010b). Establishment of a DNA-barcode library for the Nemaliales (Rhodophyta) from Canada and France uncovers overlooked diversity in the species Nemalion helminthoides (Velley) Batters. Cryptogamie, Algologie, 31: 403–421.
- Maldonado, A. (1985). Evolution of the Mediterranean basins and a detailed reconstruction of the Cenozoic paleoceanography. In Key Environments: Western Mediterranean (Margalef, R, editor), 17–59. Pergamon Press, Oxford, UK.
- Manghisi, A., Morabito, M., Bertuccio, C., Le Gall, L., Couloux, A., Cruaud, C. & Genovese, G. (2010). Is routine DNA barcoding an efficient tool to reveal introduction of alien macroalgae? A case study of Agardhiella subulata (Solieriaceae, Rhodophyta) in Cape Peloro lagoon (Sicily, Italy). Cryptogamie, Algologie, 31: 423–433.
- Myers, N., Mittermeier, R.A., Mittermeier, C.G., Da Fonseca, G.A.B. & Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature, 403: 853–858.
- Norris, R.E. (1957). Morphological studies on the Kallymeniaceae. University of California Publications in Botany, 28: 251–334.
- Norris, R.E. (1964). The morphology and taxonomy of South African Kallymeniaceae. Botanica Marina, 7: 90–129.
- Norris, R.E. & Norris, J.N. (1973). Kallymenia pertusa (Rhodophyceae, Cryptonemiales) from the Gulf of California. Phycologia, 12: 71–74.
- Robba, L., Russell, S.J., Barker, G.L. & Brodie, J. (2006). Assessing the use of the mitochondrial cox1 marker for use in DNA barcoding of red algae (Rhodophyta). American Journal of Botany, 93: 1101–1108.
- Rodriguez-Prieto, C. & De Clerck, O. (2009). Leptofauchea coralligena (Faucheaceae, Rhodophyta), a new species from the Mediterranean Sea. European Journal of Phycology, 44: 107–121.
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61: 539–542.
- Saunders, G.W. (2005). Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philosophical Transactions of the Royal Society B: Biological Sciences, 360: 1879–1888.
- Saunders, G.W. (2009). Routine DNA barcoding of Canadian Gracilariales (Rhodophyta) reveals the invasive species Gracilaria vermiculophylla in British Columbia. Molecular Ecology Resources, 9: 140–150.
- Schneider, C.W. & Searles, R.B. (1991). Seaweeds of the southeastern United States. Cape Hatteras to Cape Canaveral. Duke University Press, NC.
- Setchell, W.A. & Gardner, N.L. (1924). New marine algae from the Gulf of California. Proceedings of the California Academy of Science, Series 4, 12: 695–949.
- Thiers, B. (2013, continuously updated). Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/ih/
- Thuret, G. & Bornet, É. (1878). Études phycologiques. Analyses d’algues marines. Publiées par les soins de M. le Dr Édouard Bornet. Ouvrage accompagné de cinquante planches gravées d’après les desssins de M. Alfred Riocreux. [i–v], i–iii, 1–105, pls I–LI. G. Masson, Paris.
- Tortonese, E. (1985). Distribution and ecology of endemic elements in the Mediterranean fauna (fishes and echinoderms). In Mediterranean Marine Ecosystems (Moraitou-Apostolopoulou, M. & Kiortsis, V., editors), 57–83. Plenum Press, New York, NY.
- Turner, D. (1808–1809). Fuci; or, coloured figures and descriptions of the plants referrred by botanists to the genus Fucus. Vol. 2. M’Creery (printer), London.
- Vergés, A. (2001). El gènere Kallymenia (Kallymeniaceae, Rhodophyta) a la península Ibèrica i illes Balears. PhD Thesis. University of Girona, Spain.
- Vergés, A. & Rodríguez-Prieto, C. (2006). Anatomical characteristics and reproductive structures of Kallymenia lacerata (Kallymeniaceae, Rhodophyta) from the Mediterranean Sea. Cryptogamie, Algologie, 27: 31–43.
- Vergés, A., Sánchez, N., Peteiro, C., Polo, L. & Brodie, J. (2013a). Pyropia suborbiculata (Bangiales, Rhodophyta): first records from the northeastern Atlantic and Mediterranean of this North Pacific species. Phycologia, 52: 121–129.
- Vergés, A., Comalada, N., Sànchez, N. & Brodie, J. (2013b). A reassessment of the foliose Bangiales (Rhodophyta) in the Balearic Islands including the proposed synonymy of Pyropia olivii with Pyropia koreana. Botanica Marina, 56: 229–240.
- Villesen, P. (2007). FaBox: an online toolbox for fasta sequences. Molecular Ecology Notes, 7: 965–968.
- Wolf, M.A., Sfriso, A., Andreoli, C. & Moro, I. (2011). The presence of exotic Hypnea flexicaulis (Rhodophyta) in the Mediterranean Sea as indicated by morphology, rbcL and cox1 analyses. Aquatic Botany, 95: 55–58.
- Womersley, H.B.S. (1994). The marine benthic flora of Southern Australia. Rhodophyta-Part IIIA. Bangiophyceae and Florideophyceae (Acrochaetiales, Nemaliales, Gelidiales, Hildenbrandiales and Gigartinales sensu lato). Australian Biological Resources Study, Canberra.
- Womersley, H.B.S. & Norris, R.E. (1971). The morphology and taxonomy of Australian Kallymeniaceae (Rhodophyta). Australian Journal of Botany, Suppl. 2: 1–62.
- Wynne, M.J. (2011). A checklist of benthic marine algae of the tropical and subtropical western Atlantic: third revision. Nova Hedwigia Beihefte, 140: 7–166.
