Abstract
A phylogenetic and morphological study of green algae resembling Ulva conglobata from Japan was undertaken, along with morphological observations of the original material of U. conglobata Kjellman. The samples resembling U. conglobata included five genetically distinct species: U. fasciata, U. pertusa, U. tanneri, Ulva sp. 1 and Ulva sp. 2. The discovery of marginal denticulations in some of the original material of U. conglobata, made it possible to distinguish those species without denticulations: U. pertusa, U. tanneri and Ulva sp. 2. The morphological characteristics of Ulva sp. 1 matched those of U. conglobata, but Ulva sp. 1 was not clearly identified as U. conglobata owing to the lack of DNA sequence data of the original material. Ulva sp. 2 had lobes adhering to each other by rhizoids. This morphological feature is stable in Ulva sp. 2 and unique among Ulva species. In conjunction with the molecular data, Ulva sp. 2 was described as a new species, U. adhaerens sp. nov. This species features rhizoidal extensions in regions other than the base and an elaborate arrangement of the extensions used for adhesion. It thereby expands our knowledge of the morphogenesis of the morphologically simple genus Ulva.
Introduction
The green algal genus Ulva Linnaeus (Ulvales, Ulvophyceae) occurs worldwide, in various aquatic habitats from fresh water to the marine environment (Guiry & Guiry, Citation2013). Ulva species have either a monostromatic tubular thallus (previously placed in Enteromorpha) or a distromatic foliose thallus (Hayden et al., Citation2003). The genus currently includes approximately 100 species (Guiry & Guiry Citation2013) that have been distinguished mainly by differences in gross morphology, cell shape and size, plastid orientation, pyrenoid number per cell, and presence or absence of marginal denticulations (e.g. Koeman & van den Hoek, Citation1981; Maggs et al., Citation2007; Loughnane et al., Citation2008). However, because of their simple and plastic morphology, it is sometimes difficult to identify the species by morphological characteristics (Loughnane et al., Citation2008 and reference therein). Therefore, studies have applied molecular data to resolve the species taxonomy of Ulva (e.g. Loughnane et al., Citation2008; Kraft et al., Citation2010; O’Kelly et al., Citation2010).
Ulva conglobata Kjellman is characterized by its small size (usually 2‒4 cm high) and ball-forming distromatic thallus that consists of broad, undulate lobes (Kjellman, Citation1897). Kjellman (Citation1897) collected this species at three localities in Japan (Yokohama, Goto and Amakusa), but did not designate the type locality or a holotype specimen. Kjellman (Citation1897) mentioned that the thalli were from 50 μm to more than 75 μm in thickness and the length/width ratio of cells in section was approximately 1‒2. He also showed sketches of the base, the surface, and the sections of thalli. In addition, he observed that U. conglobata grew gregariously in intertidal habitats and that the species was most abundant in May (Kjellman, Citation1897). One forma, U. conglobata f. densa, was described based on specimens from Amakusa. This forma had smaller (about 1 cm in diameter), denser and thicker thalli (up to 150 μm), and the cells in section were vertically elongated (Kjellman, Citation1897).
However, several reports have questioned the distinctiveness of this small ball-like species. Ulva fasciata Delile [considered to be a synonym of U. lactuca Linnaeus (O’Kelly et al., Citation2010)] typically has a long strap-shaped thallus (Phillips, Citation1988), but Mshigeni & Kajumulo (Citation1979) showed that the thalli of this species were small, rosette-like and irregularly lobed in wave-exposed habitats. Ulva pertusa Kjellman [considered to be a synonym of U. australis Areschoug (Couceiro et al., Citation2011; Kirkendale et al., Citation2013)] typically has a large, expanding thallus with holes (Kjellman Citation1897). Kang & Lee (Citation2002) reported that U. pertusa and U. ‘conglobata’ from Korea were indistinguishable in ITS2 (Internal Transcribed Spacer 2 of the ribosomal cistron) sequence, and Hiraoka & Shimada (Citation2004) reported that U. pertusa and U. ‘conglobata’ from Japan crossed with each other. Considering the reports mentioned above, Hiraoka & Shimada (Citation2004) suggested that U. conglobata was not a distinct species but a morphological variation of some species. In addition, when Iima & Fukusumi (Citation1996) reported U. tanneri Hayden & Waaland from Japan for the first time, they mentioned that the thalli of this species resembled the smaller thalli of U. ‘conglobata’ and had possibly been misidentified as U. conglobata, though U. tanneri was distinguished by having an intercellular space between the cell layers.
To determine the identity of the green algae that are generally called U. ‘conglobata’, we conducted molecular analyses based on rbcL and ITS sequences, along with morphological studies which included observations of the original material of U. conglobata shown by Kjellman (Citation1897).
Materials and methods
Samples were collected in the intertidal at Tenjin-jima Marine Biological Garden (35° 13′ 20″ N, 139° 36′ 10″ E), Yokosuka, Kanagawa Prefecture, Japan on 24 April 2012 and on 23 June 2013. We collected small (< 5 cm tall) green algae with lobed and rosette-like thalli that resembled U. conglobata. The habit of all the collected samples was photographed in the field to compare species later. The fresh specimens were transported to the laboratory. Voucher herbarium specimens were prepared and deposited in the Herbarium of National Museum of Nature and Science, Tsukuba, Japan (TNS) and the Herbarium of the Faculty of Science at Hokkaido University, Sapporo, Japan (SAP): U. fasciata, TNS-AL 183430; U. pertusa, TNS-AL 183431, TNS-AL 183432; U. tanneri, TNS-AL 183433; Ulva sp. 1, TNS-AL 183434; Ulva sp. 2 (U. adhaerens), TNS-AL 183435, TNS-AL 183436, SAP 114680.
Total DNA was extracted from fresh or dried samples using Cica Geneus DNA Extraction Reagent (Kanto Kagaku, Chuo-ku, Tokyo, Japan). Plastid encoded rbcL and the nuclear encoded ITS1-5.8S-ITS2 region were PCR amplified with TaKaRa Ex Taq (TaKaRa Bio Inc., Otsu, Shiga, Japan). Dimethylsulfoxide was added for the amplification (5% of reaction volume). The PCR conditions consisted of an initial denaturation at 95°C for 60 s, followed by 40–45 cycles of denaturation at 95°C for 15 s, annealing at 50–55°C for 15–30 s and extension at 68°C for 30–120 s. The PCR products were purified with Wizard SV Gel and PCR Clean-Up System (Promega, Madison, Wisconsin, USA), and directly sequenced by use of BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Life Technologies, Carlsbad, California, USA) on a 3130xl Genetic Analyzer (Applied Biosystems). These processes were conducted following the manufacturer’s protocols. For phylogenetic analyses, the sequences of rbcL and the ITS1-5.8S-ITS2 region were obtained from two individuals of each genetic species (see results). The primers for rbcL sequences were rbcLstart, R750, F650 and rbcLend (Shimada et al., Citation2003). As the samples of Ulva sp. 2 had an intron-like region in rbcL, the sequence of the region was determined using pairs of each forward primer positioned in the 5′ exon or in the intron-like region (insF, 5′-CGTGCTTTACGTTTAGAAGATTT-3′; insF2, 5′-TCAATGGCATAAAGTTGAACAAAGA-3′; insF3, 5′-ACCGCAAGGCGGAATCATAA-3′; insF4, 5′-GGCGATCACATCTACTGGACA-3′) and a reverse primer positioned in the 3′ exon (insR, 5′-AAAGGTTGTGAGTTTACGTTTTCA-3′). We confirmed that the intron-like region of rbcL in Ulva sp. 2 was indeed removed in the cDNA; total RNA was extracted from culture thalli using RNeasy Plant mini Kit (Qiagen, Hilden, Germany), RT-PCR was conducted with PrimeScript RT reagent Kit (TaKaRa Bio Inc.), and the products were PCR amplified and sequenced as mentioned above. The primers for ITS1-5.8S-ITS2 region were 1 and 3 (Coat et al., Citation1998), ITS3 (Shimada et al., Citation2003) and ITS4 (5′-TGATATGCTTAAGTTCAGC-3′, modified from White et al., Citation1990). The ITS1 in U. pertusa had a region with adjacent poly(G) and poly(C) that was difficult to sequence, and the ITS2 in Ulva sp. 1 had intra-individual variation of nucleotide insertion. To overcome these difficulties, their sequences were confirmed with another primer pair: 18S1505 (Hayden et al., Citation2003) and ENT26S (Blomster et al., Citation1998). For molecular identification of a relatively large number of samples, we determined their short rbcL sequence (530 nucleotides), using primers that did not include the intronic region of Ulva sp. 2 in the target region: F751 (5′-ATGGAACGTGGTCAATTTGC-3′) and rbcLend (Shimada et al., Citation2003). As the short rbcL sequence of Ulva sp. 1 was identical to those of some closely related species, the ITS1 sequence was also determine for identification of the samples of Ulva sp. 1.
The sequences for phylogenetic analyses were aligned using SeaView ver.4 (Gouy et al., Citation2010), together with previously published sequences of Ulva, and three sequences of the closely related genus, Umbraulva and Ulvaria (Heesch et al., Citation2009), as out-group taxa. The rbcL sequences were aligned manually, and the alignment consisted of 1329 sites of 1142–1329 known nucleotides. The rbcL intron of Ulva sp. 2 was removed in the alignment. The ITS1 and the ITS2 sequences were aligned by MUSCLE (Edgar Citation2004) followed by manual adjustment. The 5.8S region was removed in accordance with the sequence of Chlamydomonas reinhardtii Dangeard (U66954). The ITS1 and the ITS2 regions were 270 sites of 152–216 nucleotides with gaps, and 213 sites of 173–198 nucleotides with gaps. Ambiguously aligned parts in the ITS regions were removed with TrimAl v1.2 (Capella-Gutiérrez et al., Citation2009), using the heuristic ‘automated1’ method. The trimmed ITS1 and the ITS2 regions were 151 sites of 145–151 nucleotides with gaps, and 170 sites of 166–170 nucleotides with gaps. The rbcL dataset and the combined dataset of the trimmed ITS1 and ITS2 regions were analysed under maximum likelihood (ML), maximum parsimony (MP) and neighbour joining (NJ) methods. ML analysis was performed with Treefinder (Jobb et al., Citation2004), and MP and NJ analysis was performed with SeaView ver.4 (Gouy et al., Citation2010). Appropriate nucleotide substitution models for ML analysis were selected according to the corrected Akaike information criterion (AICc), using Kakusan4 (Tanabe Citation2011). The models for the rbcL dataset were: the first codon position, TIM+G: the second codon position, JC69+G: and the third codon position, J2+G. The model for ITS1 region was TN93ef+G, and the model for ITS2 region was J2+G. NJ trees were based on Kimura’s two parameter (K2P) distance. Bootstrap analysis was performed with 1000 replicates.
Thallus morphology was observed using molecularly identified fresh materials of two (U. fasciata), four (Ulva sp. 1) or five (U. pertusa, U. tanneri and Ulva sp. 2) individuals. Sections were obtained by hand using a razor, or with the aid of a freezing microtome (Leica CM1850; Leica Microsystems, Wetzlar, Germany). To calculate thickness ratio of layers (as ratio of cell length), and length/width ratio of cells, thallus thickness, and length and width of cells were measured from the basal (the region without rhizoidal extensions), middle and upper portions of thalli. Cell sizes were measured for three cells per cell layer in a single microscope field. Plastid orientation and the number of pyrenoids per cell were observed in the surface view of the middle region of thalli. The original material of U. conglobata illustrated by Kjellman (Citation1897: tafl. 2, –) was also examined. Specimens were directly observed under a stereomicroscope to see if the ‘secondary rhizoids’ found in Ulva sp. 2 (see results) were present. In addition, some pieces of the specimens were rehydrated and observed in surface view and section.
Fig. 1. ML tree of Ulva based on rbcL sequences. Bootstrap values are indicated at branches (ML/MP/NJ). A bootstrap value of 100% is represented by *, while that below 50% is represented by -. Samples sequenced in this study are shown in bold. 1Blomster et al. (Citation1999). 2Tan et al. (Citation1999). 3Hayden et al. (Citation2003). 4Shimada et al. (Citation2003). 5Hiraoka et al. (Citation2004a). 6Hayden & Waaland (Citation2004). 7Loughnane et al. (Citation2008). 8Heesch et al. (Citation2009). 9Ichihara et al. (Citation2009). 10O’Kelly et al. (Citation2010). 11Kraft et al. (Citation2010). 12Mareš et al. (Citation2011).13Horimoto et al. (Citation2011). 14Ichihara et al. (Citation2013).
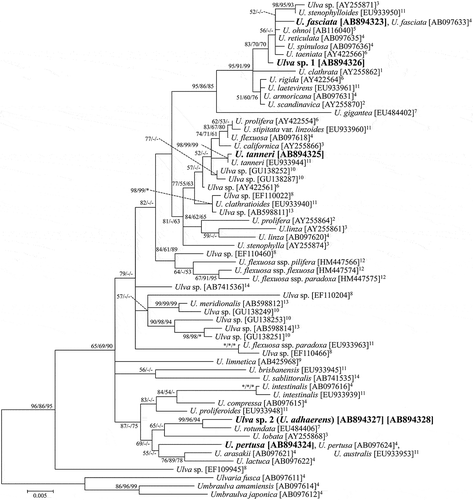
Fig. 2. ML tree of Ulva based on ITS1 and ITS2 sequences. Bootstrap values are indicated at branches (ML/MP/NJ). A bootstrap value of 100% is represented by *, while that below 50% is represented by -. Samples sequenced in this study are shown in bold. 1Coat et al. (Citation1998). 2Blomster et al. (Citation1999). 3Tan et al. (Citation1999). 4Hayden et al. (Citation2003). 5Shimada et al. (Citation2003). 6Hiraoka et al. (Citation2004a). 7Hayden & Waaland (Citation2004). 8Kraft et al. (Citation2010). 9Mareš et al. (Citation2011).
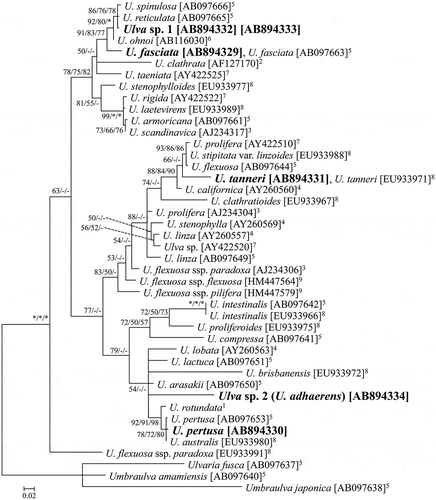
Figs 3–7. Representative fresh materials of green algae resembling Ulva conglobata. Fig. 3. U. fasciata. Fig. 4. U. pertusa. Fig. 5. U. tanneri. Fig. 6. Ulva sp. 1. Fig. 7. Ulva sp. 2 (U. adhaerens; fresh condition of the specimen designated for the holotype). Scale bars: 1 cm.
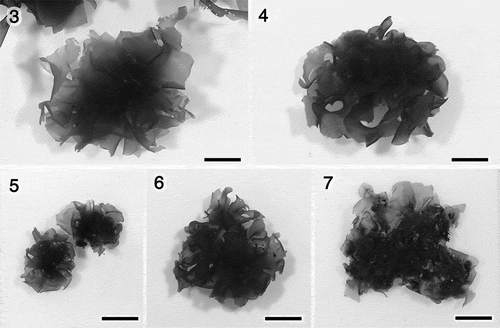
Figs 8–12. Morphology of Ulva sp. 1. Fig. 8. Marginal denticulations. Fig. 9. Surface view of the middle region. Fig. 10. Section near the base. Fig. 11. Section of the middle region. Fig. 12. Section near the margin. Scale bars: 50 μm.
Culture studies were conducted on Ulva sp. 1 and Ulva sp. 2. Swarmers were liberated from living specimens in the laboratory. The long and short axes of the swarmers were measured after fixation with a drop of 0.1% glutaraldehyde. For developmental observation, the swarmers were cultured in 30 PSU artificial seawater (Viesalt, Marine Tech, Tokyo, Japan) supplemented with a stock solution of PES medium (Provasoli’s enriched seawater; Provasoli Citation1968), at 20°C with a 14:10 h light/dark cycle (Ulva sp. 1) or 15°C with a 12:12 h light/dark cycle (Ulva sp. 2), with fluorescent light of 15–30 μmol photons m−2s−1. The swarmers settled on glass coverslips and were cultured in Petri dishes (90 mm diameter, 20 mm high). After growing to distromatic blades (approximately 1 month old), they were placed in 500 ml aerated flasks still attached to the coverslips.
Results
Molecular analysis
There were five distinct genetic taxa among the examined specimens based on rbcL and ITS sequences. The sequences of three genetic taxa were identical or almost identical to the reported sequences named U. fasciata, U. tanneri and U. pertusa, while those of the other two genetic taxa were distinct from other species in the genus (, ). For U. fasciata, U. tanneri and U. pertusa, the two individuals of each genetic species tested had identical sequences to each other in both the rbcL and ITS1-5.8S-ITS2 region. In Ulva sp. 1, the ITS2 region of each individual had intra-individual variation of one nucleotide insertion. In Ulva sp. 2, the rbcL of each individual had an intron of 2521–2522 bases; the intron had intra-individual variation of one nucleotide insertion. Those insertions were detected as overlapped signals, and were confirmed by sequencing with the forward and reverse primers with two or more separate PCRs. Total numbers of the molecularly identified samples, including those identified based on short rbcL (for all the five species) and ITS1 (for Ulva sp. 1) sequences, were as follows: U. fasciata (n = 3), U. tanneri (n = 10), U. pertusa (n = 18), Ulva sp. 1 (n = 8) and Ulva sp. 2 (n = 18). The sequences of the short rbcL and ITS1 sequences were identical among the samples of each genetic taxon.
Ulva sp. 1 in the rbcL tree () was in a well-supported clade mainly consisting of distromatic species with marginal denticulation: the group from Ulva sp. (AY255871) to U. scandinavica (AY225870; ML/MP/NJ: 95/91/99). Within the clade, Ulva sp. 1 was grouped with four denticulate species from Japan, Ulva fasciata, U. reticulata Forsskål, U. spinulosa Okamura & Segawa, U. ohnoi Hiraoka & Shimada, and three species from other localities of Pacific coasts (ML/MP/NJ: 83/70/70). In the ITS tree, Ulva sp. 1 was also in the clade of denticulate species (; from U. spinulosa (AB097666) to U. scandinavica (AJ234317); ML/MP/NJ: 78/75/82), and was grouped with the four Japanese species rather than the other species in the clade (ML/MP/NJ: 91/83/77).
Ulva sp. 2 in the rbcL tree () was in a moderately supported clade consisting of the species from U. intestinalis (AB097616) to U. lactuca (AB097622; ML/MP/NJ: 87/-/75), and was sister to U. rotundata from Ireland (ML/MP/NJ: 99/96/94). In the ITS tree (), Ulva sp. 2 was included in the clade consisting of the same members as those in the rbcL tree and one additional sequence, but this clade was supported only in the ML analysis (ML/MP/NJ: 79/-/-). The position of Ulva sp. 2 within the clade was not resolved in the ITS tree. Ulva sp. 2 was not grouped with U. rotundata in the ITS tree, though the ITS sequence named U. rotundata was obtained from a different material from that in the rbcL tree.
Morphology, phenology and habitat of field materials
The gross morphologies of the five species that resemble U. conglobata are shown in . The results of the microscopic morphological observations are summarized in . In this study there were no samples matching the characteristics of U. conglobata f. densa based on thallus size and thickness, and the section view. The microscopic morphology of the specimens matching sequences of U. fasciata, U. pertusa and U. tanneri was in agreement with the previous reports of these species (Figs S1–S8; Phillips, Citation1988; Iima & Fukusumi, Citation1996; Hiraoka et al., Citation2004b; López et al., Citation2007).
The morphology of Ulva sp. 1 is shown in . This species had microscopic marginal denticulations (). The plastids were usually covering the cells in surface view, and the number of pyrenoids was from one to three per cell (). The thickness of the layers was sometimes apparently different near the base, which was similar to the other species observed in this study (, ).
Table 1. Summary of morphological characters of the specimens resembling U. conglobata, and of the original material of U. conglobata.
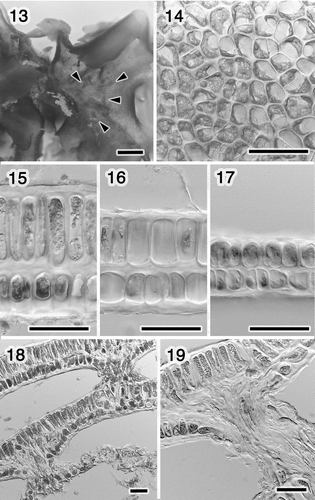
The morphology of Ulva sp. 2 (U. adhaerens) is shown in . The holdfast could not be observed, and the lobes adhered to each other by ‘secondary rhizoids’ (). Here we define ‘secondary rhizoid’ as a rhizoid generated in middle of thalli, not at the holdfast region. Marginal denticulations were not observed. The plastid orientation ranged from lateral to covering the cells in surface view (). There were one or two pyrenoids per cell (). The thickness of the layers was apparently different except in the marginal portion, and the difference was greater around the secondary rhizoids (). The rhizoidal extensions arose from both cell layers, broke through the thinner cell layer in a bundle, and adhered to another lobe (). We also observed the thalli adhering to other macroalgae by the secondary rhizoids.
Figs 13–19. Morphology of Ulva sp. 2 (U. adhaerens). Fig. 13. Close-up of thallus, showing rhizoids in a distromatic region (arrowheads). Fig. 14. Surface view of the middle region. Fig. 15. Section near the rhizoid. Fig. 16. Section of the middle region. Fig. 17. Section near the margin. Fig. 18. Section of the lobes adhering with the rhizoids. Fig. 19. Close-up of the rhizoid. Scale bars: Fig. 13, 1 mm; Figs 14–19, 50 μm.
On 24 April 2012, thalli of Ulva species were abundant in a wide range of habitats at the coast. The small thalli, which formed a turf over a large area in the upper intertidal zone, were U. tanneri. Ulva sp. 2 was found growing singly or in small clumps in the mid-intertidal zone. Only a few individuals of Ulva fasciata were found, occurring in tide pools. The thalli of U. pertusa growing singly or in small groups were found in a relatively wide range of habitats, including rocks from the upper to mid-intertidal zone as well as in tide pools, and this species was sometimes growing in association with U. fasciata, U. tanneri and Ulva sp. 2. Ulva sp. 1 was not found at that time. On 23 June 2013, thalli of Ulva species were abundant but limited to sheltered habitats of the upper intertidal zone where Ulva species had been rare in April 2012. The thalli in June were densely clumped, covering large areas and all the samples collected from several clumps were Ulva sp. 1. Only a few individuals of U. pertusa were found, but in a tide pool away from the area inhabited by Ulva sp. 1. The other three species were not found at that time.
Culture study
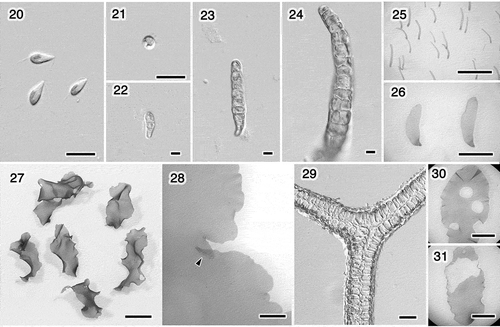
Ulva sp. 1 (). Swarmers were biflagellate, 9.5 ± 0.5 × 4.0 ± 0.3 μm in size (four strains, n = 20 per strain; ). The swarmers either settled around the release area or gathered close to the light source. The thalli that developed from the swarmers generated the same type of biflagellate swarmers, similar in size and showing a similar response to light. The settled swarmers germinated in approximately 4 days, developed into uniseriate filaments, formed tubes, then flattened and expanded into foliose thalli (), which is similar to other species of distromatic Ulva (Hayden & Waaland, Citation2002). They grew into wavy, oblong or lanceolate/oblanceolate thalli (). The margin was undulate, sometimes overlapped a little () and was occasionally denticulate. In approximately 45 days, thalli 1‒2 cm in length started releasing swarmers from the middle portion, resulting in blades with holes ().
Figs 20–31. Ulva sp. 1. Swarmers and cultures grown at 20°C with a 14: 10 h light/dark cycle. Fig. 20. Swarmers. Fig. 21. Settled swarmer. Figs 22–26. Germling. Fig. 22. 4 days. Fig. 23. 6 days. Fig. 24. 10 days. Fig. 25. Two weeks. Fig. 26. Five weeks. Fig. 27. Thalli, 52 days. Fig. 28. Margin with an overlapped portion (arrowhead), 58 days. Fig. 29. Section of the overlapped marginal portion of the thallus shown in Fig. 28. Figs 30, 31. Thallus with holes due to releasing swarmers, 44 and 45 days. Scale bars: Figs 20–24, 10 μm; Figs 25, 26, 30, 31, 5 mm; Fig. 27, 1 cm; Fig. 28, 1 mm; Fig. 29, 50 μm.
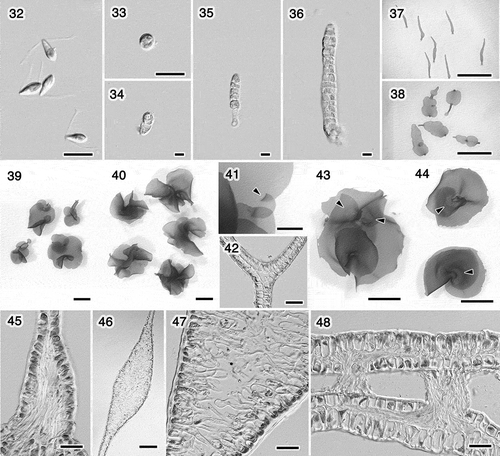
Ulva sp. 2 (U. adhaerens; ). The swarmers were biflagellate, 9.0 ± 0.5 × 4.4 ± 0.4 μm in size (five strains, n = 20 per strain; ). The swarmers settled around the release area, or gathered at the light and sometimes changed to be negatively phototactic. The thalli that developed from the swarmers generated the same type of biflagellate swarmers. The settled swarmers germinated into uniseriate filaments, formed tubes, then expanded into foliose thalli (), similar to Ulva sp. 1. In this species, however, some regions did not expand, resulting in the thalli consisting of separate lobes (). Each lobe expanded into an orbicular or reniform shape, and overlapped with another (). The margins sometimes overlapped in a manner similar to Ulva sp. 1 (). In approximately 50 days, thalli (< 1)1‒2 cm in length started releasing swarmers which were usually released from the margin of the thallus (), except where lobes were connected. Mature thalli usually had secondary rhizoids, especially near the connecting regions of the lobes () and near the base (). Rhizoidal extensions were observed in three different areas: the bases, the connecting regions of lobes, and the distromatic regions where the extensions formed the secondary rhizoids. Rhizoidal extensions at the bases were similar to the other species of Ulva, which were uniformly basipetal and adhered to the substratum (). The extensions in the connecting regions arose from both layers and extended inward, but did not emerge from the intercellular space (). The extensions in distromatic regions and the secondary rhizoids they formed were similar to those in field materials (). In culture, each adhered portion usually generated secondary rhizoids toward the other portion (). We found that thalli could generate secondary rhizoids from different surfaces within a lobe (the upper thallus in , ). The emerged rhizoidal bundles were always found adhering to something.
Figs 32–48. Ulva sp. 2 (U. adhaerens). Swarmers and cultures grown at 15°C with a 12: 12 h light/dark cycle. Fig. 32. Swarmers. Fig. 33. Settled swarmer. Figs 34–38. Germling. Fig. 34. 5 days. Fig. 35. 6 days. Fig. 36. 9 days. Fig. 37. 20 days. Fig. 38. Five weeks. Fig 39. Thalli, 53 days. Fig. 40. Thalli, 62 days. Fig. 41. Margin with an overlapped portion (arrowhead), 49 days. Fig. 42. Section of the overlapped marginal portion of the thallus shown in Fig. 41. Fig. 43. Thallus having released swarmers from marginal portion, 62 days. The adhered regions with rhizoids look paler; arrowheads. Fig. 44. Thalli adhering near the bases (arrowheads), between the different surfaces (upper) or the same surfaces (lower). Fig. 45. Basal portion with the basipetal rhizoidal extensions, 31 days. Fig. 46. Longitudinal section of the connecting region of the distromatic lobes, 47 days. Fig. 47. Close-up of Fig. 46, showing rhizoidal extensions. Fig. 48. Section of the adhering region of the thallus shown in Fig. 44 (upper), showing secondary rhizoids. Scale bars: Figs 32–36, 10 μm; Figs 37, 38, 5 mm; Figs 39, 40, 43, 44, 1cm; Fig. 4, 1 mm; Figs 42, 45, 47, 48, 50 μm; Fig. 46, 200 μm.
Morphology of the original material of U. conglobata
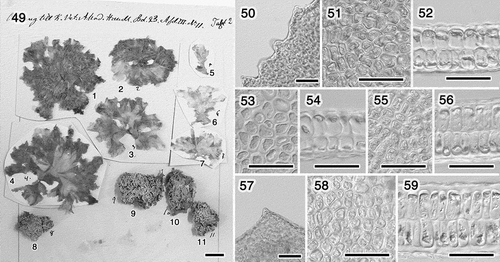
There are 11 numbered individuals on a sheet shown in the original description; specimens numbers 1–7 are U. conglobata f. typica, and specimens numbers 8–11 are U. conglobata f. densa (). We studied the microscopic morphology of specimens that were in relatively good condition: specimens 1 (), 2 (), 3 () and 4 (). We found marginal denticulations on specimens 1 and 4 (). The plastids were mostly covering the cells in the surface view on the four specimens (, , , ), which is in agreement with the sketch shown in the original description by Kjellman (Citation1897; tafl. 3, fig. 14). The pyrenoids were obscure, but we were able to observe some mainly on specimens 1 and 3. The number of pyrenoids per cell appeared to be mostly one to two (), and probably three in some cells. The section views (, , , ) were similar to the sketch shown in the original description (Kjellman, Citation1897; tafl. 3, figs 11, 12). The secondary rhizoids, which were found on Ulva sp. 2 in this study, were not found in any of Kjellman’s original material of U. conglobata f. typica, even though we scrutinized the specimens. We could not observe secondary rhizoids in the specimens of U. conglobata f. densa, due to their poor condition. The microscopic morphological characteristics of U. conglobata f. typica reported by Kjellman (Citation1897) and those observed in this study were summarized in , together with those of the entities resembling U. conglobata.
Figs 49–59. Morphology of the original material of U. conglobata. Fig. 49. Specimens of U. conglobata f. typica (1–7) and U. conglobata f. densa (8–11). Figs 50–52. Specimen no. 1. Fig. 50. Marginal denticulations. Fig. 51. Surface view. Fig. 52. Section. Figs 53, 54. Specimen no. 2. Fig. 53. Surface view. Fig. 54. Section. Figs 55, 56. Specimen no. 3. Fig. 55. Surface view. Fig. 56. Section. Figs 57–59. Specimen no. 4. Fig. 57. Marginal denticulation. Fig. 58. Surface view. Fig. 59. Section. Scale bars: Fig. 49, cm; Figs 50–59, 50 μm.
Species description
Based on the molecular analysis and the morphological features of Ulva sp. 2, we propose it as a new species of Ulva, U. adhaerens sp. nov.
Ulva adhaerens Matsumoto & Shimada, sp. nov. (, , , )
Fig. 60. Ulva adhaerens. Photograph of the holotype herbarium specimen, TNS-AL 183435. Scale bar: 1 cm.
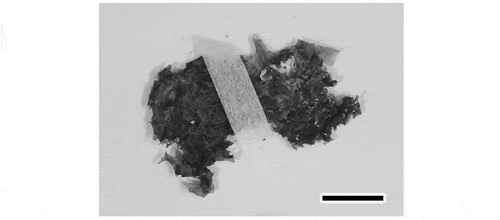
Diagnosis: Thalli distromatic foliose, lobed, on bedrock or macroalgae, growing singly or in clumps; lobes expanding only a few centimetres, 50–110 μm thick, adhering to each other by rhizoids generated in distromatic regions; thickness of the cell layers much different around the rhizoids; rhizoidal extensions in distromatic regions arising from both cell layers toward interlayer space, breaking through the thinner cell layer in a bundle, and adhering to another lobe; pyrenoids one or two per cell; plastid orientation ranging from lateral to covering the cells in surface view; marginal denticulations not observed. Plastid-encoded rbcL gene sequence = DDBJ accession AB894327, AB894328, nuclear-encoded ITS1-5.8S-ITS2 region sequence = DDBJ accession AB894334.
Holotype: Specimen TNS-AL 183435 deposited in TNS (photograph shown in ); collected on 24 Apr. 2012.
Isotypes: Specimens TNS-AL 183436 deposited in TNS; specimens SAP 114680, deposited in the Herbarium of the Faculty of Science at Hokkaido University, Sapporo, Japan (SAP).
Type locality: Tenjin-jima, Sajima, Yokosuka, Kanagawa Prefecture, Japan: 35° 13′ 20″ N, 139° 36′ 10″ E.
Etymology: adhaerens refers to the lobes adhering to each other by rhizoids.
Japanese name: Kasane-aosa
Discussion
The results showed that green algal samples resembling Ulva conglobata in gross morphology included at least five genetically distinct species: U. fasciata, U. pertusa, U. tanneri, Ulva sp. 1 and Ulva sp. 2 (U. adhaerens). Based on these results and examination of the original material of U. conglobata, it is possible to distinguish some of these species by microscopic morphology. According to the morphological observation of the original material and the sketches in the original description, U. conglobata is considered to have marginal denticulations and plastids mostly covering the cells in surface view. Marginal denticulation is considered a heritable characteristic, as this trait repeatedly appeared in culture only on particular species and its presence was associated with the phylogeny (Hiraoka et al., Citation2004a). Therefore we can distinguish the species without denticulation, namely, U. pertusa, U. tanneri and U. adhaerens from U. conglobata. Their distinctiveness from U. conglobata is also supported by other characteristics. Ulva pertusa had a thicker thallus and lateral plastid orientation. Ulva adhaerens showed rhizoids in distromatic regions. The margin of U. tanneri was often corrugated and sometimes looked as if it had denticulations, perhaps because this species forms foliose thalli by rupturing distromatic saccate thalli. However, U. tanneri had an interlayer space that is absent in the sketches of U. conglobata (Kjellman Citation1897, tafl. 3, figs 11, 12, 15). On the other hand, the species with denticulation, U. fasciata and Ulva sp. 1, could possibly apply to U. conglobata. These two species do not clearly differ from U. conglobata in other traits either.
In Japan, six denticulate species have been reported (Yoshida & Yoshinaga, Citation2010). The four closely related species, U. fasciata, U. reticulata, U. spinulosa and U. ohnoi, have been distinguished based on gross morphology and crossing tests (Hiraoka et al., Citation2004a). However, O’Kelly et al. (Citation2010) reported that Ulva sp. from the Hawaiian Islands showed U. reticulata morphology but had rbcL sequences of U. ohnoi and ITS1 sequence that was a little different from those of U. ohnoi or U. reticulata, which suggests the necessity for reinvestigation of the species concept. The other two species, U. armoricana Dion, de Reviers & Coat and U. scandinavica Bliding [both suggested to be synonyms of U. rigida C. Agardh (Hayden & Waaland Citation2004; Maggs et al., Citation2007)], were recently reported in Japan, and are considered non-indigenous (Shimada et al., Citation2003; Kawai et al., Citation2007). Most of those denticulate species are known to have large expanding thalli (Dion et al., Citation1998; Hiraoka et al., Citation2004a; Kawai et al., Citation2007), though U. spinulosa was described as a small, circular and irregularly lobed species (2‒3 cm high; Segawa Citation1936). Among the known forms of the denticulate species, the samples of Ulva sp. 1 observed in this study seem most similar to U. conglobata in the ball-like morphology, the thinner thallus, and the gregarious behaviour in the field, although Ulva sp. 1 was found in the upper intertidal zone while Kjellman (Citation1897) reported that U. conglobata was in mid to lower coastal areas. At this time, however, uncertainty remains as to whether or not Ulva sp. 1 conforms to U. conglobata. Given the morphological plasticity and ongoing reports of genetic entities in this genus (e.g. Heesch et al., Citation2009; Kraft et al., Citation2010; O’Kelly et al., Citation2010), DNA sequences of type specimens should be determined in order that comparisons can be made with sequence data from collected material. However, as we could not obtain the permission of the owner, DNA extraction and sequencing for the original material of U. conglobata was not possible. Besides this, it would also be useful to conduct a broad-scale study on the phylogeny, morphology, life cycle and nomenclature of the denticulate species in order to arrive at clear species limits and appropriate species names.
Ulva adhaerens was unique among Ulva species in that it produced rhizoids at distromatic regions other than the base (e.g. Bliding, Citation1968; Koeman & van den Hoek Citation1981; Yoshida, Citation1998; Kraft, Citation2007; Maggs et al., Citation2007). This ‘secondary rhizoid’ was considered as a stable characteristic, as it was almost always observed in culture. The developmental pattern seen in culture, in which some regions did not become distromatic and were filled by rhizoidal extensions, was also a unique feature in this genus. The connecting regions are considered analogous to the thallus base, as they produced rhizoidal extensions and did not mature, and as secondary rhizoids occurred nearby. The standard structure of Ulva species, which consist only of the leaf and the base with unidirectional extensions, may be interpreted by single polarity that is likely derived from the first asymmetric cell division (Spoerner et al., Citation2012). However, U. adhaerens provided unexpected insights in the morphogenesis of Ulva. Firstly, its cells can differentiate rhizoidal extensions at regions other than the base. Secondly, the thallus has two additional kinds of polarity: unevenness in the parallel direction of the surface for gathering the rhizoidal extensions, and dorsoventrality for protruding the rhizoidal bundle from the thinner layer. The dorsoventrality was constructed locally, as it could be reversed in different portions of the same thallus. In addition, since the rhizoidal bundles were always found adhering, their formation may be triggered by mechanical touch stimuli.
Ulva species basically have an isomorphic and diphasic life history with biflagellate gametes and quadriflagellate zoospores (Bliding, Citation1968; Koeman & van den Hoek, Citation1981; Tatewaki Citation1994). However, some species in various lineages have an asexual life history with biflagellate or quadriflagellate swarmers; such species are considered diploid and to have lost their sexuality secondarily (e.g. Bliding, Citation1968; Hiraoka et al., Citation2003a, c). There can also be variation of the life history within a species, that is, among the thalli that are not distinguished by DNA sequences or in morphology (Hiraoka et al., Citation2003b, Citation2011). In Ulva, the asexual swarmers and zoospores are different from gametes in size and phototaxis; they are larger than gametes, and are generally negatively phototactic while gametes are positively phototactic, though asexual swarmers sometimes show positive phototaxis soon after their release (Bliding, Citation1968; Hiraoka et al., Citation2003a, c). The phototaxis of the swarmers of Ulva sp. 1 and U. adhaerens was ambiguous. However, the swarmers were biflagellate, as large as asexual swarmers and zoospores of other species (Bliding, Citation1968; Hiraoka et al., Citation2003a), and the offspring generated the same type of swarmers. Therefore, we consider that the specimens of Ulva sp. 1 and U. adhaerens observed in this study have an asexual life cycle.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at http://10.1080/09670262.2014.994189
Supplementary Figs S1–S7.
Supplementary Figs S1-S7
Download JPEG Image (981.4 KB)Acknowledgements
We thank M. Hiraoka for teaching culture methods and for helpful comments; S. Ryman and T. Kitayama for loaning the original material of U. conglobata; K. Ichihara for field assistance and technical advice; Y. Omori and M. Nagano for field assistance; H. Nozaki and H. Kawai for helpful comments; H. Verbruggen, K. Yura and A. Kage for linguistic advice.
Additional information
Funding
References
- Bliding, C. (1968). A critical survey of European taxa in Ulvales. Part II: Ulva, Ulvaria, Monostroma, Kornmannia. Botaniska notiser, 121: 535–629.
- Blomster, J., Maggs, C.A. & Stanhope, M.J. (1998). Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles. Journal of Phycology, 34: 319–340.
- Blomster, J., Maggs, C.A. & Sanhope, M.J. (1999). Extensive intraspecific morphological variation in Enteromorpha muscoides (Chlorophyta) revealed by molecular analysis. Journal of Phycology, 35: 575–586.
- Capella-Gutiérrez, S., Silla-Martínez, J.M. & Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 25: 1972–1973.
- Coat, G., Dion, P., Noailles, M.C., De Reviers, B., Fontaine, J.M., Bergaer-Perrot, Y. & Loiseaux-De Goër, S. (1998). Ulva armoricana (Ulvales, Chlorophyta) from the coasts of Brittany (France). II. Nuclear rDNA ITS sequences analysis. European Journal of Phycology, 33: 81–86.
- Couceiro, L., Cremades, J. & Barreiro, R. (2011). Evidence for multiple introductions of the Pacific green alga Ulva australis Areschoug (Ulvales, Chlorophyta) to the Iberian Peninsula. Botanica Marina, 54: 391–402.
- Dion, P., de Reviers, B. & Coat, G. (1998). Ulva armoricana sp. nov. (Ulvales, Chlorophyta) from the coasts of Brittany (France). I. Morphological identification. European Journal of Phycology, 33: 73–80.
- Edgar, R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32: 1792–1797.
- Gouy, M., Guindon, S. & Gascuel, O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution, 27: 221–224.
- Guiry, M.D. & Guiry, G.M. (2013). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 6 December 2013.
- Hayden, H.S. & Waaland, R.J. (2002). Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences. Journal of Phycology, 38: 1200–1212.
- Hayden, H.S. & Waaland, R.J. (2004). A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific. Phycologia, 43: 364–382.
- Hayden, H.S., Blomster, J., Maggs, C.A., Silva, P.C., Stanhope, M.J. & Waaland, J.R. (2003). Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. European Journal of Phycology, 38: 277–294.
- Heesch, S., Broom, J.E.S., Neill, K.F., Farr, T.J., Dalen, J.L. & Nelson, W.A. (2009). Ulva, Umbraulva and Gemina: genetic survey of New Zealand taxa reveals diversity and introduced species. European Journal of Phycology, 44: 143–154.
- Hiraoka, M. & Shimada, S. (2004). In Biology and Technology of Economic Seaweeds (Ohno, M., editor), 14–23. Uchida Rokakuho Publishing, Tokyo.
- Hiraoka, M., Shimada, S., Ohno, M. & Serisawa, Y. (2003a). Asexual life history by quadriflagellate swarmers of Ulva spinulosa (Ulvales, Ulvophyceae). Phycological Research, 51: 29–34.
- Hiraoka, M., Dan, A., Shimada, S., Hagihira, M., Migita, M. & Ohno, M. (2003b). Different life histories of Enteromorpha prolifera (Ulvales, Chlorophyta) from four rivers on Shikoku Island, Japan. Phycologia, 42: 275–284.
- Hiraoka, M., Shimada, S., Serisawa, Y., Ohno, M. & Ebata, H. (2003c). Two different genetic strains of stalked-Ulva (Ulvales, Chlorophyta) grow on intertidal rocky shores in Ebisujima, central Japan. Phycological Research, 51: 161–167.
- Hiraoka, M., Shimada, S., Uenosono, M. & Masuda, M. (2004a). A new green-tide-forming alga, Ulva ohnoi Hiraoka et Shimada sp. nov. (Ulvales, Ulvophyceae) from Japan. Phycological Research, 52: 17–29.
- Hiraoka, M., Ohno, M., Kawaguchi, S. & Yoshida, G. (2004b). Crossing test among floating Ulva thalli forming ‘green tide’ in Japan. Hydrobiologia, 512: 239–245.
- Hiraoka, M., Ichihara, K., Zhu, W., Ma, J. & Shimada, S. (2011). Culture and hybridization experiments on an Ulva clade including the qingdao strain blooming in the Yellow Sea. PLoS ONE, 6: e19371.
- Horimoto, R., Masakiyo, Y., Ichihara, K. & Shimada, S. (2011). Enteromorpha-like Ulva (Ulvophyceae, Chlorophyta) growing in the Todoroki river, Ishigaki island, Japan, with special reference to Ulva meridionalis Horimoto et Shimada, sp. nov. Bulletin of the National Museum of Nature and Science. Series B, Botany, 37: 155–167.
- Ichihara, K., Arai, S., Uchimura, M., Fay, E.J., Ebata, H., Hiraoka, M. & Shimada, S. (2009). New species of freshwater Ulva, Ulva limnetica (Ulvales, Ulvophyceae) from the Ryukyu Islands, Japan. Phycological Research, 57: 94–103.
- Ichihara, K., Miyaji, K. & Shimada, S. (2013). Comparing the low-salinity tolerance of Ulva species distributed in different environments. Phycological Research, 61: 52–57.
- Iima, M. & Fukusumi, K. (1996). First report of Chloropelta caespitosa (Ulvaceae, Ulvales, Ulvophyceae) in Japan. Phycological Research, 44: 167–171.
- Jobb, G., von Haeseler, A. & Strimmer, K. (2004). TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evolutionary Biology, 4: 18.
- Kang, S.-H. & Lee, K.-W. (2002). Phylogenetic relationships between Ulva conglobata and U. pertusa from Jeju Island inferred from nrDNA ITS 2 sequences. Algae, 17: 75–81.
- Kawai, H., Shimada, S., Hanyuda, T., Suzuki, T. & Gamagori City Office (2007). Species diversity and seasonal changes of dominant Ulva species (Ulvales, Ulvophyceae) in Mikawa Bay, Japan, deduced from ITS2 rDNA region sequences. Algae, 22: 221–228.
- Kirkendale, L., Saunders, G.W. & Winberg, P. (2013). A molecular survey of Ulva (Chlorophyta) in temperate Australia reveals enhanced levels of cosmopolitanism. Journal of Phycology, 49: 69–81.
- Kjellman, F.R. (1897). Marina chlorophyceer från Japan. Bihang till Kungliga Svenska Vetenskaps-akademiens Handlingar, 23 (Afd. III, 11): 1–44.
- Koeman, R.P.T. & van den Hoek, C. (1981). The taxonomy of Ulva (Chlorophyceae) in the Netherlands. British Phycological Journal, 16: 9–53.
- Kraft, G.T. (2007). Algae of Australia: Marine benthic algae of Lord Howe Island and the southern Great Barrier Reef, 1: Green algae. ABRS, Canberra; CSIRO Publishing, Melbourne. 347 pp.
- Kraft, L.G.K., Kraft, G.T. & Waller, R.F. (2010). Investigations into Southern Australian Ulva (Ulvophyceae, Chlorophyta) taxonomy and molecular phylogeny indicate both cosmopolitan and endemic cryptic species. Journal of Phycology 46: 1257–1277.
- López, S.B., Fernández, I.B., Lozano, R.B. & Ugarte, J.C. (2007). Is the cryptic alien seaweed Ulva pertusa (Ulvales, Chlorophyta) widely distributed along European Atlantic coasts? Botanica Marina, 50: 267–274.
- Loughnane, C.J., McIvor, L.M., Rindi, F., Stengal, D.B. & Guiry, M.D. (2008). Morphology, rbcL phylogeny and distribution of distromatic Ulva (Ulvophyceae, Chlorophyta) in Ireland and southern Britain. Phycologia, 47: 416–429.
- Maggs, C.A., Blomster, J., Mineur, F. & Kelly, J. (2007). Ulva Linnaeus. In Green Seaweeds of Britain and Ireland (Brodie, J., Maggs, C.M. & John, D.M., editors), 80–103. British Phycological Society, London.
- Mareš, J., Leskinen, E., Sitkowska, M., Skácelová, O. & Blomster, J. (2011). True identity of the European freshwater Ulva (Chlorophyta, Ulvophyceae) revealed by a combined molecular and morphological approach. Journal of Phycology, 47: 1177–1192.
- Mshigeni, K.E. & Kajumulo, A.A. (1979). Effects of the environment on polymorphism in Ulva fasciata Delile (Chlorophyta, Ulvaceae). Botanica Marina, 22: 145–148.
- O’Kelly, C.J., Kurihara, A., Shipley, T.C. & Sherwood, A.R. (2010). Molecular assessment of Ulva spp. (Ulvophyceae, Chlorophyta) in the Hawaiian Islands. Journal of Phycology, 46: 728–735.
- Phillips, J.A. (1988). Field, anatomical and developmental studies on southern Australian species of Ulva (Ulvaceae, Chlorophyta). Australian Systematic Botany, 1: 411–456.
- Provasoli, L. (1968). Media and prospects for the cultivation of marine algae. In Cultures and Collections of Algae. Proceedings of U.S.–Japan Conference, Hakone, September 1966 (Watanase, A. & Hattori, A., editors), 47–74. The Japanese Society of Plant Physiologist, Tokyo.
- Segawa, S. (1936). On the marine algae of Susaki, Prov. Izu, and its vicinity II. Scientific Papers of the Institute of Algological Research, Faculty of Science, Hokkaido Imperial University, 1: 175–197.
- Shimada, S., Hiraoka, M., Nabata, S., Lima, M. & Masuda, M. (2003). Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva. Phycological Research, 51: 99–108.
- Spoerner, M., Wichard, T., Bachhuber, T., Stratmann, J. & Oertel, W. (2012). Growth and thallus morphogenesis of Ulva mutabilis (Chlorophyta) depends on a combination of two bacterial species excreting regulatory factors. Journal of Phycology, 48: 1433–1447.
- Tan, I.H., Blomster, J., Hansen, G., Leskinen, E., Maggs, C.A., Mann, D.G., Sluiman, H.J. & Stanhope, M.J. (1999). Molecular phylogenetic evidence for a reversible morphogenetic switch controlling the gross morphology of two common genera of green seaweeds, Ulva and Enteromorpha. Molecular Biological Evolution, 16: 1011–1018.
- Tanabe, A.S. (2011). Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Molecular Ecology Resources, 11: 914–921.
- Tatewaki, M. (1994). In An Illustrated Atlas of the Life History of Algae. I. Green Algae (Hori, T., editor), 190–195. Uchida Rokakuho Publishing, Tokyo.
- White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR protocols: a guide to methods and applications (Innis, A., Gelfand, D.H., Sninsky, J.J. & White, T.J., editors), 315–322. Academic Press, New York.
- Yoshida, T. (1998). Marine Algae of Japan. Uchida Rokakuho Publishing, Tokyo. 1222 pp.
- Yoshida, T. & Yoshinaga, K. (2010). Checklist of marine algae of Japan (revised in 2010). Japanese Journal of Phycology, 58: 69–122.
