Abstract
Two non-native stalk-forming diatoms that were recently observed in the West Branch of the Farmington River, a tributary of the Connecticut River in Connecticut (USA), are characterized morphologically and barcode marker sequences were obtained for each of them. Cymbella janischii, the dominant stalk-forming species during the summer of 2012, previously had not been found in the northeastern USA. Samples of C. janischii were examined microscopically and used to obtain four sequences of the barcode marker, the V4 region of the 18S rDNA gene. Phylogenetic analysis indicated that the four independent sequences of C. janischii were distinct from, but most closely related to, published sequences of C. janischii from Idaho and C. mexicana from Texas, USA. A second non-native stalk-producing diatom, resembling Didymosphenia geminata, was found in November 2012 – June 2013 and first reported as Didymosphenia sp. Over this period, the observed cells had a compressed morphology and were consistently small compared with D. geminata. Sequences of the V4 region, obtained from three independent direct polymerase chain reactions (PCR) of single cells isolated from the Connecticut samples, indicated a close relationship to three published sequences of D. geminata from Italy, New Zealand and the USA, and to D. siberica and D. dentata from Russia. Frustules of the cells used in the PCR reactions were recovered and examined using scanning electron microscopy, providing a direct link between the observed morphology and sequence data. The morphology of the novel Connecticut Didymosphenia taxon was compared with that of other Didymosphenia taxa, being most similar to D. pumila, D. laticollis, D. grunowii and smaller cells of D. geminata. Didymosphenia sp. had a triundulate morphology with a consistent length of 40–60 µm. Given the unique morphological features of this diatom, it is proposed as a new species, Didymosphenia hullii Khan-Bureau, sp. nov.
INTRODUCTION
The freshwater stalk-forming diatom Didymosphenia geminata (Lyngb.) M. Schmidt is a well-known invasive and nuisance species with an ability to produce copious extracellular polymeric substances that form the stalks (Blanco & Ector, Citation2009; Aboal et al., Citation2012). Cymbella janischii (A.W.F. Schmidt) De Toni, another stalk-forming diatom with abundant stalk growth is often mistaken for D. geminata at the macroscopic level with tufts that are similar in appearance (Pite et al., Citation2009; Whitton et al., Citation2009). Both species are commonly referred to as rock snot. Under certain environmental conditions these species grow prolifically, forming thick mats that cover sections of the river substrate, negatively impacting other aquatic organisms (Kilroy, Citation2004; Spaulding & Elwell, Citation2007; Kumar et al., Citation2009; Morales et al., Citation2012; Zgłobicka, Citation2013; Kuhajek & Wood, Citation2014). Unlike C. janischii, D. geminata prefers oligotrophic, cold and low soluble reactive phosphorus environments, which may in part cause unusual overgrowth conditions (Krammer & Lange-Bertalot, Citation1986; Kilroy & Bothwell, Citation2011; Bothwell et al., Citation2012, Citation2014). In addition, D. geminata establishment is influenced by a structurally suitable substrate, the development of a pad which adheres to the substrate, and the orientation of the cell upon attachment (Kilroy & Bothwell, Citation2014; Kuhajek & Wood, Citation2014; Kuhajek et al., Citation2014).
Many states throughout the USA are monitoring their waterways for D. geminata because of its expanding geographic range (Kuhajek & Wood, Citation2014). In the USA, D. geminata was transported from the western states into several southeastern states, and more recently to northeastern states (Bothwell & Spaulding, Citation2008; Blanco & Ector, Citation2009; Spaulding, Citation2010). In May 2013, Massachusetts first recorded and confirmed an occurrence of D. geminata with growth lasting two months (A. Madden, MA. Div. Fisheries and Wildlife, pers. comm.).
The Connecticut Department of Energy and Environmental Protection (CT DEEP, Citation2011) started monitoring the West Branch of the Farmington River after purported D. geminata tufts were observed in 2011. In July 2012 reports of mucilaginous tufts occurring downstream of the original location in 2011 were later confirmed to be substantial growths of C. janischii. In addition, an unusual morphological population of Didymosphenia sp. was found in November 2012 (Khan-Bureau et al., Citation2014) at another location along the river. The present study characterizes the morphology of these two diatoms from the West Branch of the Farmington River in Connecticut. We show that Didymosphenia sp. is morphologically distinct from other Didymosphenia species and propose it as a new taxon. In addition, we present new sequence data of the V4 rDNA region in order to link the morphological and genetic information for these two diatoms.
MATERIALS AND METHODS
The West Branch of the Farmington River (WBFR) is located in Northwestern Connecticut, USA. The WBFR is impounded twice, first at the Colebrook River Reservoir and then at the West Branch Reservoir. The Metropolitan District Commission (MDC) operates West Branch Reservoir and has a contractual agreement to maintain a minimum discharge from the oligotrophic hypolimnion (C. Bellucci CT DEEP, pers. comm.; MDC, Citation2013). The WBFR has stable flow regimes and substrate stability because discharge from this reservoir is managed. It also has very cold and oligotrophic water, making it conducive for the growth of D. geminata as reported by Kilroy (Citation2004), Spaulding & Elwell (Citation2007), Bothwell & Kilroy (Citation2011) and Didymosphenia sp. (Khan-Bureau et al., Citation2014).
Benthic samples were collected from the West Branch of the Farmington River in July 2012 – June 2013 (Khan-Bureau et al., Citation2014). Samples of the mucilaginous tufts were taken from rock substrate, submerged vegetation, and overhanging tree branches, and placed in Whirlpac® bags. The latter were placed on ice, and transported to the lab for processing.
Light microscopy (LM)
Prior to acid-washing the samples, live samples were placed on a microscope slide with a coverslip overlain and then viewed at 200 and 400× magnifications using a BX 60 Olympus microscope. Images were digitally captured using an Olympus DP 25 camera and cellSens software then viewed to identify the taxa according to Krammer & Lange-Bertalot (Citation1986), Round et al. (Citation1990), and three online databases, the ANSP Algae Image Database (http://diatom.ansp.org/algae_image/), Diatoms of the United States (http://westerndiatoms.colorado.edu/) and the Great Lakes Image Database (http://www.umich.edu/~phytolab/GreatLakesDiatomHomePage/top.html). For permanent slide preparation the river samples were centrifuged to concentrate the diatom cells to the bottom of the microtube. The supernatant was poured off and distilled water was added. Samples were then simmered on a hot plate in a 1:1 ratio of water and 68% nitric acid to oxidize organic matter, then taken off the hotplate and allowed to cool for several minutes. Deionized water was used to rinse the samples of the acid, 4–5 times to neutralize samples, and then centrifuged to concentrate the diatom frustules (following the protocol of R. Lowe, pers. comm.). After air-drying the diatom samples overnight on coverslips, frustules were mounted on glass microscope slides in the mounting medium NAPHRAX®, heated on a hot plate and then cooled to produce permanent vouchers. The diatom frustules were examined at 600 and 1000× magnifications with a BX 60 Olympus microscope. 125 valves were measured. Images were captured using an Olympus DP 25 colour camera (2560 × 1920 pixels).
Scanning electron microscopy (SEM)
A mixture of glutaraldehyde and Bold Basal Medium (BBM) was used to fix and prepare the Didymosphenia sp. tufts for SEM analyses. The tufts were placed in the mixture and centrifuged at 600 × g to guard against stalk material damage. The supernatant was discarded and the tufts were resuspended in BBM combined with 4% glutaraldehyde overnight. The samples were placed in 2% osmium tetroxide for 2 h. The samples were dehydrated through a graded ethanol (C2H5OH) series of 30, 50, 70, 95 and 100% on ice, with the sample remaining in a shallow glass Petri dish bath for 15 m per step. Critical point drying using a Tousimis 931.GL apparatus was followed by sputter coating. The stubs were viewed with the field emission Leo/Zeiss DSM 982 and a field emission FEI Nova Nano 450 scanning electron microscope. LM and SEM image plates were created using Adobe® Creative Suite® 6 Photoshop.
Also, the 0.2 ml Eppendorf® tube residual from the original PCR product (see below) was saved for SEM verification of the isolated cell for identification purposes. The single frustule was retrieved from several individual PCR tubes, washed in deionized water, and centrifuged to ensure that the cell was not discarded and could later be found at the bottom of the tube for SEM verification. The supernatant was removed and replaced with 25 µl of deionized water and transferred onto a 25 mm, 3 μm pore size polycarbonate Millipore filter (Lang & Kaczmarska, Citation2011). The filter was placed on a SEM stub with double-sided tape. For C. janischii we followed the methodology of Morales et al. (Citation2001), and the stubs were coated for 1 min at 1.8 kV with gold/palladium using a Polaron sputter coater.
DNA extraction, PCR and cloning of Cymbella janischii
One water sample from July 2012 was used for molecular characterization of C. janischii. This sample was centrifuged, rinsed with deionized water, and then split into three replicate 50 µl microtubes. DNA extraction was accomplished using the MoBio PowerLyser™ Soil Extraction kit. PCR amplification of the V4 region of the 18S rDNA gene was achieved using primers D512F and D978R (Zimmermann et al., Citation2011). The PCR temperature profile included an initial denaturation step at 9°C (2 min), then five cycles consisting of denaturation at 94°C (45 s), annealing at 52°C (45 s) and elongation at 72°C (1 min), followed by 35 cycles in which the annealing temperature was lowered to 50°C, and a final elongation at 72°C (10 min). Resulting PCR products were visualized on a Syber Safe stained agarose gel, then quantified with a Nano Drop spectrophotometer. Cloning of PCR products was performed using Invitrogen TOPO® TA Cloning® Kit. Plasmid Prep followed using the QIAprep® Spin Miniprep Kit.
Direct PCR of single cells of Didymosphenia
To best match DNA sequences with a specific morphotype, direct PCR was performed on cells that later were used for SEM imaging. Initially several cells from fresh samples, were isolated using a micropipette and placed in a 0.2 ml PCR tube. From these tubes, one individual cell of Didymosphenia sp. was placed in three replicate PCR tubes and washed 3–5 times (Lang & Kaczmarska, Citation2011). After the final wash and centrifugation, the supernatant was removed and replaced by 1 μl of sterile water. The samples were then heated at 95°C on a thermocycler for 10 min prior to PCR to open the frustules for DNA extraction (Lang & Kaczmarska, Citation2011). PCR amplification of the V4 region of the 18S rDNA gene was achieved using the primers D512F and D978R of Zimmermann et al. (Citation2011). The PCR mix consisted of 10 μl GoTaq® Green Master Mix, 0.5 μl of each primer (Zimmermann et al., Citation2011), and sterile deionized water for a final volume of 20 μl in the PCR tubes, each containing a single Didymosphenia sp. cell. The PCR temperature profile used for the amplification of C. janischii DNA was also employed here.
Sequencing of Didymosphenia sp. and C. janischii
The cloned fragments and cleaned PCR fragments were directly sequenced using the amplification primers of Zimmermann et al. (Citation2011). The sequencing cycle comprised 27 cycles of denaturing at 96°C for 30 s, annealing at 50°C for 15 s, and extension at 60°C for 4 min, using the Big Dye™ Terminator Cycle Sequencing Reaction Kit (Applied Biosystems, Foster City, California, USA). Products of cycle sequencing were cleaned using ethanol precipitation and analysed on an ABI 3100 DNA Sequencer™ (Applied Biosystems, Foster City, California, USA). Contigs of individual reads were assembled in Geneious© (Geneious, Citation2013), to produce consensus sequences. These were compared with published data in GenBank, using the BLASTn tool, to obtain information on the closest matches. The sequences were aligned against a sampling of the sequences presented in Kermarrec et al. (Citation2011) and the sequence of Cocconeis stauroneiformis (W. Sm.) Okuno AB430614 (Sato et al., Citation2008). Sequences were aligned manually using Geneious© (Geneious, Citation2013). Confidence of branch support was assessed using MrBayes (Huelsenbeck & Ronquist, Citation2001; Ronquist & Huelsenbeck, Citation2003). Bayesian analyses were run for 106 × 3 generations with one cold chain and 3 heated chains, and sampling every 1000 generations. The first 10% of the trees were discarded as burn-in. Alignments will be available from www.treebase.org.
RESULTS
LM and SEM analysis
Cells of C. janischii were an average of 130 μm long, consistent with the published range of 105–383 μm (Bahls, Citation2007) (Supplementary figs 1–4). Several LM images were taken of live single and recently divided cells of Didymosphenia sp. prior to preparing the samples for acid cleaning (–). Sexual reproduction was not observed over this eight month time period. The valve and girdle views of Didymosphenia sp. illustrate the cell size variation of this population (–). Comparisons were made of LM images of smaller D. geminata cells, which ranged in size from 57–161 μm from plates in Spaulding (Citation2010) and Metzeltin & Lange Bertalot (Citation2014), and Didymosphenia sp. cells (–) as shown in Supplementary table 1. The smaller D. geminata cells vary significantly in size whereas the Didymosphenia sp. valves in all observed cells of this population ranged consistently from 40–60 μm over eight months (–).
Figs 1–6. LM images of field collected Didymosphenia hullii. , , : Cells attached to their stalks illustrate cell division. Scale bars = 20 μm (, , ); 10 μm (); 50 μm (); 40 μm ().
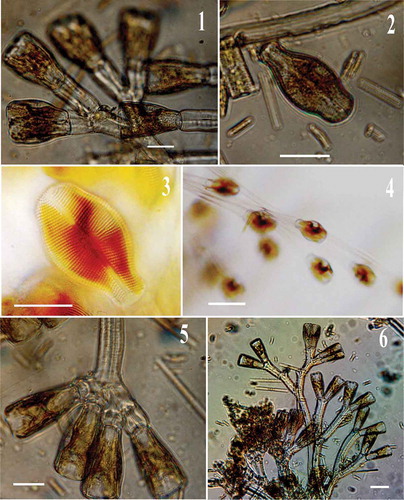
Figs 7–15. LM images of Didymosphenia hullii cells in valve view showing size variation. : LM image of D. hullii cell in girdle view. Scale bars = 10 μm.
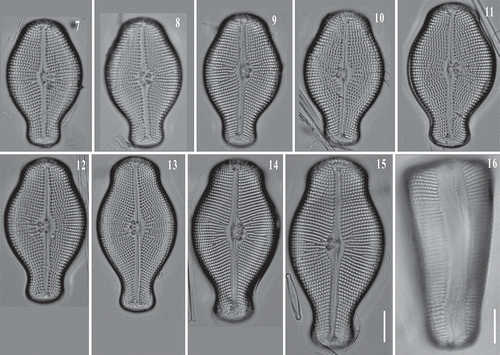
Figs 17–25. LM images of Didymosphenia geminata cells in valve view showing size variation. : Courtesy of S. Spaulding, –: Courtesy of D. Metzeltin. Reprinted from Metzeltin & Lange-Bertalot (Citation2014, Iconographia Diatomologica 25) with the permission of Koeltz Scientific Books. : D. hullii cell in valve view. Scale bars = 10 μm.
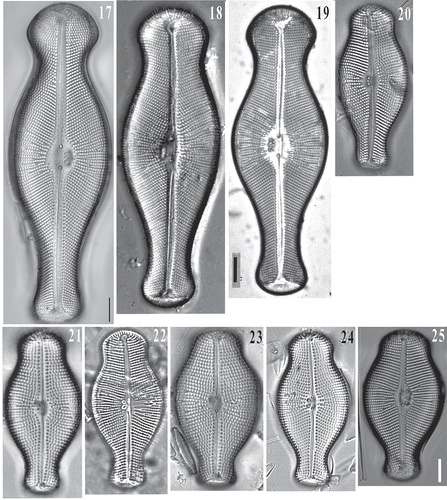
Didymosphenia sp. frustules were successfully retreived after the single-cell PCR reactions were completed, placed on a Millipore filter on a stub and viewed with SEM. Although the recovered frustules were fractured, they were adequate for identification (–). These frustules correspond to the cells used to generate sequences of Didymosphenia sp. in the phylogenetic tree (, see below). Also, SEM images of the Didymosphenia sp. cells attached to their stalks further demonstrate the small cell size and the compressed valve morphology (–). This is in contrast to cells of D. geminata found at other North American sites, including Massachusetts, which are more robust and up to 137 µm (Kilroy, Citation2004; Spaulding, Citation2010). SEM features of the apical pore field, stigmata, striae, shape, areolae and length of the frustule can be used to distinguish this taxon from other members of Didymosphenia (–, ). Didymosphenia sp. is 40–60 μm in length, with cells of 38 µm and 68 µm occuring less frequently. The width is 26.5–30.5 μm with 1–4 stigmata and 9–11 striae in 10 µm. The areolae have deep inclined walls and are surrounded by spine-like projections (spines) with dendritic slits below the spines, and are most similar to the areolae of D. geminata, D. clavaherculis (Ehrenb.) Metzeltin & Lange-Bert., and D. laticollis Metzeltin & Lange-Bert. and as described by Metzeltin & Lange-Bertalot (Citation1995, Citation2014), although the cell shape and size of D. clavaherculis and D. laticollis are distinct from that of Didymosphenia sp.
Figs 26–28. SEM images of single valves of Didymosphenia hullii retrieved after PCR reactions. : The valve is fractured but still identifiable. : High magnification image of a cell with two stigmata. Scale bars = 20 μm (, ); 5 μm ().
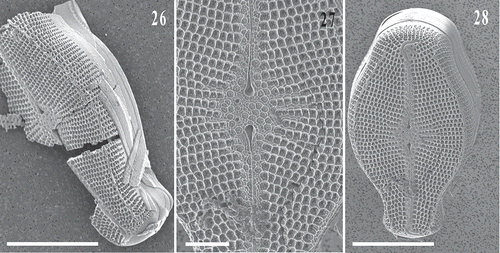
Fig. 29. Bayesian phylogenetic tree based the V4 region of the 18S rDNA of selected published sequences of Cymbellales, plus newly obtained sequences from three isolated single cells of Didymosphenia sp. and four sequences of Cymbella janischii (GenBank accession numbers are included in the taxon labels). The current taxonomic families are indicated. Bayesian posterior probabilities (BPP × 100) values indicate node support. Scale bar = expected number of substitutions/site.
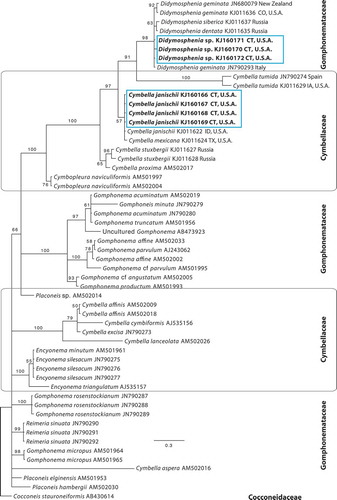
Figs 30–38. SEM images of Didymosphenia hullii on stalks from the West Branch of the Farmington River showing bifurcated stalks with cells attached. Scale bars = 20 µm (, , ); 50 µm (, , ); 40 µm (); 5 µm (); 200 µm ().
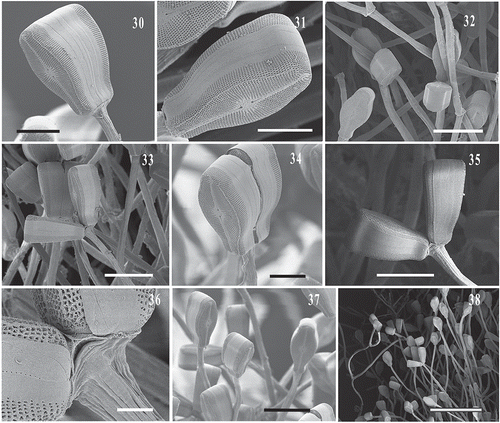
Figs 39–44. SEM images of Didymosphenia hullii. : Internal view of the valve displaying three stigmata. : View of apical pore field, footpole. , : Central valve view with striae, and 1 and 4 stigmata, respectively. : Internal view of central valve showing two stigmata and uniseriate striae. : External views of frustule girdle and valve morphology. Scale bars = 20 μm (); 5 μm (, ); 10 μm (, ); 30 μm ().
Table 1. Comparison of traits of selected Didymosphenia taxa and their localities. Morphology of areolae determined using images and descriptions in Metzeltin & Lange-Bertalot (Citation1995, Citation2014) and Mrozińska et al. (Citation2006).
The D. geminata-like diatom that occurs in the West Branch of the Farmington River has similar autecology and physico-chemical preferences to D. geminata (Khan-Bureau et al., Citation2014). Unlike the diatom that was found in Massachusetts (Supplementary ), Didymosphenia sp. is morphologically distinct from D. geminata and other species in this genus, warranting unique species-level status.
DNA barcode analysis
Three independent V4 rDNA sequences of Didymosphenia sp., resulting from the three single cell isolations, plus four sequences from the cloned material of C. janischii were obtained. The V4 sequences were 334–410 bp in length, and were compiled into a final alignment with selected related published diatom sequences to produce an alignment of 1816 nucleotides in length. The four sequences of C. janischii from Connecticut were identical across the barcode region and were resolved closest to sequences of C. janischii from Idaho, USA, and C. mexicana (Ehrenb.) Cleve (). These sequences grouped next most closely with those of C. tumida (Bréb. ex Kütz.) Van Heurck, C. proxima Reimer, and D. geminata, but were distinct from other Cymbella representatives. The three new V4 sequences from Didymosphenia sp. differed by just one nucleotide. These were identified as close matches to three published sequences of D. geminata from Italy (JN790293), New Zealand (JN680079), Colorado, USA (KJ011637), D. dentata Skvortsov & C.I. Meyer (Skvortzow & Meyer, Citation1928) from Russia (KJ011635) and D. siberica (Grunow) M. Schmidt from Russia (KJ011637), and were more distantly related to C. janischii from Idaho, USA (KJ011622) and from this study, C. mexicana from Texas USA (KJ011624), and C. tumida from Spain (JN790274).
Didymosphenia hullii Khan-Bureau, sp. nov. (–, –, –)
Description: Didymosphenia hullii Khan-Bureau sp. nov. forms colonies of cells on long stalks. The cells are heteropolar, with a rounder, shorter and more compressed headpole and footpole than the distinctly capitate D. geminata. Valves of D. hullii (–) are compact with transapically expanded headpoles and the concavity at the constriction of the headpole is shallow and short. The footpole is slightly capitate, though blunt. The footpole has an apical pore field of very small spherical perforations that are present where the stalk growth originates. The frustule comprises the epivalve and hypovalve through four girdle bands patterned with raised pustules. From the girdle view, the headpole is broad and tapers to the footpole similar to a wedge or a V shape. The valve has a length consistently in the range of 40–60 μm with cells slightly smaller at 38 μm and larger to 68 μm occassionally observed, the width range of 26.5–30.5 μm, and 1–4 stigmata present. The central area is inflated and elliptical with 9–11 striae in 10 μm that are radiate and have irregular short and long lengths. Large pentagonal and square shaped depressions with pores (areolae) are present throughout the valve in complex deep wells that are surrounded by spines. At the distal end, the raphe quickly curves or forms a hook shape, but does not go through the apical pore field. This taxon has both asymmetrical apical and transpical axis.
Holotype: (LM) and (SEM) are made from the holotype from the population on slide CONN00178537, partially illustrated here in – (University of Connecticut Herbarium), and –. Samples were collected by D. Khan-Bureau, 29 November 2012.
Isotypes: Population on slide HCUCB D-00791 (Herbario Criptogámico, Universidad Católica, San Pablo, Cochabamba, Bolivia).
Type locality: The West Branch of the Farmington River, a tributary of the Connecticut River in Barkhamsted, Connecticut, USA (41.960° N, 73.017° W). Samples were taken from the epilithon.
Habitat: Abundant growth occurred on a wide range of large cobbles and boulders covering the river substrate, bank to bank.
Etymology: The species is named in honour of the late David Hull MD, Director of Transplant at Hartford Hospital in Connecticut. He enjoyed nature and aspired to understand the many facets of science.
Didymosphenia hullii is similar in size to D. pumila Metzeltin & Lange-Bert., D. grunowii Lange-Bert. & Metzeltin, D. laticollis Metzeltin & Lange-Bert. (Citation2014) (the Sweden population), D. tatrensis Mrozińska, Czerwik-Marcinkowska & Gradziński, and smaller populations of D. siberica (Grunow) M. Schmidt but differs in lacking a smooth external valve surface as is characteristic of these species. Didymosphenia hullii also differs in stria density, number of stigmata, and areola structure (Metzeltin & Lange-Bertalot, Citation1995, Citation2014; Mrozińska et al., Citation2006) (). The rough external valve face and areolae of D. hullii are most similar to that of D. geminata and D. clavaherculis, the only two Didymosphenia species currently found in the USA, and D. laticollis known from Russia. Valves of D. hullii fall into the lower end of the size range of D. geminata but valves of D. hullii observed from cells collected across eight months (–) were compact with transapically expanded head poles, and the concavity at the constriction of the head pole is shallow and shorter in D. hullii compared with D. geminata and other Didymosphenia taxa. Unlike D. tatrensis, D. siberica, D. pumila and D. grunowii the areolae of D. hullii have deep inclined walls with dendritic slits within the valves which are surrounded by spines below the interior walls (Mrozińska et al., Citation2006; Metzeltin & Lange-Bertalot, Citation2014). These species are the smaller of the Didymosphenia taxa. They are compressed and their basal poles are much less elongated than D. geminata, and they differ in valve length and width, stria density, and number of stigmata (). Didymosphenia siberica was reported by Dawson (Citation1973) and Stoermer et al. (Citation1986) as having only one isolated stigma internally and referred to this as a raised convolution. Subsequently, Metzeltin & Lange-Bertalot (Citation1995) described D. siberica with 1–3 stigmata and D. pumila having 1–2 stigmata, whereas D. hullii has 1–4 stigmata internally.
DISCUSSION
In this study we characterized two recently reported nuisance stalk-forming diatoms in Connecticut, thus contributing information on the morphology, variation and phylogenetic relationships of cymbelloid diatoms. The morphology of the Cymbella species from Connecticut is consistent with that of C. janischii. In the USA, C. janischii is described as endemic to the Pacific Northwest. Outside of the Pacific Northwest, C. janischii had only been observed in four other states, Arizona, Colorado, New York and Oklahoma (Bahls, Citation2007), until recently reported in Connecticut in July 2012 (Khan-Bureau et al., Citation2014). Suzawa et al. (Citation2011) confirmed blooms of the non-native C. janischii in Japan and reported that it was introduced from North America. Phylogenetic analysis of four independent V4 sequences of C. janischii from Connecticut indicate that this species is related to published sequences of C. janischii and C. mexicana, and more distantly related to C. tumida (Cymbellaceae) and D. geminata (Gomphonemataceae). The C. janischii sequences also indicate that this species is distantly related to most sequences from the family Cymbellaceae, such as Cymbella affinis Kütz. and C. cymbiformis (Ehrenb.) Grunow. These relationships are consistent with a recent multi-locus investigation (Nakov et al., Citation2014) and provide additional evidence that the taxonomy of the families Gomphonemataceae and Cymbellaceae requires re-evaluation (Kociolek & Stoermer, Citation1988; Nakov & Theriot, Citation2009; Kermarrec et al., Citation2011; Graeff & Kociolek, Citation2013; Nakov et al., Citation2014).
We also described the new taxon, Didymosphenia hullii. Twenty-two species (and several varieties and subspecies of D. geminata) are currently known within the genus Didymosphenia (Metzeltin & Lange-Bertalot, Citation2014). The type material of the best-known species, D. geminata, was not readily available until recently (D. Metzeltin, pers. comm.) and morphological data are still limited even though it has been almost 200 years since Lyngbye first described Didymosphenia geminata as Echinella geminata (Lyngbye, Citation1819; Whitton et al., Citation2009; Blanco & Ector, Citation2013). Most early reports were based on drawings and light micrographs (Blanco & Ector, Citation2013). Only one species of Didymosphenia, D. geminata, occurs commonly within the geographic boundaries of the continental USA, specifically in the western states, although D. clavaherculis was documented in Alaska (Spaulding, Citation2010). New reports of D. geminata in the USA have come from midwestern and eastern states (Kilroy, Citation2004; Spaulding & Elwell, Citation2007; Blanco & Ector, Citation2009; Kumar et al., Citation2009; Kilroy & Unwin, Citation2011; Bothwell et al., Citation2012), as well as Canada (Kirkwood et al., Citation2007; Gillis & Chalifour, Citation2009; Lavery et al., Citation2014) and South America (Kilroy & Unwin, Citation2011; Segura, Citation2011; Morales et al., Citation2012; Rivera et al., Citation2013; Sastre et al., Citation2013; Reid & Torres, Citation2014). The diatom reported from Connecticut is morphologically distinct from other species of Didymosphenia, including D. geminata reported from neighbouring Massachusetts (Supplementary ), and is thus recognized as a distinct species.
Despite the economic and ecological importance of Didymosphenia there were only two accessions of this genus in GenBank, both reporting sequences of the 18S gene of D. geminata, until Nakov et al. (Citation2014) provided additional sequences from D. geminata, D. dentata and D. siberica. None of the existing D. geminata sequences are from the type locality, thus we cannot state which D. geminata sequence represents the species. We targeted the V4 region of the 18S gene to make the new data from D. hullii comparable to the published sequences of D. geminata and related taxa. The results of the present study indicate that D. geminata and D. hullii are more closely related to C. janischii than to Gomphonema and other genera in Gomphonemataceae, further illustrating the non-monophyly of Cymbellaceae and Gomphonemataceae as noted by other authors (Kociolek & Stoermer, Citation1988; Nakov & Theriot, Citation2009; Kermarrec et al., Citation2011; Graeff & Kociolek, Citation2013; Nakov et al., Citation2014).
As was seen among sequences of the Cymbella janischii clade (but not for other species of Cymbella), and from the data from the V4 region we were unable to separate D. hullii, D. geminata, D. siberica or D. dentata sequences due to very low sequence variation, indicating that these taxa are very closely related and perhaps evolutionarily young. The lack of variation across this short barcode region, coupled with a lack of published sequences from the other Didymosphenia species means that we are currently unable to resolve the relationship among species of Didymosphenia. A more variable marker may better differentiate among taxa and facilitate identification of new or cryptic species of diatoms. However, finding a suitable species level marker has proven challenging for some algae and specifically the diatoms (Evans et al., Citation2007; Hall et al., Citation2010; Hamsher et al., Citation2011; Zimmermann et al., Citation2011; Luddington et al., Citation2012; Kermarrec et al., Citation2014). Nakov et al. (Citation2014) recently examined phylogenetic signal in alternative diatom barcode genes, including the plastid rbcL gene (which we were unsuccessful in obtaining from our samples), for the Cymbellales. Although the selected markers provided resolution across the order it is not possible to assess if they would be useful within the genus Didymosphenia because just one representative from each of three species of Didymosphenia was used in that study.
Future work is needed to identify suitable phylogenetic markers for the species of Didymosphenia. Molecular data are not currently available for most species, and most of the sequences available are not associated with a specific morphology and do not represent the taxonomic type. Also needed are studies that include samples of single species from distinct geographic locations in order to understand the levels of sequence variation within and among species. Together, these studies would provide a better understanding of the geographic ranges and help connect physiological preferences and tolerances to particular diatom species.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at http://dx.doi.org/10.1080/09670262.2015.1126361
Supplementary table 1: Taxon list and source page numbers of images used for morphological comparisons of D. geminata and D. hullii.
Supplementary figs 1–4: LM images offield-collected C. janischii cells showing size distribution. Fig. 3: Central valve illustrates the distinct radiated striae with striae more closely together and longer pointed areolae in the central area, differentiating between other larger Cymbella taxa and C. janischii. Figs 1–3: Scale bars = 10 μm. Fig. 4: LM image of cell on its stalk prior to acid cleaning. Scale bar = 20 μm.
Supplementary fig. 5: LM image of D. geminata from Massachusetts. Scale bar = 10 μm.
Supplementary Table 1
Download PDF (103.9 KB)Supplementary Fig S5
Download TIFF Image (30 MB)Supplementary Figs S1-S4
Download TIFF Image (22.6 MB)ACKNOWLEDGEMENTS
We thank the Connecticut Institute of Water Resources (CTIWR) for funding this research.
We thank K. Fučíková, S. Olm, C. Lo, M. Letsch, H. McManus and acknowledge the late F. Trainor for helpful discussion. This work made use of the computer cluster maintained by the Bioinformatics Facility (Biotechnology/Bioservices Center) at the University of Connecticut. We are grateful for D. Metzeltin’s expert opinion and H. Lange-Bertalot for the use of their D. geminata images. Thanks to S. Spaulding for access to her D. geminata image. We thank Three Rivers Community College’s Dean of Information Technology S. Goetchius, K. Barfield, S. Cohen, D. Jewett, and retired President G. S. Jones for providing continual support for this project, J. Sgro, John Carroll University, for providing his input and continual support, for the expert assistance of M. Cantino and S. Daniels of the EM lab at UCONN, NSF 1126100 supporting the purchase of the Nova Nano SEM, V. Kask scientific illustrator in Biological Central Services at UCONN, J. Morrison, East Hartford, USGS, L. Matthews VT DEC, M. Becker and C. Bellucci, CT DEEP for their assistance. We thank the reviewers for their helpful comments.
DISCLOSURE STATEMENT
No potential conflict of interest was reported by the author(s).
Additional information
Notes on contributors
Diba A. Khan-Bureau
D.A. Khan-Bureau: original concept, LM and SEM and molecular analyses, drafting and editing the manuscript; E.A. Morales: reviewed LM and SEM images and edited the manuscript; L. Ector reviewed the LM and SEM images and edited the manuscript; M.S. Beauchene, provided water quality data and historical information and edited the manuscript; L.A. Lewis reviewed molecular, SEM, and LM analyses, and edited the manuscript.
Eduardo A. Morales
D.A. Khan-Bureau: original concept, LM and SEM and molecular analyses, drafting and editing the manuscript; E.A. Morales: reviewed LM and SEM images and edited the manuscript; L. Ector reviewed the LM and SEM images and edited the manuscript; M.S. Beauchene, provided water quality data and historical information and edited the manuscript; L.A. Lewis reviewed molecular, SEM, and LM analyses, and edited the manuscript.
Luc Ector
D.A. Khan-Bureau: original concept, LM and SEM and molecular analyses, drafting and editing the manuscript; E.A. Morales: reviewed LM and SEM images and edited the manuscript; L. Ector reviewed the LM and SEM images and edited the manuscript; M.S. Beauchene, provided water quality data and historical information and edited the manuscript; L.A. Lewis reviewed molecular, SEM, and LM analyses, and edited the manuscript.
Michael S. Beauchene
D.A. Khan-Bureau: original concept, LM and SEM and molecular analyses, drafting and editing the manuscript; E.A. Morales: reviewed LM and SEM images and edited the manuscript; L. Ector reviewed the LM and SEM images and edited the manuscript; M.S. Beauchene, provided water quality data and historical information and edited the manuscript; L.A. Lewis reviewed molecular, SEM, and LM analyses, and edited the manuscript.
Louise A. Lewis
D.A. Khan-Bureau: original concept, LM and SEM and molecular analyses, drafting and editing the manuscript; E.A. Morales: reviewed LM and SEM images and edited the manuscript; L. Ector reviewed the LM and SEM images and edited the manuscript; M.S. Beauchene, provided water quality data and historical information and edited the manuscript; L.A. Lewis reviewed molecular, SEM, and LM analyses, and edited the manuscript.
REFERENCES
- Aboal, M., Marco, S., Chaves, E., Mulero, I. & García-Ayala, A. (2012). Ultrastructure and function of stalks of the diatom Didymosphenia geminata. Hydrobiologia, 695: 17–24.
- Bahls, L.L. (2007). Cymbella janischii – giant endemic diatom of the Pacific Northwest: morphology, ecology and distribution compared to Cymbella mexicana. Northwest Science, 81: 284–292.
- Blanco, S. & Ector, L. (2009). Distribution, ecology and nuisance effects of the freshwater invasive diatom Didymosphenia geminata (Lyngbye) M. Schmidt: a literature review. Nova Hedwigia, 88: 347–422.
- Blanco, S. & Ector, L. (2013). The nomenclatural history of Didymosphenia geminata (Lyngbye) M. Schmidt in Schmidt et al. (Bacillariophyta) and related taxa. Acta Nova, 6: 157–164.
- Bothwell, M.L. & Spaulding, S.A. (2008). Synopsis of the 2007 International Workshop on Didymosphenia geminata. In Proceedings of the 2007 International Workshop on Didymosphenia geminata, Montréal, Québec (Bothwell, M.L. & Spaulding, S.A., editors), pp. xiii–xxxi. Canadian Technical Report of Fisheries and Aquatic Sciences, 2795.
- Bothwell, M.L. & Kilroy, C. (2011). Phosphorus limitation of the freshwater benthic diatom Didymosphenia geminata determined by the frequency of dividing cells. Freshwater Biology, 56: 565–578.
- Bothwell, M.L., Kilroy, C., Taylor, B.W., Ellison, E.T., James, D.A., Gillis, C.-A., Bladon, K.D. & Silins, U. (2012). Iron is not responsible for Didymosphenia geminata bloom formation in phosphorus-poor rivers. Canadian Journal of Fisheries and Aquatic Sciences, 69: 1723–1727.
- Bothwell, M.L., Taylor, B.W. & Kilroy, C. (2014). The Didymo story: the role of low dissolved phosphorus in the formation of Didymosphenia geminata blooms. Diatom Research, 29: 229–236.
- CT DEEP – Connecticut Department of Energy and Environmental Protection (2011). http://www.ct.gov/deep/cwp/view.asp?Q=476204&A=4013.
- Dawson, P.A. (1973). Further observations on the genus Didymosphenia M. Schmidt–D. siberica (Grun.) M. Schm. British Phycological Journal, 8: 197–201.
- Evans, K.M., Wortley, A.H. & Mann, D.G. (2007). An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist, 158: 349–364.
- Geneious (2013). Version 7.0.5, created by Biomatters. Available from http://www.geneious.com/.
- Gillis, C.-A. & Chalifour, M. (2009). Changes in the macrobenthic community structure following the introduction of the invasive algae Didymosphenia geminata in the Matapedia River (Québec, Canada). Hydrobiologia, 647: 63–70.
- Graeff, C.L. & Kociolek, J.P. (2013). New or rare species of cymbelloid diatoms (Bacillariophyceae) from Colorado (USA). Nova Hedwigia, 97: 87–116.
- Hall, J.D., Fučíková, K., Lo, C., Lewis, L.A. & Karol, K.G. (2010). An assessment of proposed DNA barcodes in freshwater green algae. Cryptogamie, Algologie, 31: 529–555.
- Hamsher, S.E., Evans, K.M., Mann, D.G., Poulíčková, A. & Saunders, G.W. (2011). Barcoding diatoms: exploring alternatives to COI-5P. Protist, 162: 405–422.
- Huelsenbeck, J.P. & Ronquist, F. (2001). MrBayes: Bayesian inference of phylogeny. Bioinformatics, 17: 754–755.
- Kermarrec, L., Ector, L., Bouchez, A., Rimet, F. & Hoffmann, L. (2011). A preliminary phylogenetic analysis of the Cymbellales based on 18S rDNA gene sequencing. Diatom Research, 26: 305–315.
- Kermarrec, L., Franc, A., Rimet, F., Chaumeil, P., Frigerio, J.M., Humbert, J.F. & Bouchez, A. (2014). A next-generation sequencing approach to river biomonitoring using benthic diatoms. Freshwater Science, 33: 349–363.
- Khan-Bureau, D.A., Beauchene, M.S., Ector, L., Morales, E.A. & Lewis, L.A. (2014). Observations of two nuisance stalk-forming diatoms (Bacillariophyta) from a river in Connecticut, Northeastern U.S.A.: Didymosphenia sp. and Cymbella janischii (A. Schmidt) De Toni. BioInvasions Records, 3: 139–149.
- Kilroy, C. (2004). A new alien diatom, Didymosphenia geminata (Lyngbye) Schmidt: its biology, distribution, effects and potential risks for New Zealand fresh waters. Available at: http://www.biosecurity.govt.nz/files/pests/didymo/didymo-preliminary-org-ianov-04.pdf.
- Kilroy, C. & Bothwell, M. (2011). Environmental control of stalk length in the bloom-forming, freshwater benthic diatom Didymosphenia geminata (Bacillariophyceae). Journal of Phycology, 47: 981–989.
- Kilroy, C. & Unwin, M. (2011). The arrival and spread of the bloom-forming, freshwater diatom, Didymosphenia geminata, in New Zealand. Aquatic Invasions, 3: 249–262.
- Kilroy, C. & Bothwell, M.L. (2014). Attachment and short-term stalk development of Didymosphenia geminata: effects of light, temperature and nutrients. Diatom Research, 29: 237–248.
- Kirkwood, A.E., Shea, T., Jackson, L.J. & McCauley, E. (2007). Didymosphenia geminata in two Alberta headwater rivers: an emerging invasive species that challenges conventional views on algal bloom development. Canadian Journal of Fisheries and Aquatic Sciences, 64: 1703–1709.
- Kociolek, J.P. & Stoermer, E.F. (1988). A preliminary investigation of the phylogenetic relationships among the freshwater, apical pore field-bearing Cymbelloid and Gomphonemoid diatoms (Bacillariophyceae). Journal of Phycology, 24: 377–385.
- Krammer, K. & Lange-Bertalot, H. (1986). Bacillariophyceae 1. Teil: Naviculaceae. In Süßwasserflora von Mitteleuropa (Ettl, H., Gerloff, J., Heynig, H. & Mollenhauer, D., editors), Band 2/1. Gustav Fisher Verlag, Stuttgart & New York. 876 pp.
- Kuhajek, J.M. & Wood, S.A. (2014). Novel techniques for the short-term culture and laboratory study of Didymosphenia geminata. Diatom Research, 29: 293–301.
- Kuhajek, J.M., Lemoine, M., Kilroy, C., Cary, S.C., Gerbeaux, P. & Wood, S.A. (2014). Laboratory study of the survival and attachment of Didymosphenia geminata (Bacillariophyceae) in water sourced from rivers throughout New Zealand. Phycologia, 53: 1–9.
- Kumar, S., Spaulding, S.A., Stohlgren, T.J., Hermann, K.A., Schmidt, T.S. & Bahls, L.L. (2009). Potential habitat distribution for the freshwater diatom Didymosphenia geminata in the continental US. Frontiers in Ecology and Environment, 7: 415–420.
- Lang, I. & Kaczmarska, I. (2011). A protocol for a single-cell PCR of diatoms from fixed samples: method validation using Ditylum brightwellii (T. West) Grunow. Diatom Research, 26: 43–49.
- Lavery, J.M., Kurek, J., Rühland, K.M., Gillis, C.A., Pisaric, M.F.J. & Smol, J.P. (2014). Exploring the environmental context of recent Didymosphenia geminata proliferation in Gaspésie, Quebec, using paleolimnology. Canadian Journal of Fisheries and Aquatic Sciences, 71: 616–626.
- Luddington, I.A., Kaczmarska, I. & Lovejoy, C. (2012). Distance and character-based evaluation of the V4 region of the 18S rRNA gene for the identification of diatoms (Bacillariophyceae). PLoS ONE 7: e45664.
- Lyngbye, H.C. (1819). Tentamen Hydrophytologiae Danicae Continens omnia Hydrophyta Cryptogama Daniae, Holsatiae, Faeroae, Islandiae, Groenlandiae hucusque cognita, Systematice Disposita, Descripta et iconibus illustrata, Adjectis Simul Speciebus Norvegicis. Hafniae. 248 pp., 70 pls.
- MDC - Metropolitan District Commission (2013). http://www.themdc.com/
- Metzeltin, D. & Lange-Bertalot, H. (1995). Kritische Wertung der Taxa in Didymosphenia (Bacillariophyceae). Nova Hedwigia, 60: 381–405.
- Metzeltin, D. & Lange-Bertalot, H. (2014). The genus Didymosphenia M. Schmidt. A critical evaluation of established and description of 11 new taxa. Iconographia Diatomologica, 25: 1–293.
- Morales, E.A., Siver, P.A. & Trainor, F.R. (2001). Identification of diatoms (Bacillariophyceae) during ecological assessments: comparison between light microscopy and scanning electron microscopy techniques. Proceedings of the Academy of Natural Sciences of Philadelphia, 151: 95–103.
- Morales, E.A., Rivera, S.F., Veizaga, A. & Fiorini, R. (2012). Didymosphenia geminata (Lyngbye) M. Schmidt (Bacillariophyta), una especie invasora y potencial amenaza para ecosistemas acuáticos bolivianos. Acta Nova, 5: 327–343.
- Mrozińska, T., Czerwik-Marcinkowska, J. & Gradziński, M. (2006). A new species of Didymosphenia (Bacillariophyceae) from the Western Carpathian Mountains of Poland and Slovakia. Nova Hedwigia, 83: 499–510.
- Nakov, T. & Theriot, E.C. (2009). Preliminary molecular phylogeny of the Cymbellales (Bacillaryophyceae). Program and Abstracts. 20th North American Diatom Symposium, Iowa Lakeside Lab, September 23–27, 2009, Milford Iowa, pp. 28–29.
- Nakov, T., Ruck, E.C., Galachyants, Y., Spaulding, S.A. & Theriot, E.C. (2014). Molecular phylogeny of the Cymbellales (Bacillariophyceae, Heterokontophyta) with a comparison of models for accommodating rate variation across sites. Phycologia, 53: 359–373.
- Pite, D.P., Lane, K.A., Hermann, A.K., Spaulding, S.A. & Finney, B.P. (2009). Historical abundance and morphology of Didymosphenia species in Naknek Lake, Alaska. Acta Botanica Croatica, 68: 183–197.
- Reid, B. & Torres, R. (2014). Didymosphenia geminata invasion in South America: ecosystem impacts and potential biogeochemical state change in Patagonian rivers. Acta Oecologica, 54: 101–109.
- Rivera, P., Basualto, S. & Cruces, F. (2013). Acerca de la diatomea Didymosphenia geminata (Lyngbye) M. Schmidt: su morfología y distribución en Chile. Gayana Botánica, 70: 154–158.
- Ronquist, F. & Huelsenbeck, J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572–1574.
- Round, F.E., Crawford, R.M. & Mann, D.G. (1990). The Diatoms: Biology and Morphology of the Genera. Cambridge University Press, Cambridge. 747 pp.
- Sastre, A.V., Santinelli, N.H., Bauer, G.A., Ayestarán, M.G. & Uyua, N.M. (2013). First record of the invasive diatom Didymosphenia geminata (Lyngbye) Schmidt in a Patagonian Andean river of Argentina. BioInvasions Records, 2: 11–17.
- Sato, S., Mann, D.G., Matsumoto, S. & Medlin, L.K. (2008). Pseudostriatella (Bacillariophyta): a description of a new araphid diatom genus based on observations of frustule and auxospore structure and 18S rDNA phylogeny. Phycologia, 47: 371–391.
- Segura, P. (2011). A slimy invader blooms in the rivers of Patagonia. Science, 331: 18.
- Skvortzow, B.W. & Meyer, C.I. (1928). A contribution to the diatoms of Baikal Lake. Proceedings of the Sungaree River Biological Station, 1: 1–55.
- Spaulding, S.A. (2010). Didymosphenia geminata. In Diatoms of the United States. From http://westerndiatoms.colorado.edu/taxa/species/didymosphenia_geminata. Accessed on 10 June 2015.
- Spaulding, S.A. & Elwell, L. (2007). Increase in nuisance blooms and geographic expansion of the freshwater diatom Didymosphenia geminata. White Paper. Recommendations for response. Federation of Fly Fishers and USA Environmental Protection Agency, Livingston, MT. 33 pp.
- Stoermer, E.F., Yu-Zao, Q. & Ladewski, T.B. (1986). A quantitative investigation of shape variation in Didymosphenia (Lyngbye) M. Schmidt (Bacillariophyta). Phycologia, 25: 494–502.
- Suzawa, T., Seino, S. & Mayama, S. (2011). Blooms of Cymbella janischii (A.W.F.Schmidt) De Toni accompanied by Gomphoneis minuta (Stone) Kociolek & Stoermer from the upper stream of the Chikugo River, Kyushu, Japan: possibility of new alien diatom species. Diatom, 27: 58–64.
- Whitton, B.A., Ellwood, N.T.W. & Kawecka, B. (2009). Biology of the freshwater diatom Didymosphenia: a review. Hydrobiologia, 630: 1–37.
- Zgłobicka, I. (2013). Aspects of structural biology of Didymosphenia geminata (Lyngb.) M. Schmidt (Bacillariophyta). International Journal on Algae, 15: 293–312.
- Zimmermann, J., Jahn, R. & Gemeinholzer, B. (2011). Barcoding diatoms: evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Organisms, Diversity and Evolution, 11: 173–192.
