ABSTRACT
Ergosterol and 7-dehydroporiferasterol are the predominant sterols in the membranes of the alga Chlamydomonas reinhardtii Dangeard. Ergosterol is primarily found in most fungi, which produce their end-product sterols by way of the intermediate lanosterol. In contrast, plants rarely make ergosterol and plant sterols are made via the intermediate cycloartenol. The cycloartenol-lanosterol bifurcation has been used as a means to evolutionarily categorize species based on their sterol production. Use of bioinformatics has revealed that the green alga, C. reinhardtii is probably producing sterols using a pathway very similar to that of higher plants. The Chlamydomonas genome was searched for genes that encoded proteins exhibiting high similarity to sterol biosynthetic proteins in the higher plant Arabidopsis thaliana or the yeast Saccharomyces cerevisiae. Genes with the greatest similarity were chosen and annotated. To establish whether these genes were expressed, the presence of their transcripts was determined and quantitative RT-PCR was performed. It was observed that many of the transcripts coordinately increased in abundance after deflagellation, which induces an increase in membrane biosynthesis. This work demonstrates that C. reinhardtii has all of the genes necessary for the biosynthesis of ergosterol and 7-dehydroporiferasterol and that this alga uses the higher plant pathway to make a sterol normally associated with fungi. Evidence is also presented that other organisms commonly known as algae, including chlorophytes, haptophytes, rhodophytes, bacillariophytes, a phaeophyte and a cryptomonad, are likely to utilize the same biosynthetic pathway, making sterols via a cycloartenol intermediate.
Introduction
Sterols are a class of lipids found in the membranes of eukaryotic organisms. These compounds have four fused rings and intercalate between the phospholipids that make up the membrane bilayer. The positioning and structure of the sterol influences the fluidity and permeability of the membrane (Solomon et al., Citation2008). All sterols are synthesized from the triterpene precursor squalene. A variety of sterols are made in different organisms, with ergosterol being the most common sterol found in fungi and cholesterol being the most abundant sterol in vertebrates (Benveniste, Citation2004). Plants synthesize several sterols including stigmasterol, campesterol and sitosterol, but usually have very low levels of ergosterol and 7-dehydroporiferasterol (Benveniste, Citation2004). In contrast, the two most abundant sterols found in the green alga, Chlamydomonas reinhardtii Dangeard, are ergosterol and 7-dehydroporiferasterol (Gealt et al., Citation1981).
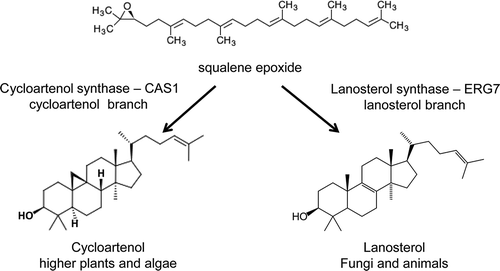
While all sterols are derived from squalene, two major variations in the cyclization step to form the sterol nucleus are known. Non-photosynthetic species make lanosterol from squalene-epoxide (), a reaction catalysed by lanosterol synthase. On the other hand, higher plants make cycloartenol (), a molecule with five rings including a cyclopropane ring. If cycloartenol is made, the cyclopropane ring is eventually cleaved through the action of cyclopropyl sterol isomerase (CPI1). Cycloartenol and lanosterol are considered to be stereoisomers (Nes et al., Citation1990). In the past this ‘bifurcation of the sterol pathway’ has been used to distinguish between those organisms which produced their sterols by way of ‘oxygenic photosynthesis’ making cycloartenol and those species that make lanosterol as the first cyclized intermediate (Nes et al., Citation1990; Desmond & Gribaldo, Citation2009). However, recently, it has been discovered that end-product sterols are also synthesized by way of lanosterol synthase in dicotyledonous plants (Ohyama et al., Citation2009) and there is now evidence that Arabidopsis thaliana makes both lanosterol synthase and cycloartenol synthase (Ohyama et al., Citation2009).
Fig. 1. Schematic diagram showing the biochemical reactions from squalene epoxide to cycloartenol and lanosterol. In plants, sterols are synthesized via the five ring intermediate, cycloartenol. In animals and fungi, sterols are made via the four ring intermediate lanosterol. Lanosterol synthase (ERG7) and cycloartenol synthase (CAS1) synthesize these reactions.
Little is known about the synthesis of sterols in algae in general or in C. reinhardtii in particular. However it has been known for a long time that the two most abundant sterols in C. reinhardtii are ergosterol and 7-dehydroporiferasterol. In 1978, the first sterol mutants in C. reinhardtii were isolated by selecting for single colonies resistant to nystatin (Bard et al., Citation1978). In 1981, a chromatographic analysis was performed to identify the major sterols and fatty acids in C. reinhardtii (Gealt et al., Citation1981) and in 1982, sterol synthesis was studied during the cell cyle by Janero & Barrnett (Citation1982). In 1999, Salimova et al. identified three sterol mutants that were deficient in C24(28)-reductase (Salimova et al., Citation1999). Their work also identified a possible pathway for the last few steps in the synthesis of ergosterol and 7-dehydroporiferasterol. However, the pathway proposed by Salimova et al. (Citation1999) does not show potential substrates and precursors for earlier biosynthetic steps of the ergosterol pathway. In 2012, Miller et al. showed that ergosterol in C. reinhardtii is made using a pathway that differs from fungi (Miller et al., Citation2012).
With over 95% of the C. reinhardtii genome now sequenced (Merchant et al., Citation2007; Blaby et al., Citation2014), it is possible to identify genes encoding the proteins involved in sterol biosynthesis (http://phytozome.jgi.doe.gov/pz/portal.html). This sequence information was used in our previous study to demonstrate that ERG3 was part of the sterol biosynthetic pathway (Brumfield et al., Citation2010). In that work the C. reinhardtii ERG3 gene was used to restore ergosterol biosynthesis in the ERG3 knockout of Saccharomyces cerevisiae by functional complementation. However, ERG3 encodes a protein involved in a latter step of sterol biosynthesis (Brumfield et al., Citation2010). In this work, we have identified the genes involved in the conversion of the precursor, squalene epoxide, to the final sterols in C. reinhardtii. We show that while C. reinhardtii makes ergosterol, a sterol often associated with fungi, it is made using the higher plant biosynthetic pathway. A pathway for sterol biosynthesis in C. reinhardtii is also proposed.
Materials and methods
C. reinhardtii strains and growth
Strain CC-124 mt− was obtained from the Chlamydomonas Resource Center (http://chlamycollection.org/). Cultures were grown on Tris Acetate Phosphate (Sueoka, Citation1960) (TAP) medium plates. They were inoculated into 100 ml of TAP medium (Sueoka, Citation1960) and set to grow with continuous light (100 µmol photons m−2 s−1) and shaking for 2–3 d. Aliquots of the culture were transferred to Minimal media (MIN) (Sueoka, Citation1960), which has no carbon source other than CO2, in a larger flask or carboy and bubbled with high CO2 (5% CO2 in air) for several days. Cells were then collected for further analysis: pH shock or RNA preparation.
Bioinformatics and sequence analysis
Information about genes found in C. reinhardtii was gathered from searching the genomic database, which yielded gene models, scaffold and chromosome locations (http://phytozome.jgi.doe.gov/pz/portal.html). Ergosterol biosynthetic enzymes were obtained from S. cerevisiae sequences on the NCBI protein database. The C. reinhardtii EST database using the BLAST program provided alignments with the S. cerevisiae amino acid sequences (http://www.ncbi.nlm.nih.gov/BLAST) (Altschul et al., Citation1997). In order to determine the correct gene, E-values were considered. The gene with the highest similarity was chosen as the best choice for the gene. Splice sites of the exon/intron and open reading frames were identified using the method described by Silflow et al. (Citation1995). These programs included hyperlinks to the EXPASY server (http://ca.expasy.org/tools/#translate). The Pfam database was used to identify possible domains in the protein sequences (http://pfam.sanger.ac.uk). PSORT (Prediction of Protein Sorting Signals and Localization Sites in Amino Acid Sequences), ChloroP and TargetP were used to predict the localization of specific proteins (http://psort.nibb.ac.jp/). Transmembrane helix predictions were made using TMpred (Hofmann & Stoffel, Citation1993) and SOSUI (Hirokawa et al., Citation1998). The S. cerevisiae genome database provided information about the function of the enzymes in yeast (http://www.yeastgenome.org/). The A. thaliana genome gave additional information about sterol biosynthetic genes in higher plants (http://www.arabidopsis.org).
Genomic sequences for Oryza sativa, Physcomitrella patens, Volvox carterii, Coccomyxa subellipsoidea, Chlorella variabilis, Auxenochlorella protothecoides, Helicosporidium sp. ATCC 50920, Ostrecoccus lucimarinus, Micromonas pusilla CCMP1545, Galdieria sulphuraria, Chondrus crispus, Cyanidioschyzon meolae, Emiliania huxleyi CCMP1516, Chrysochromulina sp. CCMP291, Phaeodactylum tricornutum, Thalassiosira pseudonana, Ectocarpus siliculosus, Guillardia theta CCMP2712, Aureococcus anophagefferens were retrieved from NCBI (http://www.ncbi.nlm.nih.gov), Pico PLAZA (http://bioinformatics.psb.ugent.be/plaza/versions/pico-plaza/) and the JGI database (http://genome.jgi-psf.org). Protein alignments were generally made using the pBLAST servers at the PLAZA or NCBI websites. However in some cases we searched the nucleotide sequences using tBLASTn.
Multiple sequence alignment programs from the European Bioinfomatics Institute EMBL-EBI server (http://www.ebi.ac.uk/clustalw/) were used for sequence analysis and alignment data. The Prosite database from EBI server (http://ca.expasy.org/prosite/) was used to identify putative consensus motifs for specific domains. The C. reinhardtii genomic database (version 5.5) provided information about the genomic sequence, intron-exon structure, as well as information regarding potential membrane spanning regions of the protein coding sequence (https://phytozome.jgi.doe.gov/pz/portal.html).
Flagellar pH shock
Chlamydomonas reinhardtii strain CC-124 mt− cultures were grown on Tris-Acetate Phosphate (TAP) agar plates, and aseptically inoculated after 5 d into Tris-Acetate Phosphate media for 2 d with continuous shaking under light. TAP cultures were transferred to Minimal media (MIN) and incubated at 28°C for 48 h under continuous light and CO2 with continuous shaking. Cultures were harvested by centrifugation in a Beckman J2-21 centrifuge at 4°C, 480g for 10 min in a JA-10 rotor. For the control sample, the cell pellet was resuspended in 30 ml of fresh MIN medium and incubated at 28°C on a shaker under light.
Acid shock for removal of flagella
The pellet was resuspended in 50 ml of 10 mM Tris-HCl buffer, pH 7.8 + 7% sucrose (w/v) at 4°C. Cells were deflagellated by addition of 0.5 N acetic acid to pH 4.5 and held at acid pH for 45 s, following which the pH was raised to 7.8 with 0.5 N KOH (Witman et al., Citation1972).
Regeneration of flagella
After the initial pH shock, the cell aliquots lacking the flagella were resuspended in 300 ml of MIN media and incubated at 28°C on a shaker under light. Immediately, a 30 ml aliquot was removed for the ‘time 0’ sample. At 30 min intervals, 30 ml aliquots were removed from the shaker, up to 240 min, to isolate the RNA for Quantitative PCR analysis. The data derived from these samples was used to measure the relative expression of ergosterol genes during reflagellation.
Total RNA isolation
Triazol reagent (Invitrogen, Carlsbad, California, USA) was used to extract RNA from C. reinhardtii. Cells were harvested and transferred to 50 ml conical centrifuge tubes. Cells were then spun at 1000 g for 5 min at a final temperature of 4°C. The pellet was resuspended in 1.0 ml of Triazol and transferred to a 1.5 ml Eppendorf tube. Cells were then vortexed, incubated for 1 h at room temperature and frozen until all of the samples were collected. Later, these samples were thawed and 200 µl of chloroform was added. To mix the sample, the tubes were again vortexed and incubated at room temperature for 5 min. The samples were centrifuged at 12 500 g for 15 min at 4°C. The aqueous phase of approximately 600 µl was removed and transferred into a fresh 1.5 ml Eppendorf tube, 0.5 ml of isopropanol was added and the samples were incubated for 30 min. The samples were then centrifuged at 12 500 g for 15 min at 4°C, the supernatant was removed and the RNA pellet was washed with 1.0 ml 75% ethanol, then centrifuged at 6000 g for 5 min. The supernatant was removed and the pellet was air dried for an hour. The RNA was finally treated with Ambion Turbo Dnase (Invitrogen, Carlsbad, California, USA). The manufacturer’s protocol was followed and the samples were then treated by an RNA clean-up using Qiagen’s RNeasy Clean-up Kit (Qiagen, Valencia, California, USA). Total RNA concentration was determined using the Nanodrop 2000 series.
Quantitative RT (Real Time) PCR
Three micrograms of total RNA was used as template for synthesis of cDNA. SuperscriptFirst Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, California, USA) was used to synthesize the cDNA according to the manufacturer’s instruction. An aliquot of 100 ng cDNA was used as the template with SYBR Select (Applied Biosystems, Foster City, California, USA) for quantitative PCR in an ABI Prism 7000 sequence detection system following the manufacturer’s instructions (Applied Biosystems, Foster City, California, USA). Primers are listed in Supplementary Table 1. The qPCR data were analysed using ΔΔCT for comparative quantification. Expression level of each gene was assayed for up- or down-regulation using Cblp as the normalizer. The normalized gene of interest was compared with the control (cells that had not undergone deflagellation). Primers specific for the gene CBLP (Chlamydomonas beta subunit-like polypeptide) were used as control primers for equal loading (Im & Grossman, Citation2001). The CBLP specific sequences were: CBLPF; 5’-CTTCTCGCCCATGACCAC-3’ and CBLPR 5’-CCCACCAGGTTGTTCTTCAG-3’.
Table 1. Orthologues of sterol biosynthetic genes in Saccharomyces cerevisiae, Arabidopsis thaliana and Chlamydomonas reinhardtii.
Results
Chlamydomonas has a complete set of genes for sterol biosynthesis
To identify the genes involved in sterol biosynthesis in C. reinhardtii, the C. reinhardtii genome was searched using tBLASTn for proteins that would match either the yeast S. cerevisiae, or A. thaliana sterol biosynthetic proteins. Saccharomyces cerevisiae and A. thaliana were used as these are the best studied systems in fungi and higher plants. Orthologues of all the sterol biosynthesis genes present in A. thaliana were found in C. reinhardtii (). Most of the C. reinhardtii genes also have a corresponding gene (or genes) in yeast. The exceptions are cycloartenol synthase (CAS1), cyclopropyl isomerase (CPI1) and C(8,7) sterol isomerase (HYD1), all of which are absent in yeast. Yeast uses the lanosterol pathway and has lanosterol synthase (LAS) instead of CAS1 (Benveniste, Citation2004). CAS1, CPI1 and HYD1 are found only in organisms that use the cycloartenol biosynthetic pathway and are not found in S. cerevisiae. In yeast, the demethylation step is catalysed by a complex encoded by ERG25/ERG26/ERG27/ERG28 (Mo et al., Citation2004). In A. thaliana, orthologues of ERG25 and ERG26 can be found. However, no A. thaliana gene can be found with a significant match to the yeast ERG27 (Desmond & Gribaldo, Citation2009). Chlamydomonas reinhardtii also has orthologues to ERG25, ERG26 and ERG28, and like A. thaliana does not have an orthologue to ERG27 in yeast. Genes encoding the other proteins of the sterol pathway are present in C. reinhardtii, and they exhibit a strong identity to the A. thaliana orthologue and, in general, a good identity to the yeast orthologue (). Only one gene encoding each protein was found in the haploid C. reinhardtii genome. Some of the proteins involved in sterol biosynthesis in C. reinhardtii have not been studied, but recently the C-5 sterol desaturase (Brumfield et al., Citation2010) and the sterol C-24 methyltranferase (Haubrich et al., Citation2015) were characterized. The identification for each gene of the C. reinhardtii sterol pathway is given in using phytozome version 10.3 (http://phytozome.jgi.doe.gov/pz/portal.html).
Table 2. Chlamydomonas reinhardtii ergosterol genes and genome location.
Chlamydomonas reinhardtii synthesizes sterols via the cycloartenol intermediate
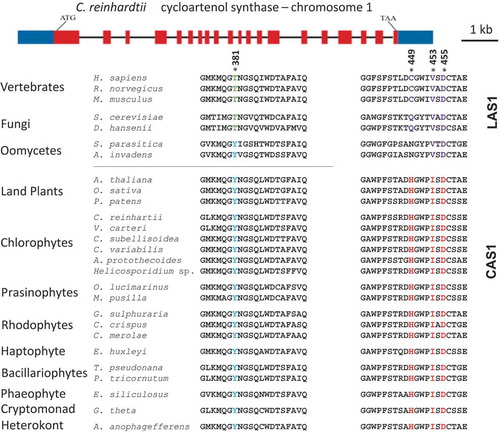
There are two major variations in the cyclization step of squalene-epoxide. The pathway used by fungi and vertebrates results in the synthesis of sterols via lanosterol, while higher plants make cycloartenol as the intermediate (Desmond & Gribaldo, Citation2009). The proteins that make lanosterol or cycloartenol are similar, sharing as much as a 35% sequence identity. Summons et al. (Citation2006) have identified key amino acids that are diagnostic for determining whether a protein is likely to catalyse the cyclization to cycloartenol or lanosterol (). Amino acids at position 381, 449 and 453, based on the Homo sapiens sequence, are conserved among lanosterol synthases but are different in cycloartenol synthases. In contrast, amino acid D455 is thought to be a highly conserved catalytic residue and is conserved in all lanosterol and cycloartenol synthases (Summons et al., Citation2006). Looking at these specific amino acids, the C. reinhardtii sequence aligns well with the higher plant sequence for cycloartenol synthase (). The C. reinhardtii gene is predicted to encode a protein that has the amino acids that match the CAS group of proteins and not the LAS clade (). The C. reinhardtii CAS1 gene is found on chromosome 1. It has 20 exons and sequence alignment reveals a 34% similarity between lanosterol synthase, ERG7, in yeast and 58% sequence similarity with cycloartenol synthase from A. thaliana (). This alignment predicts that C. reinhardtii synthesizes sterols via a cycloartenol intermediate ().
Fig. 2. Structure of the CAS1 gene of Chlamydomonas reinhardtii and multiple sequence alignment of key amino acids in both lanosterol synthase and cycloartenol synthase across species. * = conserved residues. Amino acids 381, 449 and 453 are highly conserved residues, D455 is the catalytic residue. The numbering of the amino acids is based on the Homo sapiens sequence. The crystal structure of the oxidosqualene cyclase in H. sapiens has been published (Thoma et al., Citation2004).
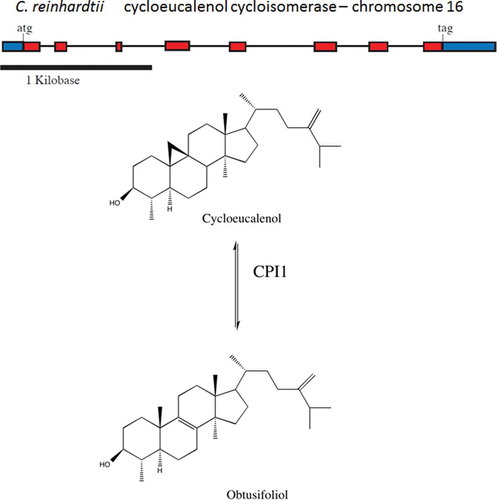
The presence of CPI1 and HYD1 also indicate the use of the cycloartenol bifurcation pathway in C. reinhardtii
Chlamydomonas reinhardtii also has genes predicted to encode cyclopropyl sterol isomerase (CPI1) and the C(8,7) sterol isomerase (HYD1). These genes are found primarily in higher plants and their presence strongly indicates the use of the cycloartenol pathway as a means to produce sterols. CPI1 functions to open the cyclopropane ring in cycloeucalenol to catalyse the production of obtusifoliol (Heintz & Benveniste, Citation1974; Rahier et al., Citation1989; Rahier & Karst, Citation2014) () while HYD1 catalyses the Δ8 sterol into its Δ7 sterol isomer (Souter et al., Citation2002). Until recently, this gene was found only in higher plants. In C. reinhardtii, CPI1 is located on chromosome 16 (, ). The genomic DNA spans 3.6 kb and the cDNA is 0.8 kb in size. The CPI1 gene has 8 exons, and the protein is predicted to have 7 transmembrane domains. The C. reinhardtii CPI1 gene has sequenced ESTs which span the entire transcribed message. HYD1 is found on chromosome 12 (). The presence of both CPI1 and HYD1 as well as CAS1 in the C. reinhardtii genome supports the idea that C. reinhardtii uses the higher plant pathway to synthesize sterols.
Table 3. Identification of CPI1 in other organisms.
CAS1, CPI1 and HYD1 are also found in other algae
The presence of CAS1, CPI1 and HYD1 in C. reinhardtii raised the question of how prevalent these genes are in other photosynthetic protists. While the number of algal genomes that have been sequenced is still limited, it appears that CAS1 (), CPI1 () and HYD1 (data not shown) are present in all algal genomes analysed. This implies that these organisms all have the capability of synthesizing sterols via a cycloartenol intermediate. The algae investigated included the chlorophytes Volvox carterii, Coccomyxa subellipsoidea, Chlorella variabilis, Auxenochlorella protothecoides and Helicosporidium; the prasinophytes Ostrecoccus lucimarinus and Micromonas pusilla; the rhodophytes Galdieria sulphuraria, Chondrus crispus and Cyanidioschyzon merolae; the haptophytes Emiliania huxleyi and Chrysochromulina; the bacillariophytes Phaeodactylum tricornutum and Thalassiosira pseudonana; the phaeophyte Ectocarpus siliculosus; the cryptomonad Guillardia theta; the oomycetes Saprolegnia parasitica and Aphanomyces invadans; and the heterokont Aureococcus anophagefferens. A search of the NCBI database revealed that all of these organisms contain an orthologue of CPI1 (). In addition, most of the organisms had an orthologue of HYD1 although some the alignments were not as strong. The only other organisms that have these genes are land plants () and Dictyostelium (Desmond & Gribaldo, Citation2009). In addition, each of the algae and all of the plants were predicted to encode an orthologue of CAS1 that contained the key amino acids that distinguish between a protein that catalyses the synthesis of lanosterol versus cycloartenol (). The CAS1 from all of the algae are expected to catalyse the formation of cycloartenol based on their predicted amino acid sequences (Summons et al., Citation2006; Desmond & Gribaldo, Citation2009). It is also notable that none of the algae had an orthologue that matched lanosterol synthase. The presence of CAS1 and CPI1 in these genomes support the hypothesis that all of these algae synthesize sterols via cycloartenol.
The CAS1/LAS1 proteins from the oomycetes S. parasiticaand A. invadens were also included in this analysis as they are also grouped with the heterokonts. The proteins from these two oomycetes more closely aligned with LAS1 activity as they have a valine at position 453, the amino acid that most closely predicts lanosterol versus cycloartenol activity (Desmond & Gribaldo, Citation2009). In addition we were unable to find a gene or protein that significantly aligned with CPI1 in either of the oomycetes.
The sterol biosynthetic genes identified in the C. reinhardtii genome are actively transcribed
There is substantial evidence that the genes of the sterol biosynthetic pathway are actively transcribed in C. reinhardtii. There are sequenced ESTs in the C. reinhardtii genome that match each of the genes presented in . In addition, next generation sequence data confirms that all of the genes in this proposed pathway are expressed ( and http://genomes.mcdb.ucla.edu/Cre454/). In fact, there was nearly complete coverage for all of the sterol genes. We also investigated the expression of these genes during reflagellation. Chlamydomonas reinhardtii will drop its flagella when exposed for a short time to a low external pH (pH 4.5). Once the external pH is returned to normal (pH 6.5–7.5), the flagella will regenerate over a period of a few hours (Harris, Citation1989). Since the flagellar membrane contains sterols (Gealt et al., Citation1981), we measured the expression of the sterol biosynthetic genes during the reflagellation. In this study, we were able to obtain evidence for the expression of each gene tested (). It was also confirmed that each gene in the pathway was expressed during reflagellation ( and http://genomes.mcdb.ucla.edu/Cre454/). This analysis showed that the genes thought to be involved in sterol biosynthesis are actively transcribed during the regrowth of the flagella in reflagellation.
Table 4. Evidence for the expression of C. reinhardtii sterol biosynthetic genes.
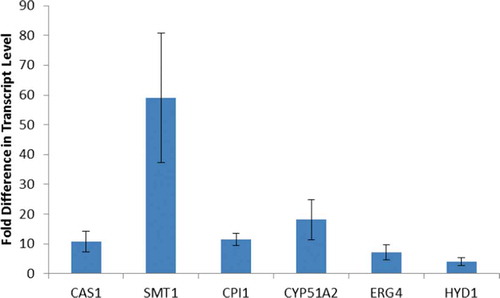
Some of the genes involved in sterol biosynthesis showed increases in expression during reflagellation (). We investigated the expression levels of the genes that synthesize the metabolic reactions after the cyclization of squalene and found that the messages for CAS1, SMT1, CPI1, CYP51A2 and HYD1 all increase in expression at least 10-fold during reflagellation (). The data in show the average increase in the expression levels 1 hour after deflagellation. This increase in expression continues for at least 2–3 hours. A comparison of the transcript levels for all seven genes at 30-minute intervals is shown in Supplementary Fig. 1. These data provide further support for the role of these genes in sterol biosynthesis as C. reinhardtii makes new membranes during reflagellation.
Fig. 4. Transcript levels of sterol biosynthetic genes during reflagellation. Fold increase in expression for seven genes of the sterol biosynthetic pathway 1 hour after deflagellation compared with control cells that did not undergo deflagellation. The data are the mean (+/- standard error) of three biological replicates, each done in duplicate. The primers used in this qRT-PCR study are listed in Supplementary .
Discussion
Previous studies have reported on the lipid composition of C. reinhardtii membranes, but little was known about the enzymatic mechanisms by which these lipids are produced in this organism. This paper presents a bioinformatics analysis of the genes involved in ergosterol biosynthesis in C. reinhardtii. In a review by Benveniste (Citation2004), an analysis and description of each gene in sterol biosynthesis found in A. thaliana was presented and hallmark experiments used to analyse each specific enzyme were described. Comparing the material presented by Benveniste (Citation2004) and the two pathways for sterol biosynthesis in S. cerevisiae and A. thaliana, we gathered information to construct a putative pathway in C. reinhardtii for further study and experimentation.
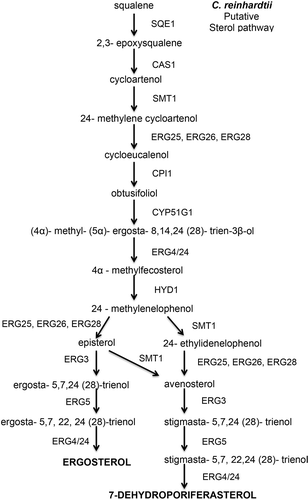
The presence of the three genes CAS1, CPI1 and HYD1 in the C. reinhardtii genome strongly suggests that this alga uses the higher plant branch of the sterol biosynthetic pathway with cycloartenol as an intermediate step. Cycloartenol synthase catalyses the reaction from 2,3-epoxysqualene to cycloartenol. This critical step is a major evolutionary branching point between photosynthetic eukaryotes and non-photosynthetic organisms like yeast (Benveniste, Citation2004; ). For example, in S. cerevisiae, 2,3-epoxysqualene is cyclized to lanosterol by a 2,3-lanosterol synthase, or ERG7 (Benveniste, Citation1986). The hypothesis that C. reinhardtii uses the cycloartenol pathway is in agreement with the work of Miller et al. (Citation2012) who presented mass spectrometrics indicating ergosterol in C. reinhardtii was made differently than in yeast. In their work they observed most of the intermediates for ergosterol and 7-dehydroporiferasterol shown in . Notably they found cycloartenol but not lanosterol (Miller et al., 2012).
Fig. 5. Putative sterol pathway in Chlamydomonas reinhardtii showing the reactants and enzymes that catalyse the steps to synthesize ergosterol in C. reinhardtii.
Functional complementation of yeast mutants lacking lanosterol synthase with A. thaliana cycloartenol synthases led to the discovery of the A. thaliana gene (Corey et al., Citation1993). Very interestingly, A. thaliana cycloartenol synthase has been shown to produce lanosterol when point mutations are present (Y410T or H477N) (Herrera et al., Citation1998; Hart et al., Citation1999; Segura et al., Citation2002; Benveniste, Citation2004; Lodiero et al., Citation2005). Lanosterol synthase and cycloartenol synthase are quite similar (more than 35% similarity between the genes). Summons et al. (Citation2006) identified key amino acid differences between lanosterol synthase and cycloartenol synthase. Amino acids at position 381, 449 and 453 (numbered according to the human protein) are conserved among lanosterol and cycloartenol synthase, while amino acid D455 is thought to be a highly conserved catalytic residue (Summons et al., Citation2006). Looking at these specific amino acids, C. reinhardtii sequence aligns well with the higher plant sequence for cycloartenol synthase ().
When cycloartenol is made, another enzyme, cyclopropyl sterol isomerase (CPI1), is needed to break the cyclopropane ring. The gene encoding CPI1 is also present in the C. reinhardtii genome with a 45% identity to the higher plant gene (; ). HYD1, which also participates in the cycloartenol branch of sterol biosynthesis, is also present in the C. reinhardtii genome. Thus cycloartenol synthase (CAS1), cyclopropyl sterol isomerase (CPI1) and C(8,7) sterol isomerase (HYD1) have all been identified in the C. reinhardtii genome, supporting the idea that the algal model system does follow a sterol biosynthetic pathway very similar to that of higher plants. A proposed sterol biosynthetic pathway in C. reinhardtii is shown in .
A survey of other algal species indicates that they are also likely to use the cycloartenol branch of the pathway. The genomes of all of the photosynthetic organisms analysed so far have the CPI1 gene and CAS1 instead of LAS1 (; ). Our results confirm and extend the work of Desmond & Gribaldo (Citation2009) who looked at the CAS1/LAS1 sequences for a number of photosynthetic and non-photosynthetic organisms. In the photosynthetic organisms they checked, they found that the enzymes that cyclized squalene had the amino acids that better matched CAS1 than LAS1. It would appear that the cycloartenol pathway is widespread in algae, as CAS1 and CPI1 were found in all of the eukaryotic algae with sequenced genomes, which now includes a wide variety of species (; ). Organisms that did not have the cycloartenol pathway were the oomycetes, including Saprolegnia parasitica and Aphanomyces invadans which are non-photosynthetic heterokonts. The gene encoding the cyclization step in these oocytes aligned more closely with LAS1 than CAS1. In addition we were unable to identify CYP1 in either organism. Since the photosynthetic heterokonts Phaeodactylum tricornutum, Thalassiosira pseudonana, Ectocarpus siliculosus and Aureococcus anophagefferens all have CAS1 and CPI1, this raises the possibility that the cycloartenol pathway is found in most or all organisms that formed an endosymbiotic association with a photosynthetic bacterium. It is not clear whether alveolates have the cycloartenol pathway. Sterols consistent with the higher plant pathway were found in the alveolate (Chromera velia) although CAS1, CPI1 and HYD1 were not specifically identified (Leblond et al., Citation2012).
While it has been several decades since ergosterol and 7-dehydroporiferasterol were identified in the cell and flagella membranes in C. reinhardtii (Gealt et al., Citation1981), there have been very few studies on the regulation of sterol biosynthesis in this alga. Here evidence is presented that the genes of the sterol biosynthetic pathway are expressed and that many of them are up-regulated during reflagellation (; Supplementary Fig. 1). Our results are consistent with those of Albee et al. (Citation2013) who observed an increase in expression of many of the genes involved in the sterol biosynthesis pathway after deflagellation. They observed an increase in expression of the sterol pathway genes after farnesyl pyrophosphate synthase but did not see an increase in expression in the genes encoding proteins of the 2-C-methyl-D-erythritol 4-phosphate pathway (Albee et al., Citation2013). The identification of the sterol genes in C. reinhardtii will enhance our knowledge of lipid biosynthesis in the alga and allow for the study of interplay between diacylglycerol biosynthesis and sterol biosynthesis. For example, in other organisms, several processes in phospholipid biosynthesis are regulated by ergosterol. Phospholipid composition is affected by the transfer rate of phospholipids by phosphoinositol-transfer protein when stimulated by ergosterol (Szolderits et al., Citation1989). Ergosterol is also known to upregulate the activity of phosphatidylinositol kinase (Dahl & Dahl, Citation1985; Dahl et al., Citation1987), and stimulate the methylation of phosphatidylethanolamine to phosphatidylcholine (Kawasaki et al., Citation1985). There is also evidence for crosstalk between ergosterol and other lipid metabolic pathways as ceramide production is down-regulated when 3-hydroxy-3-methylglutaryl-CoA reductase, a critical enzyme in ergosterol biosynthesis, is inhibited by lovastatin in mammalian cells (Storey et al., Citation1998). In addition, it has been found that ergosterol acts as an anti-cancer agent, by inhibiting breast cancer cell growth in vitro (Subbiah & Abplanalp, Citation2003).
Because metabolic engineering in yeast has opened so many doors to understanding the ergosterol biosynthetic pathway (Wriessnegger & Pichler, Citation2013), this may lead to future industrial and pharmaceutical use of ergosterol from genetically engineered C. reinhardtii. Future studies can be performed to carefully characterize these sterol genes and identify their significance to other lipid biosynthetic pathways in this model system.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at http://dx.doi.org/10.1080/09670262.2016.1225318
Supplementary table 1. Chlamydomonas ergosterol qPCR primers.
Supplementary fig. 1. Transcript levels of sterol biosynthetic genes for the first four hours after deflagellation. RNA samples were taken every 30 minutes after cells were placed into the media allowing reflagellation. The data shows the fold increase in expression after deflagellation compared with control cells that did not undergo deflagellation. The data are an average of three biological replicates, each done in duplicate. The primers used in this qRT-PCR study are listed in Supplementary Table 1.
Supplemental Figure 1
Download TIFF Image (2.7 MB)Supplemental Table 1
Download MS Word (12.8 KB)Acknowledgements
The authors would like to thank Bratati Mukherjee and Meredith Blackwell for all their helpful advice in revising the article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Albee, A.J., Kwan, A.L., Lin, H., Granas, D., Stormo, G.D. & Dutcher, S.K. (2013). Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3: Genes |Genomes |Genetics, 3: 979–991.
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25: 3389–3402.
- Bard, M., Wilson, K. & Thompson, R. (1978). Isolation of sterol mutants in Chlamydomonas reinhardi: chromatographic analyses. Lipids, 13: 533–539.
- Benveniste, P. (1986). Sterol biosynthesis. Annual Review of Plant Physiology, 37: 275–308.
- Benveniste, P. (2004). Biosynthesis and accumulation of sterols. Annual Review of Plant Biology, 55: 429–457.
- Blaby, I.K., Blaby-Haas, C.E., Tourasse, N., Hom, E.F.Y., Lopez, D., Aksoy, M., Grossman, A., Umen, J., Dutcher, S., Porter, M., King, S., Witman, G.B., Stanke, M., Harris, E.H., Goodstein, D., Grimwood, J., Schmutz, J., Vallon, O., Merchant, S.S. & Prochnik, S. (2014). The Chlamydomonas genome project: a decade on. Trends in Plant Science, 19: 672–680.
- Brumfield, K.M., Moroney, J.V., Moore, T.S., Simms, T.A. & Donze, D. (2010). Functional characterization of the Chlamydomonas reinhardtii ERG3 ortholog, a gene involved in the biosynthesis of ergosterol. PLoS ONE, 5: e8659.
- Corey, E., Matsuda, S. & Bartel, B. (1993). Isolation of an Arabidopsis thaliana gene encoding cycloartenol synthase by functional expression in a yeast mutant lacking lanosterol synthase by the use of a chromatographic screen. Proceedings of the National Academy of Sciences USA, 90: 11628–11632.
- Dahl, C., Biemann, H.P. & Dahl, J. (1987). A protein kinase antigenically related to pp60v-src possibly involved in yeast cell cycle control: positive in vivo regulation by sterol. Proceedings of the National Academy of Sciences USA, 84: 4012–4016.
- Dahl, J. S. & Dahl, C. E. (1985). Stimulation of cell proliferation and polyphosphoinositide metabolism in Saccharomyces cerevisiae GL7 by ergosterol. Biochemical and Biophysical Research Communications, 133: 844–850.
- Desmond, E. & Gribaldo, S. (2009). Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biology and Evolution, 1: 364–381.
- Gealt, M., Adler, J. & Nes, W. (1981). The sterols and fatty acids from purified flagella of Chlamydomonas reinhardi. Lipids, 16: 133–136.
- Harris, E. H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego, CA. 780 pp.
- Hart, E., Hua, L., Daer, L., Wilson, W. & Pang, J. (1999). Directed evolution to investigate steric control of enzymatic oxidosqualene cyclization. Journal of the American Chemical Society, 121: 9887–9888.
- Haubrich, B.A., Collins, E.K., Howard, A.L., Wang, Q., Snell, W.J., Miller, M.B., Thomas, C.D., Pleasant, S.K. & Nes, W.D. (2015). Characterization, mutagenesis and mechanistic analysis of an ancient algal sterol C24-methyltransferase: implications for understanding sterol evolution in the green lineage. Phytochemistry, 113: 64–72.
- Heintz, R. & Benveniste, P. (1974). Plant sterol metabolism. Enzymatic cleavage of the 9beta, 19beta-cyclopropane ring of cyclopropyl sterols in bramble tissue cultures. Journal of Biological Chemistry, 249: 4267–4274.
- Herrera, J., Bartel, B., Wilson, W. & Matsuda, S. (1998). Cloning and characterization of the Arabidopsis thaliana lupeol synthase gene. Phytochemistry, 49: 1905–1911.
- Hirokawa, T., Boon-Chieng, S. & Mitaku, S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics, 14: 378–379.
- Hofmann, K. & Stoffel, W. (1993). TMbase – a database of membrane spanning proteins segments. Biological Chemistry Hoppe-Seyler, 374: 166.
- Im, C.S. & Grossman, A. (2001). Identification and regulation of high light-induced genes in Chlamydomonas reinhardtii. The Plant Journal, 30: 301–313.
- Janero, D.R. & Barrnett, R. (1982). Sterol synthesis in Chlamydomonas reinhardtii 137+ cell cycle variations. Biochimica et Biophysica Acta, 710: 242–247.
- Kawasaki, S., Ramgopal, M., Chin, J. & Bloch, K. (1985). Sterol control of the phosphatidylethanolamine-phosphatidylcholine conversion in the yeast mutant GL7. Proceedings of the National Academy of Sciences USA 82: 5715–5719.
- Leblond, J. D., Dodson, J., Khadka, M., Holder, S. & Seipelt, R. L. (2012). Sterol composition and biosynthetic genes of the recently discovered photosynthetic alveolate, Chromera velia (chromerida), a close relative of apicomplexans. Journal of Eukaryotic Microbiology, 59: 191–197.
- Lodiero, S., Schulz-Gasch, T. & Madsuda, P.T. (2005). Enzyme redesign: two mutations cooperate to convert cycloartenol synthase to an accurate lanosterol synthase. Journal of the American Chemical Society, 127: 14132–14133.
- Merchant, S.S., Prochnik, S.E., Vallon, O., Harris, E.H., Karpowicz, S.J., Witman, G. B., Terry, A., Salamov, A., Fritz-Laylin, L.K., Marechal-Drouard, L., Marshall, W.F., Qu, L.H., Nelson, D.R., Sanderfoot, A.A., Spalding, M.H., Kapitonov, V.V., Ren, Q., Ferris, P., Lindquist, E., Shapiro, H., Lucas, S.M., Grimwood, J., Schmutz, J., Cardol, P., Cerutti, H., Chanfreau, G., Chen, C.L., Cognat, V., Croft, M.T., Dent, R., Dutcher, S., Fernandez, E., Fukuzawa, H., Gonzalez-Ballester, D., Gonzalez-Halphen, D., Hallmann, A., Hanikenne, M., Hippler, M., Inwood, W., Jabbari, K., Kalanon, M., Kuras, R., Lefebvre, P.A., Lemaire, S.D., Lobanov, A.V., Lohr, M., Manuell, A., Meier, I., Mets, L., Mittag, M., Mittelmeier, T., Moroney, J.V., Moseley, J., Napoli, C., Nedelcu, A.M., Niyogi, K., Novoselov, S.V., Paulsen, I.T., Pazour, G., Purton, S., Ral, J.P., Riano-Pachon, D.M., Riekhof, W., Rymarquis, L., Schroda, M., Stern, D., Umen, J., Willows, R., Wilson, N., Zimmer, S.L., Allmer, J., Balk, J., Bisova, K., Chen, C.J., Elias, M., Gendler, K., Hauser, C., Lamb, M.R., Ledford, H., Long, J.C., Minagawa, J., Page, M. D., Pan, J., Pootakham, W., Roje, S., Rose, A., Stahlberg, E., Terauchi, A.M., Yang, P., Ball, S., Bowler, C., Dieckmann, C.L., Gladyshev, V.N., Green, P., Jorgensen, R., Mayfield, S., Mueller-Roeber, B., Rajamani, S., Sayre, R.T., Brokstein, P., Dubchak, I., Goodstein, D., Hornick, L., Huang, Y.W., Jhaveri, J., Luo, Y., Martinez, D., Ngau, W. C., Otillar, B., Poliakov, A., Porter, A., Szajkowski, L., Werner, G., Zhou, K., Grigoriev, I. V., Rokhsar, D.S. & Grossman, A.R. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science, 318: 245–250.
- Miller, M.B., Haubrich, B.A., Wang, Q., Snell, W.J. & Nes, W.D. (2012). Evolutionarily conserved Delta(25(27))-olefin ergosterol biosynthesis pathway in the alga Chlamydomonas reinhardtii. Journal of Lipid Research, 53: 1636–1645.
- Mo, C., Valachovic, M. & Bard, M. (2004). The ERG28-encoded protein, Erg28p, interacts with both the sterol C-4 demethylation enzyme complex as well as the late biosynthetic protein, the C-24 sterol methyltransferase (Erg6p). Biochimica et Biophysica Acta, 1686: 30–36.
- Nes, W.D., Norton, R.A., Crumley, F.G., Madigan, S.J. & Katz, E.R. (1990). Sterol phylogenesis and algal evolution. Proceedings of the National Academy of Sciences USA, 87: 7565–7569.
- Ohyama, K., Suzuki, M., Kikuchi, J., Saito, K. & Muranaka, T. (2009). Dual biosynthetic pathways to phytosterol via cycloartenol and lanosterol in Arabidopsis. Proceedings of the National Academy of Sciences USA, 106: 725–730.
- Rahier, A. & Karst, F. (2014). Plant cyclopropylsterol-cycloisomerase: key amino acids affecting activity and substrate specificity. Biochemical Journal, 459: 289–299.
- Rahier, A., Taton, M. & Benveniste, P. (1989). Cycloeucalenol-obtusifoliol isomerase. Structural requirements for transformation or binding of substrates and inhibitors. European Journal of Biochemistry, 181: 615–626.
- Salimova, E., Boschetti, A., Eichenberger, W. & Lutova, L. (1999). Sterol mutants of Chlamydomonas reinhardtii: characterisation of three strains deficient in C24(28) reductase. Plant Physiology and Biochemistry, 37: 241–249.
- Segura, M., Lodeiro, S., Meyer, M., Patel, A. & Matsuda, S. (2002). Directed evolution experiments reveal mutations at cycloartenol synthase residue His477 that dramatically alter catalysis. Organic Letters, 4: 4459–4462.
- Silflow, C.D., Kathir, P. & Lefebvre, P.A. (1995). Molecular mapping of genes for flagellar proteins in Chlamydomonas. Methods in Cell Biology, 47: 525–530.
- Solomon, E.P., Berg, L.R. & Martin, D.W. (2008). Biology. 8th ed. Thomson-Brooks/Cole, Belmont, CA. 1376 pp.
- Souter, M., Topping, J., Pullen, M., Friml, J., Palme, K., Hackett, R., Grierson, D. & Lindsey, K. (2002). Hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell, 14: 1017–1031.
- Storey, M.K., Byers, D.M., Cook, H.W. & Ridgway, N.D. (1998). Cholesterol regulates oxysterol binding protein (OSBP) phosphorylation and Golgi localization in Chinese hamster ovary cells: correlation with stimulation of sphingomyelin synthesis by 25-hydroxycholesterol. The Biochemical Journal, 336: 247–256.
- Subbiah, M.T. & Abplanalp, W. (2003). Ergosterol (major sterol of baker’s and brewer’s yeast extracts) inhibits the growth of human breast cancer cells in vitro and the potential role of its oxidation products. International Journal of Vitamin and Nutrition Research, 73: 19–23.
- Sueoka, N. (1960). Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proceedings of the National Academy of Sciences USA, 46: 83–91.
- Summons, R.E., Bradley, A.S., Jahnke, L.L. & Waldbauer, J.R. (2006). Steroids, triterpenoids and molecular oxygen. Philosophical Transactions of the Royal Society B Biological Sciences, 361: 951–968.
- Szolderits, G., Hermetter, A., Paltauf, F. & Daum, G. (1989). Membrane properties modulate the activity of a phosphatidylinositol transfer protein from the yeast, Saccharomyces cerevisiae. Biochimica et Biophysica Acta, 986: 301–309.
- Thoma, R., Schulz-Gasch, T., D’Arcy, B., Benz, J., Aebi, J., Dehmlow, H., Hennig, M., Stihle, M. & Ruf, A. (2004). Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature, 432: 118–122.
- Witman, G.B., Carlson, K., Berliner, J. & Rosenbaum, J.L. (1972). Chlamydomonas flagella 1. Isolation and electrophoretic analysis of microtubules, matrix, membranes and amstigonemes. Journal of Cell Biology 54: 507–539.
- Wriessnegger, T. & Pichler, H. (2013). Yeast metabolic engineering – targeting sterol metabolism and terpenoid formation. Progress in Lipid Research, 52: 277–293.