Abstract
The volvocine lineage is a monophyletic grouping of unicellular, colonial and multicellular algae, and a model for studying the evolution of multicellularity. In addition to being morphologically diverse, volvocine algae boast a surprising amount of organelle genomic variation. Moreover, volvocine organelle genome complexity appears to scale positively with organismal complexity. However, the organelle DNA architecture at the origin of colonial living is not known. To examine this issue, we sequenced the plastid and mitochondrial DNAs (ptDNA and mtDNA) of the 4-celled alga Tetrabaena socialis, which is basal to the colonial and multicellular volvocines.
Tetrabaena socialis has a circular-mapping mitochondrial genome, contrasting with the linear mtDNA architecture of its relative Chlamydomonas reinhardtii. This suggests that a circular-mapping mtDNA conformation emerged at or near the transition to group living in the volvocines, or represents the ancestral state of the lineage as a whole. The T. socialis ptDNA is very large (>405 kb) and dense with repeats, supporting the idea that a shift from a unicellular to a colonial existence coincided with organelle genomic expansion, potentially as a result of increased random genetic drift. These data reinforce the idea that volvocine algae harbour some of the most expanded plastid chromosomes from the eukaryotic tree of life. Circular-mapping mtDNAs are turning out to be more common within volvocines than originally thought, particularly for colonial and multicellular species. Altogether, volvocine organelle genomes became markedly more inflated during the evolution of multicellularity, but complex organelle genomes appear to have existed at the very beginning of colonial living.
INTRODUCTION
The volvocine green algae are a model lineage for exploring the origins and evolution of multicellularity, particularly at the genomic level (Prochnik et al., Citation2010; Hanschen et al., Citation2016). Its members span the gamut of cellular complexity and include single-celled species (e.g. Chlamydomonas reinhardtii), colonial forms (e.g. the 4-celled Tetrabaena socialis and the 16-celled Gonium pectorale), organisms with partially differentiated cell types (e.g. Pleodorina starrii), and complex multicellular taxa with germ-soma differentiation (e.g. Volvox carteri) (Nishii & Miller, Citation2010). It is estimated that colonial volvocine algae first arose between 209–260 million years ago (MYA), and that soon after its appearance the lineage underwent rapid radiation, giving rise to various multicellular groups (Herron et al., Citation2009).
Volvocine algae form a monophyletic lineage (Kirk, Citation2005; Prochnik et al., Citation2010) within the Reinhardtinia clade of the Chlamydomonadales (Nakada et al., Citation2008). The phylogenetic relationships among volvocines have been investigated extensively (Coleman, Citation1999; Nozaki et al., Citation2000; Herron & Michod, Citation2008; Nakada et al., Citation2008), and the lineage can be divided into three families: the Volvocaceae (e.g. P. starrii and V. carteri), the Goniaceae (e.g. G. pectorale) and the Tetrabaenaceae (e.g. T. socialis), which is the deepest-branching of the three families (Nozaki et al., Citation2000). The 4-celled T. socialis is one of the ‘simplest’ known colonial volvocine algae (Nozaki, Citation1986; Arakaki et al., Citation2013), and is therefore well suited for studying the early steps of multicellularity, but neither its organelle nor nuclear genomes have been characterized.
Whole mitochondrial and plastid DNA (mtDNA and ptDNA) sequence data exist for a wide range of volvocine taxa, including C. reinhardtii (Boer et al., Citation1985; Maul et al., Citation2002), G. pectorale (Hamaji et al., Citation2013), P. starrii (Smith et al., Citation2013) and V. carteri (Smith & Lee, Citation2009), as well as for several non-volvocine chlamydomonadalean species (Smith et al., Citation2010a; Del Vasto et al., Citation2015). Population-genetic statistics and detailed genome conformation results are also available for various volvocines (Moore & Coleman, Citation1989; Vahrenholz et al., Citation1993; Laflamme & Lee, Citation2003; Smith & Lee, Citation2010). Together these data have revealed a tremendous diversity in organelle genome architecture. For example, some volvocine algae, including C. reinhardtii and the multicellular Pandorina morum, have linear mtDNAs with defined telomeres (Moore & Coleman, Citation1989; Vahrenholz et al., Citation1993), whereas G. pectorale, P. starrii and V. carteri have circular-mapping mtDNAs (Smith & Lee, Citation2009; Hamaji et al., Citation2013; Smith et al., Citation2013), which in certain instances contain large numbers of mitochondrial repeats (Smith & Lee, Citation2009). Similarly, the V. carteri ptDNA is dense with repeats and at ~525 kilobases (kb) is more than twice the size of that from C. reinhardtii (~204 kb), and among the largest plastid genomes ever observed. Conversely, Reinhardtinia species from the non-photosynthetic genus Polytomella have lost their plastid genomes entirely (Smith & Lee, Citation2014).
The forces responsible for genomic expansion/contraction within volvocine algae – and eukaryotes in general – are multifaceted and complex (Lynch & Conery, Citation2003; Lynch et al., Citation2007). Nevertheless, volvocine organelle genome complexity appears to scale positively with organismal complexity (Smith et al., Citation2013), whereby colonial and multicellular species have larger, more repeat-rich organelle DNAs than unicellular species. It has been hypothesized that this trend is a consequence of smaller effective population sizes (Ne) and/or lower organelle mutation rates (μ) in multicellular versus unicellular volvocines (Smith & Lee, Citation2010; Smith et al., Citation2013).
To further explore the evolution of volvocine organelle genomic complexity and the relationship between cell number and genome size, we sequenced the mitochondrial and plastid genomes of the basal 4-celled T. socialis. The mtDNA mapped as a circular molecule and both the mtDNA and ptDNA are inflated with non-coding nucleotides, particularly the plastid genome, which is larger than the ptDNAs from some volvocines with more complex cellular organizations. These data underscore the propensity for plastid genome inflation within colonial and multicellular volvocines and chlamydomonadalean algae in general. Complex organelle genomes appear to have existed at the very beginning of multicellularity within this model lineage.
MATERIALS AND METHODS
Culture, library preparation and sequencing
Tetrabaena socialis strain NIES-571 (previously known as Gonium sociale) was made axenic by repeated steps of isolating and transferring a single cell onto sterile solid media (Pringsheim, Citation1946). Axenic cultures were grown in Volvox medium (Nozaki, Citation1986; Arakaki et al., Citation2013) and DNA was extracted as previously described (Miller et al., Citation1993). RNA was isolated with Thermo Fisher Trizol reagent (catalogue #15596-026) from cultures that were 2 h into the dark phase of their life cycle, when both dividing and non-dividing cells were observed. Isolated RNA was preserved using the Gentegra RNA stabilization reagent (catalogue #GTR-100B) and subsequently re-suspended according to the manufacturer’s recommendations. The Qiagen RNeasy kit (catalogue #74104) and Qiagen DNase (catalogue #79254) were used to remove DNA.
Paired-end Illumina sequencing libraries (180 bp and 600 bp insert sizes) were prepared from total DNA using the Illumina TruSeq v2 DNA Sample Preparation Kit (catalogue #FC-121-2001). The 600 bp libraries were sequenced on Illumina MiSeq (v3 chemistry – 2 × 300 bp) and HiSeq 2500 platforms (v3 chemistry – 2 × 100 bp); the 180 bp library was used to create an overlap library on the HiSeq 2500 platform. Mate-pair data were generated using the Nextera Mate-Pair DNA Preparation kit with size selection for 6 kb using a Sage Bioscience BluePippen 0.75% Gel Cassette and pulse-field separation. An RNA library was generated with the TruSeq stranded-mRNA Sample Preparation Kit according to the manufacturer’s recommendations. A total of ~150 million reads were generated on an Illumina Hiseq 2500 (v4 chemistry – 2 × 125 bp).
Organelle genome assembly
Multiple genome assemblers were tested: the CLC Genomics Workbench assembler (Qiagen, Hilden, Germany) and the string-graph assembler (SGA) (Simpson & Durbin, Citation2012) gave the longest and most intact ptDNA and mtDNA contigs respectively. Organellar DNA-derived contigs were identified by 6-frame translated nucleotide BLAST searches (Altschul et al., Citation1990) of the C. reinhardtii, G. pectorale, P. starrii and V. carteri organelle-encoded proteins against the entire assemblies. For the CLC assembly, all of the paired-end data from the 180 bp (HiSeq) and 600 bp (HiSeq and MiSeq) insert libraries were used. The assembly was performed with a word-size of 60 and bubble size of 80. For the SGA assembly, only the data from the 600 bp (HiSeq) insert library were used; assembly parameters were the same as those used for Caenorhabditis elegans by Simpson & Durbin (Citation2012) (ropeBWT indexing), but with a more stringent minimum overlap (77 bp) and branch length of 350. CLC and SGA were used to generate contigs (for CLC scaffolding with PE data was performed) while SSPACE (Boetzer et al., Citation2011) was used for scaffolding of mate-pair data using bowtie mapping (without SSAKE extension) and default parameters. Post-assembly improvements were made using GapFiller (Boetzer & Pirovano, Citation2012) with 10 iterations and default parameters. TopHat2 (Kim et al., Citation2013) was used to align RNA sequencing (RNA-seq) reads to the mtDNA assembly of T. socialis. The expression of the rtl gene was subsequently examined for evidence of expression.
Genome annotation and repeat analysis
The T. socialis organellar genomes were initially annotated using MFannot (Burger & Lang, Citation2007) and then further polished using BLAST-derived DNA and amino-acid alignments with genes from available chlamydomonadalean organelle DNAs. Gene structures were curated with evidence from multiple gene alignments performed with MUSCLE (Edgar et al., Citation2004). Gene synteny was assessed using MAUVE (Darling et al., Citation2004), and genome maps were generated with OrganellarGenomeDraw (Lohse et al., Citation2013). Organelle DNAs repeats were identified with Reputer (Kurtz & Schleiermacher, Citation1999) using a minimum length of 30 bp and Hamming-distance of 3. BEDTools (Quinlan & Hall, Citation2010) was then used to mask the repeats identified with Reputer; masked regions were counted to provide an estimate of the total percentage of repetitive sequences. In addition to Reputer, Emboss Palindrome (http://www.ebi.ac.uk/Tools/emboss/) was used to identify palindromic repeats and their putative hairpin-loop structures. Jdotter (Brodie et al., Citation2004) was used to perform self-similarity dot-plots with 1000-pixel resolution and a 50 bp sliding window.
RESULTS
The circular-mapping mitochondrial genome of Tetrabaena socialis
The T. socialis mitochondrial genome (GenBank accession KX232644) assembled as an intact 27.7 kb, AT-rich (56%) circular molecule, which makes it ~7.5–15 kb larger than those of C. reinhardtii, G. pectorale and P. starrii, but ~7.5 kb smaller than that of V. carteri (; ). Telomeric-like sequences, such as long inverted repeats, were not identified in the T. socialis mtDNA assembly (), further supporting its circular-mapping structure (Bendich, Citation2007). The mitochondrial gene content of T. socialis (when ignoring non-standard open-reading frames) mirrors that of other volvocine algae (), but the arrangement and polarity of the genes are unique (). Indeed, the genes on circular-mapping chlamydomonadalean mtDNAs typically have the same transcriptional polarity (Smith et al., Citation2013; Del Vasto et al., Citation2015), but on the T. socialis mtDNA genes can be found on both strands of the genome ().
Table 1. Organismal and organelle features in the Volvocine algae.
Fig. 1. The mitochondrial genome of T. socialis maps as a 27 751 bp circular molecule. The transcriptional polarity of genes can proceed in a clockwise (inside of circle) or anti-clockwise (outside of circle) direction. Inner grey circle denotes AT content. Image generated using OrganellarGenomeDraw.
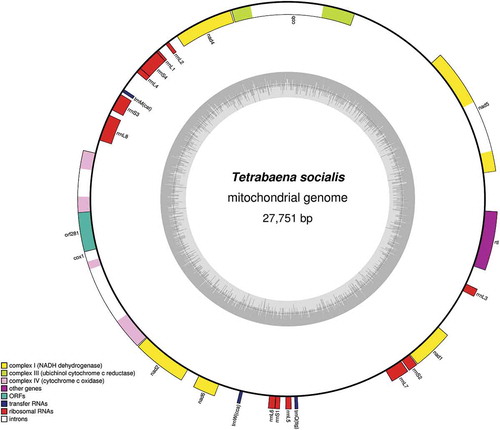
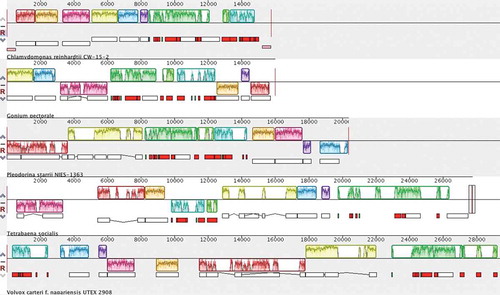
Fig. 2. Syntenic rearrangements in the mitochondrial genomes of volvocine algae. Synteny blocks (highlighted in the same colour) were determined using MAUVE. All sequences besides C. reinhardtii are circular and thus start and end sites are arbitrary. The green and turquoise blocks mostly contain tRNA genes, rRNA coding modules, and the nad1 gene; their order is generally conserved among volvocine algae with the exception of T. socialis. The arrangements of cox1 (yellow blocks), cob (red blocks) and nad5 (mostly found within green blocks) are highly variable among volvocine algae.
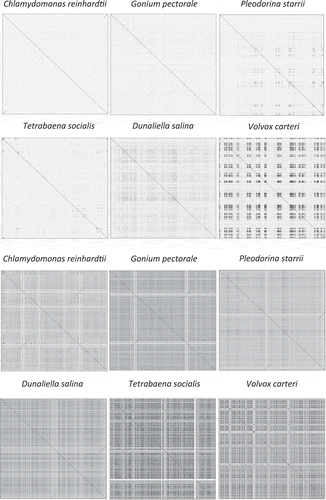
More than half of the mitochondrial genome (55%) comprises intergenic (34%) and intronic (21%) DNA, which is significantly greater than the non-coding mtDNA contents of C. reinhardtii (~20%), G. pectorale (27%) and P. starrii (43%) (Smith et al., Citation2013). As many as 40% of the repetitive elements in the T. socialis mtDNA are palindromic, which is a common theme among chlamydomonadalean mtDNAs, including those of V. carteri (Smith & Lee, Citation2009) and Dunaliella species (Del Vasto et al., Citation2015). However, unlike the V. carteri and D. salina mtDNAs, there is a relatively low degree of sequence identity among the different palindromic elements (). Tetrabaena socialis possesses more mitochondrial introns (5 in total; all of group-I affiliation) than any other Reinhardtinia mtDNA investigated to date. The introns are located within nad5 (1), cob (1) and cox1 (3); to the best of our knowledge, the second intron of cox1 has not been identified in any other member of this lineage ().
Fig. 3. Self-similarity dot-plots of organelle genomes from volvocine algae and Dunaliella salina. Mitochondrial and plastid genomes are shown on the top and bottom rows, respectively. Images generated with JDotter using 50 bp sliding window and resolution of 1000 bases/pixel. The mtDNA sequence of T. socialis shows little evidence of the high-similarity repeats found in V. carteri. The ptDNA of volvocines show a high degree of self-similarity that scales with total ptDNA sequence size.
The T. socialis mtDNA contains two non-standard genes: an intron-encoded LAGLIDADG endonuclease and a freestanding reverse transcriptase-like (rtl) gene. Similar non-standard genes have been identified in other volvocine mitochondrial genomes (Smith & Lee, Citation2009; Hamaji et al., Citation2013; Smith et al., Citation2013), including C. reinhardtii and C. incerta (Boer et al., Citation1985; Popescu & Lee, Citation2007), both of which have a freestanding rtl gene with a similar length to that of T. socialis (1119 bp and 1127 bp vs. 1269 bp). The P. starrii and V. carteri mtDNAs also contain rtl, but unlike in T. socialis and C. reinhardtii/incerta the rtl gene a maturase domain, is located within an intron and is >2,200 bp. The rtl gene is missing altogether from the G. pectorale mitochondrial genome. Of ~150 million RNA-seq reads generated for T. socialis, only 49 mapped to the rtl gene (covering 456 bp of the 3′ end of the gene), suggesting that rtl is poorly expressed.
The giant plastid genome of Tetrabaena socialis
The T. socialis ptDNA is >405 kb and distended with repeats, making it almost twice the size of the C. reinhardtii ptDNA, despite both genomes having almost identical gene contents (). Large, repeat-rich plastid genomes, like those from volvocine algae, are often challenging to assemble into a single contig (Smith & Lee, Citation2009; de Vries et al., Citation2014; Del Vasto et al., Citation2015), and the T. socialis ptDNA was no exception. The ptDNA sequence is currently contained in a single assembly of 405.5 kb (GenBank Accession KX232643), which harbours the typical volvocine plastid gene content.
Fig. 4. Map of the 405 557 bp chloroplast genome of Tetrabaena socialis. The transcriptional polarity of genes can proceed in a clockwise (inside of circle) or anti-clockwise (outside of circle) direction. Image generated using OrganellarGenomeDraw.
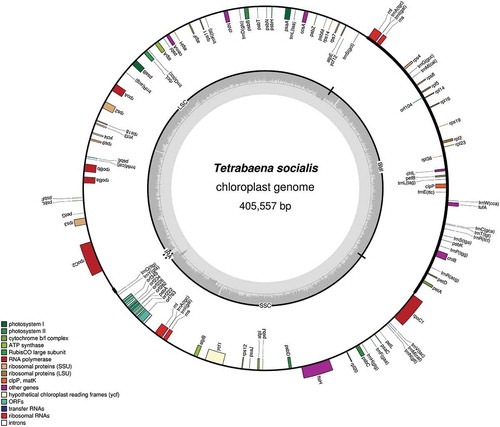
Like its V. carteri counterpart, the T. socialis plastid genome contains long uninterrupted stretches of intergenic DNA, that together represent 309 kb (~76%) of the genome – 210.1 kb of which are comprised of AT-rich palindromic repeats. In fact, the T. socialis intergenic regions alone are larger than all other available volvocine ptDNAs with the exception of that from V. carteri (Smith et al., Citation2013). Introns, which abound in some large organelle genomes (Del Vasto et al., Citation2015), account for just 2% of the T. socialis ptDNA, and are restricted to psbA, which has 7 introns and 6 intronic open-reading frames, mostly representing homing endonucleases. Consequently, the T. socialis psbA gene is surprisingly long (11.5 kb), much longer than its intron-lacking counterparts in V. carteri and G. pectorale (~1 kb).
DISCUSSION
Volvocine mitochondrial genome evolution: going in circles
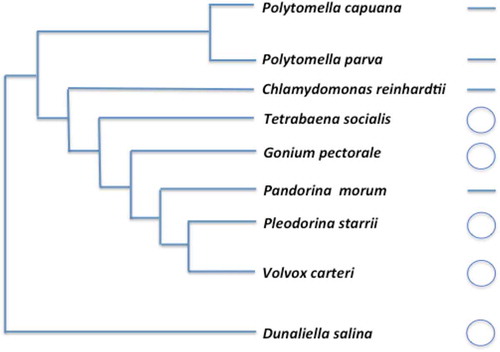
One of the first green algal mtDNAs to be characterized in detail was that of C. reinhardtii (Boer et al., Citation1985), and its linear structure went against the prevailing notion of the time that mtDNAs were circular. Although it is now well established that most green algae have circular-mapping mtDNAs, the Reinhardtinia clade remains a hotspot for linear mitochondrial genomes (Smith et al., Citation2010a). At least ten distinct Reinhardtinia species have linear or linear fragmented mtDNAs, and the most recent common ancestor of the Reinhardtinia clade is thought to have had a linear mtDNA (Fan & Lee, Citation1998; Laflamme & Lee, Citation2003; Smith et al., Citation2010a). In recent years, however, circular-mapping mtDNAs have been found in various Reinhardtinia algae from the volvocine lineage, including G. pectorale, V. carteri and now T. socialis (Smith & Lee, Citation2009; Hamaji et al., Citation2013; Smith et al., Citation2013). In fact, the only colonial or multicellular volvocine known to have a linear mtDNA is P. morum (Moore & Coleman, Citation1989). At the very least, these data suggest that a circular-mapping mtDNA existed at or just before the emergence of colonial living in the volvocines, and perhaps much sooner. If true, this would imply that there was a transition from a circular to a linear mtDNA architecture within the P. morum lineage. depicts the mtDNA conformations for various Reinhardtinia lineages.
Fig. 5. mtDNA conformations for various Reinhardtinia lineages. Relationships were derived from previous phylogenetic analyses (Coleman, Citation1999; Nozaki et al., Citation2000, Citation2014; Nakada et al., Citation2008;). Note that the mtDNA genome of Polytomella parva is fragmented into two linear segments.
Linear mtDNAs exist in a diversity of eukaryotic groups, and like the volvocines some of these groups harbour both linear and circular mitochondrial chromosomes (Rycovska et al., Citation2004; Smith & Keeling, Citation2015). The evolutionary processes responsible for changes in mitochondrial genome conformation are poorly understood, but they can place unique challenges on the recipient. For instance, linear mitochondrial genomes are more vulnerable to degradation by exonucleases and need to overcome the ‘end-replication problem’ (Nosek & Tomáška, Citation2003). However, given the frequency with which mitochondrial genomes can shift from circular to linear and back within some groups, it can be assumed that the mechanisms used to replicate either form exist in these lineages (Nosek & Tomáška, Citation2003), and that such transitions are probably non-adaptive. Moreover, the architecture of many circular-mapping mtDNAs is thought to be much more complicated than a simple closed circle (Bendich, Citation2007) – although detailed gel electrophoresis data do support the idea that some chlamydomonadalean mtDNAs are in fact circular monomeric molecules (Laflamme & Lee, Citation2003) rather than complex branched structures (Bendich, Citation2010).
The existence and function of rtl (a freestanding reverse transcriptase-like gene) in the linear mitochondrial genome of C. reinhardtii (and C. incerta) has puzzled researchers (Boer & Gray, Citation1988; Popescu & Lee, Citation2007), and led some to hypothesize that rtl is involved in mitochondrial telomere maintenance (Vahrenholz et al., Citation1993). This hypothesis, however, is inconsistent with the presence of a freestanding rtl in the mtDNA of T. socialis, which is circular-mapping and lacks telomeres. The T. socialis rtl gene is transcribed (based on Illumina RNA-seq data) and shows reasonable sequence similarity to that of C. reinhardtii at the amino acid level (), suggesting that the gene is functional and has been evolutionarily conserved for >200 MYA. The P. starrii and V. carteri mtDNAs also have an rtl gene, but it is found within a group II intron and contains a maturase domain, and thus has an obvious intronic origin and intron-related function. It remains to be determined if the same is true of the freestanding rtl genes of volvocine algae.
Organelle genome expansion
A lot has been written about organelle genome expansion in chlamydomonadalean algae (Smith & Lee, Citation2010; Del Vasto et al., Citation2015), as well as in other chlorophycean lineages, including the chaetopeltidalean taxon Floydiella terrestris, which has a 521 kb ptDNA (Brouard et al., Citation2010). The volvocine lineage in particular has emerged as a model for testing hypotheses on the evolution of organelle genome size, such as the mutational hazard hypothesis (MHH) (Smith & Lee, Citation2008, Citation2010; Smith et al., Citation2013). The MHH predicts that mutationally hazardous DNA has a greater tendency to accumulate in a system with a low mutation rate (μ) and a small effective genetic population size (Ne), where levels of random genetic drift are high, rather than in one where μ and Ne are large and drift is small (Lynch & Conery, Citation2003; Lynch et al., Citation2007). Studies of volvocine organelle DNAs have generally supported the MHH. For instance, the highly expanded organelle DNAs of V. carteri are predicted to have a very low Neμ (Smith & Lee, Citation2010), and there appears to be a negative scaling of organelle genome non-coding DNA content with Neμ (i.e. silent-site genetic variation) across the volvocine lineage (Smith et al., Citation2013).
The data presented here on the T. socialis organelle genomes reinforce the idea that colonial and multicellular volvocine algae have a propensity for organelle genomic expansion, more so than their unicellular counterparts. The tendency towards inflated organelle genomes is probably due to a multitude of forces, one of which could be elevated levels of random genetic drift. Indeed, one would expect that the transition from a unicellular to a colonial/multicellular mode of existence would result in a reduction in effective population size. It will be interesting to see if the T. socialis organelle DNAs turn out to have low predicted levels of silent-site genetic diversity, reflecting a low Neμ – as previously found in V. carteri (Smith & Lee, Citation2010). There is also the possibility that the most recent common ancestor of volvocine algae had an inflated plastid genome and that the various volvocine lineages existing today have undergone varying degrees of ptDNA reduction.
Despite having large non-coding DNA contents, the T. socialis organelle DNAs are relatively devoid of introns – a trend that is observed throughout the volvocine lineage. Conversely, some chlamydomonadalean organelle genomes from species outside of the volvocine line, such as Dunaliella salina and D. viridis, are magnets for introns (Del Vasto et al., Citation2015), sometimes containing more than one intron per gene (Smith et al., Citation2010b). Why colonial and multicellular volvocine algae have been so susceptible to amassing repetitive non-coding DNA, particularly in their plastids, yet have avoided intron accumulation is an intriguing question and worthy of further investigation.
SUPPLEMENTARY INFORMATION
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at http://dx.doi.org/10.1080/09670262.2016.1198830
Supplementary fig. 1. Introns within the T. socialis organelle genes. The image was generated from MUSCLE alignments of T. socialis, D. salina, C. reinhardtii, G. pectorale, P. starrii and V. carteri genes. Only genes for which T. socialis contains introns are included. Boxes are introns within T. socialis genes. An accurate alignment of the psbA gene was not possible given the large difference in size. Intron presence does not reflect phylogenetic relationship.
Supplementary fig. 2. Multiple sequence alignment of reverse-transcriptase-like protein for T. socialis, C. incerta and C. reinhardtii.
Supplementary fig 2
Download MS Word (950.6 KB)Supplementary fig 1
Download PDF (52.9 KB)DISCLOSURE STATEMENT
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors
Jonathan Featherston
J. Featherston original concept, sequencing, analysis and drafting manuscript; Y. Arakaki culturing, nucleic acid extractions and edited manuscript; H. Nozaki original concept, culturing, nucleic acid extraction and edited manuscript; P. Durand original concept, funded research, drafting and editing manuscript; D. Smith original concept, analysis, drafting and editing manuscript.
Yoko Arakaki
J. Featherston original concept, sequencing, analysis and drafting manuscript; Y. Arakaki culturing, nucleic acid extractions and edited manuscript; H. Nozaki original concept, culturing, nucleic acid extraction and edited manuscript; P. Durand original concept, funded research, drafting and editing manuscript; D. Smith original concept, analysis, drafting and editing manuscript.
Hisayoshi Nozaki
J. Featherston original concept, sequencing, analysis and drafting manuscript; Y. Arakaki culturing, nucleic acid extractions and edited manuscript; H. Nozaki original concept, culturing, nucleic acid extraction and edited manuscript; P. Durand original concept, funded research, drafting and editing manuscript; D. Smith original concept, analysis, drafting and editing manuscript.
Pierre M. Durand
J. Featherston original concept, sequencing, analysis and drafting manuscript; Y. Arakaki culturing, nucleic acid extractions and edited manuscript; H. Nozaki original concept, culturing, nucleic acid extraction and edited manuscript; P. Durand original concept, funded research, drafting and editing manuscript; D. Smith original concept, analysis, drafting and editing manuscript.
David R. Smith
J. Featherston original concept, sequencing, analysis and drafting manuscript; Y. Arakaki culturing, nucleic acid extractions and edited manuscript; H. Nozaki original concept, culturing, nucleic acid extraction and edited manuscript; P. Durand original concept, funded research, drafting and editing manuscript; D. Smith original concept, analysis, drafting and editing manuscript.
REFERENCES
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215: 403–410.
- Arakaki, Y., Kawai-Toyooka, H., Hamamura, Y., Higashiyama, T., Noga, A., Hirono, M., Olson, B.J.S.C. & Nozaki, H. (2013). The simplest integrated multicellular organism unveiled. PLoS ONE, 8: e81641.
- Bendich, A.J. (2007). The size and form of chromosomes are constant in the nucleus, but highly variable in bacteria, mitochondria and chloroplasts. BioEssays, 29: 474–483.
- Bendich, A.J. (2010). The end of the circle for yeast mitochondrial DNA. Molecular Cell, 39: 831–832.
- Boer, P.H. & Gray, M.W. (1988). Genes encoding a subunit of respiratory NADH dehydrogenase (ND1) and a reverse transcriptase-like protein (RTL) are linked to ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. European Molecular Biology Organization Journal, 7: 3501–3508.
- Boer, P.H., Bonen, L., Leet, R.W. & Gray, M.W. (1985). Genes for respiratory chain proteins and ribosomal RNAs are present on a 16-kilobase-pair DNA species from Chlamydomonas reinhardtii mitochondria. Proceedings of the National Academy of Sciences USA, 82: 3340–3344.
- Boetzer, M. & Pirovano, W. (2012). Toward almost closed genomes with GapFiller. Genome Biology, 13: R56.
- Boetzer, M., Henkel, C.V., Jansen, H.J., Butler, D. & Pirovano, W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics (Oxford, England), 27: 578–579.
- Brodie, R., Roper, R.L. & Upton, C. (2004). JDotter: a Java interface to multiple dotplots generated by dotter. Bioinformatics, 20: 279–281.
- Brouard, J., Otis, C., Lemieux, C. & Turmel, M. (2010). The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the chlorophyceae. Genome Biology and Evolution, 2: 240–256.
- Burger, G. & Lang, B.F. (2007). Mitochondrial introns: a critical view. Trends in Genetics, 23: 119–125.
- Coleman, A.W. (1999). Phylogenetic analysis of ‘Volvocacae’ for comparative genetic studies. Proceedings of the National Academy of Sciences USA, 96: 13892–13897.
- Darling, A.C.E., Mau, B., Blattner, F.R. & Perna, N.T. (2004). Mauve : multiple alignment of conserved genomic sequence with rearrangements. Genome Research, 14: 1394–1403.
- De Vries, J., Habicht, J., Woehle, C., Huang, C., Christa, G., Wägele, H., Nickelsen, J., Martin, W.F. & Gould, S.B. (2014). Is ftsH the key to plastid longevity in sacoglossan slugs? Genome Biology and Evolution, 5: 2540–2548.
- Del Vasto, M., Figueroa-Martinez, F., Featherston, J., Gonzalez, M.A., Reyes-Prieto, A., Durand, P.M. & Smith, D.R. (2015). Massive and widespread organelle genomic expansion in the green algal genus Dunaliella. Genome Biology and Evolution, 7: 656–663.
- Edgar, R.C., Drive, R.M. & Valley, M. (2004). MUSCLE : multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32: 1792–1797.
- Fan, J. & Lee, R.W. (1998). Mitochondrial genome of the colorless green alga Polytomella parva : two linear DNA molecules with homologous inverted repeat termini. Molecular Biology and Evolution, 19: 999–1007.
- Hamaji, T., Smith, D.R., Noguchi, H., Toyoda, A., Suzuki, M., Kawai-Toyooka, H., Fujiyama, A., Nishii, I., Marriage, T. Olsen, B.J.S.C. & Nozaki, H. (2013). Mitochondrial and plastid genomes of the colonial green alga Gonium pectorale give insights into the origins of organelle DNA architecture within the Volvocales. PLoS ONE, 8: e57177.
- Hanschen, E.R., Marriage, T.N., Ferris, P.J., Hamaji, T., Toyoda, A., Fujiyama, A., Neme, R., Noguchi, H., Minakuchi, Y., Suzuki, M., Kawai-Toyooka, H., Smith, D.R., Sparks, H., Anderson, J., Bakarić, R., Luria, V., Karger, A., Kirschner, M.W., Durand, P.M. Michod, R.E., Nozaki, H. & Olson, B.J.S.C. (2016). The Gonium pectorale genome demonstrates evolution of multicellularity. Nature Communications, 7( 11370).
- Herron, M.D. & Michod, R.E. (2008). Evolution of complexity in the volvocine algae: transitions in individuality through Darwin’s eye. Evolution; International Journal of Organic Evolution, 62: 436–451.
- Herron, M.D., Hackett, J.D., Aylward, F.O. & Michod, R.E. (2009). Triassic origin and early radiation of multicellular volvocine algae. Proceedings of the National Academy of Sciences USA, 106: 3254–3258.
- Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R. & Salzberg, S.L. (2013). TopHat2 : accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 14: R36.
- Kirk, D.L. (2005). A twelve-step program for evolving multicellularity and a division of labor. BioEssays, 27: 299–310.
- Kurtz, S. & Schleiermacher, C. (1999). REPuter : fast computation of maximal repeats in complete genomes. Bioinformatics, 15: 426–427.
- Laflamme, M. & Lee, R.W. (2003). Mitochondrial genome conformation among CW-group chlorophycean algae. Journal of Phycology, 39: 213–220.
- Lohse, M., Drechsel, O., Kahlau, S. & Bock, R. (2013). OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research, 41:1–7.
- Lynch, M. & Conery, J.S. (2003). The origins of genome complexity. Science, 302: 1401–1404.
- Lynch, M., Koskella, B. & Schaack, S. (2007). Mutation pressure and the evolution of organelle genomic architecture. Science, 311: 1727–1730.
- Maul, J.E., Lilly, J.W., Cui, L., Claude, W., Miller, W., Harris, E.H. & Stern, D.B. (2002). The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. The Plant Cell, 14: 2659–2679.
- Miller, S.M., Schmitt, R. & Kirk, D.L. (1993). Jordan, an active Volvox transposable element similar to higher plant transposons. The Plant Cell, 5: 1125–38.
- Moore, L. & Coleman, A. (1989). The linear 20 kb mitochondrial genome of Pandorina morum (Volvocaceae, Chlorophyta). Plant Molecular Biology, 14: 459–465.
- Nakada, T., Misawa, K. & Nozaki, H. (2008). Molecular phylogenetics and evolution molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Molecular Phylogenetics and Evolution, 48: 281–291.
- Nishii, I. & Miller, S.M. (2010). Volvox: simple steps to developmental complexity? Current Opinion in Plant Biology, 13: 646–653.
- Nosek, J. & Tomáška, L. (2003). Mitochondrial genome diversity: evolution of the molecular architecture and replication strategy. Current Genetics, 44: 73–84.
- Nozaki, H. (1986). Sexual reproduction in Gonium sociale (Chlorophyta, Volvacales). Phycologia, 25: 29–35.
- Nozaki, H., Misawa, K., Kajita, T., Kato, M., Nohara, S. & Watanabe, M.M. (2000). Origin and evolution of the colonial volvocales (Chlorophyceae) as inferred from multiple, chloroplast gene sequences. Molecular Phylogenetics and Evolution, 17: 256–268.
- Nozaki, H., Yamada, T.K., Takahashi, F., Matsuzaki, R. & Nakada, T. (2014). New “missing link” genus of the colonial volvocine green algae gives insights into the evolution of oogamy. BioMed Central Evolutionary Biology, 14: 1–10.
- Popescu, C.E. & Lee, R.W. (2007). Mitochondrial genome sequence evolution in Chlamydomonas. Genetics, 175: 819–826.
- Pringsheim, E. (1946). Pure Cultures of Algae. Cambridge: Cambridge University Press.
- Prochnik, S.E., Umen, J., Nedelcu, A.M., Hallmann, A., Miller, S.M., Nishii, I., Ferris, P. Kuo, A., Mitros, T., Fritz-Laylin, L.K., Hellsten, U., Chapman, J., Simakov, O., Rensing, S.A., Terry, A., Pangilinan, J., Kapitonov, V., Jurka, J., Salamov, A., Shapiro, H., Schmutz, J., Grimwood, J., Lindquist, E., Lucas, S., Grigoriev, I.V., Schmitt, R., Kirk, D. & Rokhsar, D.S. (2010). Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science, 329: 223–226.
- Quinlan, A.R. & Hall, I.M. (2010). BEDTools : a flexible suite of utilities for comparing genomic features. Bioinformatics, 26: 841–842.
- Rycovska, A., Valach, M., Tomaska, L., Bolotin-Fukuhara, M. & Nosek, J. (2004). Linear versus circular mitochondrial genomes : intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology, 150: 1571–1580.
- Simpson, J.T. & Durbin, R. (2012). Efficient de novo assembly of large genomes using compressed data structures. Genome Research, 22: 549–56.
- Smith, D.R. & Keeling, P.J. (2015). Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proceedings of the National Academy of Sciences USA, 112: 10177–10184.
- Smith, D.R. & Lee, R. (2008). Nucleotide diversity in the mitochondrial and nuclear compartments of Chlamydomonas reinhardtii: investigating the origins of genome architecture. BioMed Central Evolutionary Biology, 8: 156.
- Smith, D.R. & Lee, R.W. (2009). The mitochondrial and plastid genomes of Volvox carteri: bloated molecules rich in repetitive DNA. BioMed Central Genomics, 10: 132.
- Smith, D.R. & Lee, R.W. (2010). Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Molecular Biology and Evolution, 27: 2244–2256.
- Smith, D.R. & Lee, R.W. (2014). A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiology, 164: 1812–1819.
- Smith, D.R., Hua, J. & Lee, R. (2010a). Evolution of linear mitochondrial DNA in three known lineages of Polytomella. Current Genetics, 56: 427–438.
- Smith, D.R., Lee, R., Cushman, J., Magnuson, J., Duc, T. & Jurgen, P. (2010b). The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA. BioMed Central Plant Biology, 10: 83.
- Smith, D.R., Hamaji, T., Olson, B.J.S.C., Durand, P.M., Ferris, P., Michod, R.E., Featherston, J., Nozaki, H. & Keeling, P.J. (2013). Organelle genome complexity scales positively with organism size in Volvocine green algae. Molecular Biology and Evolution, 30:793–797.
- Vahrenholz, C., Riemen, G., Pratje, E., Dujon, B. & Michaelis, G. (1993). Mitochondrial DNA of Chlamydomonas reinhardtii: the structure of the ends of the linear 15.8-kb genome suggests mechanisms for DNA replication. Current Genetics, 24: 241–247.
