ABSTRACT
The araphid diatom genus Hyalosynedra is a very common component of marine benthic communities, occurring as an epiphyte or colonizing hard substrata, and has a worldwide distribution. The hyaline appearance of the valve and the high striation density, which is indiscernible under a light microscope, makes the identification of most species difficult and very few studies have investigated their morphology, phylogeny or ecology in detail. In an extensive study of diatom communities from the hypersaline Mar Menor coastal lagoon (SE Spain), several new taxa were found. Herein we propose Hyalosynedra lanceolata sp. nov. based on morphological observations (light and electron microscopy) and molecular data (phylogenetic and sequence divergence analyses). The species has distinctive morphological characters (radiate colony, biseriate striation, lanceolate sternum and two long laminar lobed chloroplasts) that are new for the genus and have been included in a proposal to extend the genus description. Our results also show that H. toxoneides does not belong to Hyalosynedra, but appears to be more closely related to Thalassionema and Thalassiothrix. However, further molecular and morphological research is needed to clarify its taxonomic position.
Introduction
Some studies on the phylogenetic relationships and diversity of marine diatoms have recently appeared (e.g. Li et al., Citation2015, Citation2016; Theriot et al., Citation2015). However, in general, benthic marine species are still poorly known, even in the Mediterranean Sea where several new species have been described recently (e.g. Car et al., Citation2012; Álvarez-Blanco, Citation2014; Lobban et al., Citation2015; Belando et al., Citation2016; Carballeira et al., Citation2017). In particular, very little attention has been paid to hypersaline coastal systems, which support fairly diverse and relatively unknown communities (Tomás, Citation1988; Clavero, Citation2004; Belando et al., Citation2017). The araphid diatom genus Hyalosynedra is a common component of marine benthic communities. It grows as an epiphyte or colonizes hard substrata such as rocks (e.g. Round et al., Citation1990; Álvarez-Blanco et al., Citation2014) and has a worldwide distribution. It was one of the seven genera that were derived from the division of Synedra based on morphological grounds by Williams & Round (Citation1986) and Round et al. (Citation1990): Catacombas, Hyalosynedra, Tabularia, Ctenophora, Neosynedra, Synedropsis and Ulnaria.
Until quite recently, Hyalosynedra has been regarded as monospecific with H. laevigata (Grunow) Williams & Round as the only species. However, it has been suggested that several Synedra species with ornamentation indiscernible in the light microscope (LM) probably belong to it (Williams & Round, Citation1986; Round et al., Citation1990). Currently, the genus comprises four species: three transferred from Synedra, H. laevigata, H. hyalina (Grunow) Álvarez-Blanco & Blanco, and H. toxoneides (Castracane) Coca, Chang, Wang & Wang, plus the recently described H. sublaevigata Álvarez-Blanco & Blanco. Most of these taxonomic changes have been based on morphological characters, and only Coca et al. (Citation2017) have provided molecular data, which they used to propose transferring Synedra toxoneides Castracane to Hyalosynedra.
Recent phylogenetic studies on araphid pennate diatom genera have shown that Hyalosynedra clusters in the same clade as Tabularia, Catacombas, Ulnaria, Synedropsis, Grammonema, Ctenophora and Centronella (Li et al., Citation2015, Citation2016). All have rimoportulae. However, these studies only included one Hyalosynedra strain and did not include Synedra toxoneides and Thalassionema spp., which have been found to be closely related to Hyalosynedra in previous works (Medlin et al., Citation2008; Kooistra et al., Citation2009; Lobban & Ashworth, Citation2014). Coca et al. (Citation2017) have recently proposed transferring Synedra toxoneides to the genus Hyalosynedra, but it has appeared as a sister to Thalassionema in other studies (Medlin et al., Citation2008; Kooistra et al., Citation2009). As only one or two strains of Hyalosynedra have been included in previous phylogenetic trees, and given the variability in the results reported in different studies, relationships among these groups remain uncertain. Moreover, very few of these taxa have been studied morphologically in detail. Hence, much remains to be investigated before we can unravel the relationships among these araphid genera.
During an extensive study into epiphytic diatom communities from the hypersaline Mar Menor coastal lagoon (Belando et al., Citation2016, Citation2017), we found specimens of a strain that potentially belonged to the genus Hyalosynedra, but did not match any existing Hyalosynedra species descriptions, and another taxon that seemed to be H. toxoneides, but did not exactly match the diagnosis of the genus. Based on morphological and phylogenetic research, we aimed to assess whether the strain found in the Mar Menor corresponded to a new Hyalosynedra species, and to clarify the phylogenetic relationships both within the genus and between Hyalosynedra and closely related genera.
Materials and methods
Study area, sampling and sample processing
Samples were collected from two sites in the Mar Menor lagoon (Murcia Region, SE Spain), which is one of the largest hypersaline coastal lagoons in the Mediterranean Sea. In this shallow system (mean depth of 3.5 m; 7 m maximum), temperatures range from 10 to 31°C, and salinity from 42 to 47. Despite the lagoon’s regional, national and international protection, it is subjected to several anthropogenic pressures: discharges of metals and pharmaceuticals through ephemeral streams, diffuse pollution from agriculture fields, and tourism-related impacts (e.g. Marín-Guirao et al., Citation2007; Moreno-González et al., Citation2014). The predominant macrophytes at the sampling sites were Cymodocea nodosa, which forms monospecific or mixed meadows with Caulerpa prolifera, and other macroalgae such as Cladophora dalmatica.
The samples used to conduct the morphological study of H. lanceolata and S. toxoneides were collected from C. nodosa leaves at a depth of 0.3–1 m in July 2008 and July 2012. Several extra samples were collected from the leaves of C. nodosa and C. dalmatica from January to September in different years (2009–2012). Biological samples were taken at a relatively unaltered site (El Ciervo Island, 37°39.595′N, 00°44.435′W) and a contaminated one (El Beal wadi, 37°39.975′N, 00°48.750′W). The physico-chemical background of the water at the sampling sites is summarized in Belando et al. (Citation2017). Hyalosynedra lanceolata was isolated in culture in July 2012. Cells were isolated with micropipettes under an inverted microscope and placed in f/2 culture medium (SAG Culture Collection). Cultures were maintained at 20°C in a photoperiod of 16:8 light–dark and 35 μmol photons m–2 s–1.
Morphological observations
Morphological characterizations were based on natural populations and on H. lanceolata cultures. The colonies and chloroplasts of live cells of H. lanceolata, and also the cleaned material of both taxa, were examined under a light microscope (Leica DMRB, Wetzlar, Germany). Field and cultured material was then cleaned with 33% H2O2 solution (70°C, 2 h), filtered (0.2 μm nylon membrane filter, Millipore), washed with distilled water and resuspended in 96% ethanol. For the LM observations, the cleaned material was air-dried on glass cover slips and mounted using Naphrax (refractive index 1.69, Brunel Microscopes Ltd, UK). A Leica DC-500 camera was used for the light micrographs. For scanning electron microscopy, the glass cover slips with the cleaned material of each taxon were mounted on stubs and were gold-coated. Electron micrographs were taken with a JEOL-6100 scanning electron microscope (SEM) that operated at 20 kV.
Molecular methods and phylogenetic analyses
Hyalosynedra lanceolata DNA was extracted from the pelleted culture material by the CTAB extraction method described by Doyle & Doyle (Citation1987), and was stored frozen at −20°C until the PCR reaction was carried out. For the phylogenetic analysis, the nuclear-encoded small subunit ribosomal DNA (SSU rDNA) and the large subunit of RUBISCO (rbcL) of the chloroplast were amplified. The primers used for amplification and sequencing were selected from Alverson et al. (Citation2007): SSU (primers SSU1/SSU568–, SSU301+/SSU1147–, SSU1004+/ITS1DR); and rbcL (rbcL66+/rbcL 1255–). Amplification reactions were conducted in 50 µl volumes, which contained approximately 20 ng of genomic DNA, 0.2 mM of each dNTP, 2.5 mM of MgCl2, 2 units of Taq polymerase (Biotools, Madrid, Spain), the buffer provided by the manufacturer, the combinations of primers at a final concentration of 0.4 mM and ddH2O to the final volume. Polymerase chain reactions (PCR) were performed in a thermocycler (Eppendorf mastercycler gradient, Hamburg, Germany) according to the following programme, as outlined in Alverson et al. (Citation2007): 94°C for 3:30 min, 35 cycles of 94°C for 50 s, 53C for 50 s and 1 min at 72°C. A final 8-minute step at 72°C was included to terminate the amplification products. Finally, 2 ml of the amplification products were visualized on 1.5% agarose gel and successful amplifications were cleaned with the GenElute PCR clean-up kit (Sigma–Aldrich). For sequencing, the purified PCR products were reacted with the BigDye terminator cycle sequencing ready reaction (Applied Biosystems, Foster City, California) using the amplification primers.
Maximum likelihood (ML) and Bayesian inference (BI) analyses were performed with 61 SSU rDNA and 53 rbcL sequences, including those taxa found to be closely related to Hyalosynedra in previous phylogenetic studies, and also those selected by BLAST searches. Two Bolidomonas pacifica strains were used as the outgroup in accordance with previous araphid diatoms phylogenetic trees (e.g. Li et al., Citation2015). The GenBank accession numbers of all the taxa are listed in Supplementary table S1.
Sequences were checked for inaccurate base calling using Chromas Lite, v. 2.01 (Technelysium Pty. Ltd). The consensus sequences of the rbcL gene fragments were aligned with CLUSTALX (Thompson et al., Citation1997), and the SSU rDNA sequence alignments were performed with the SSU-Align package (Nawrocki, Citation2009; Nawrocki et al., Citation2009). SSU-Align performs secondary structure alignments based on the covariance model (CM) (Cannone et al., Citation2002; Alverson et al., Citation2007; Theriot et al., Citation2009). Sequences were aligned to the consensus CM of Eukarya included in the SSU-Align package. The alignment columns with a low posterior probability (PP), which generally corresponded to large loops for which positional homology and co-varying nucleotides were not easy to assign, were removed by the SSU-Mask routine from the SSU-Align package. Bioedit (Hall, Citation1999) was employed to make minor manual adjustments of alignments. Bayesian analyses were performed by MrBayes, v. 3.1 (Ronquist & Huelsenbeck, Citation2003). Two simultaneous runs were initiated by starting from random trees. To ensure that the two runs converged onto a stationary distribution, analyses were run until the average standard deviation of the split frequencies was 0.01. Convergence was evaluated using the potential scale reduction factor (PSRF), and 1 000 000 generations were run by sampling every 100th generation at the following settings: Nst = 6, rates = gamma. Burnin (the number of starting generations ruled out from further analyses) was set at 200 000 generations after visually inspecting the likelihood values in Excel. A 50% majority rule consensus tree was constructed using the ‘sumt’ command of MrBayes.
ML phylogenetic trees were conducted with 1000 bootstrap replicates by the rapid Bootstrap analysis in v.1.5b1 of RAxMLGUI (Silvestro & Michalak, Citation2012). The best-scoring ML trees were chosen as the final trees and bootstrap values were added to nodes. Both analyses were performed with GTR + G + I (general time reversible model of DNA substitution), because it was selected as the optimal evolution model for the two genes by JModeltest (Guindon & Gascuel, Citation2003; Darriba et al., Citation2012). Trees were edited with Figtree, v.1.3.
To investigate four monophyly hypotheses, the following groups were constrained in different trees: strains of Synedra toxoneides (=Hyalosynedra toxoneides), S. toxoneides+Hyalosynedra, S. toxoneides+Thalassionema and S. toxoneides+Thalasiothrix. RAxMLGUI was used to generate ML trees from the unconstrained data set and constrained trees, and to also calculate the per-site log likelihood values of the resulting best trees. The AU test in the Consel programme (Shimodaira & Hasegawa, Citation2001) was implemented to test the four monophyly hypotheses. In the AU test, the analysed tree topology was compared with a set of trees generated by a multiscale bootstrap technique of per site log likelihoods. If the tested tree topology falls outside the 95% confidence interval of the generated trees, the hypothesis can be rejected (if P<0.05).
The uncorrected p-distances of the SSU rDNA data set were also used to study genetic divergences among species, which were calculated with MEGA 7 (Kumar et al., Citation2016).
Results
Species description
Hyalosynedra lanceolata Belando, Jiménez & Aboal sp. nov. (–)
Description: Cells forming radiate colonies attached to substrata by a mucilage pad. Two long lobed laminar chloroplasts cover the whole length of the cell. Valves linear to linear-lanceolate with rounded poles and a lanceolate sternum. Frustules 20.7–266 µm long, 1.3–3.7 (4.0–5.5) µm width at the centre, and 1.0–2.7 in the apices. Striae biseriate, parallel, 47–52 in 10 µm. Striae uninterrupted at the junction of the valve face and mantle margin. Striae opening externally by two rows of alternate pores and internally by a rounded foramen. Valve ends with plain areas and lateral poroids where the striation ceases. One rimoportula at each pole of the valve, rounded externally and like a parrot’s beak internally. Apical pore field an ocellulimbus composed of three rows of poroids, which are larger than the pores of the striae; a few small spines overhang it. Valves smooth internally, with slightly depressed apices and pores at both sides of the rimoportula. Cingulum composed of two copulae and a pleura with a single row of poroids.
Figs 1–11. Light micrographs of Hyalosynedra lanceolata sp. nov. Figs 1–6. Colonies and chloroplasts in live cells. Figs 7–11. Cleaned specimens showing the lanceolate sternum and the wide cell size range. Scale: Figs 1–2 = 40 µm, Figs 3–6 = 20 µm, Figs 7–11 = 5 µm.
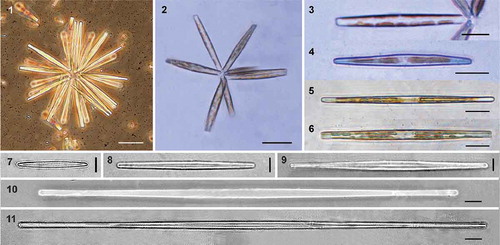
Figs 12–17. Scanning electron micrographs of Hyalosynedra lanceolata sp. nov. Fig. 12. Giant cells (200–300 µm long) with almost linear valves. Fig. 13. Internal view of valves, less than 100 µm long and with a lineal-lanceolate shape. Fig. 14. Colony of large cells and very small others showing the wide cell size range in culture. Figs 15–17. Cleaned specimens showing the lanceolate sternum. Scale = 5 µm, except Fig. 14 = 50 µm.
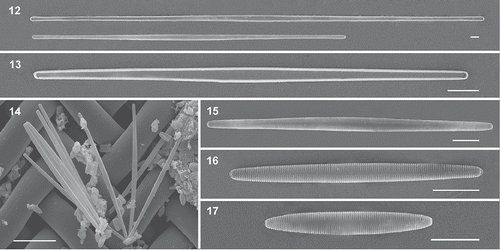
Figs 18–25. Scanning electron micrographs of Hyalosynedra lanceolata sp. nov. Fig. 18. Detail of the biseriate striation with alternate areolae and the lanceolate sternum at the centre of valves (up to one-third total width). Fig. 19. Apical part of cells showing areolae and a thin sternum. Fig. 20. Detail of the external openings of rimoportula and the rows of pores in ocellolimbus. Figs 21–22. Internal view of the smooth valves showing small rounded pores on both sides and the aperture of rimoportulae. Figs 23–24. Detail of valvocopula, copula and pleura with a row of pores (one apex in each image). Fig. 25. Cingular view of cells with the three girdle bands. Scale: Figs 18–24 = 1 µm, Fig. 25 = 10 µm.
Holotype: Permanent slide of natural material deposited at the Herbarium Universitatis Murcicae-Bacillariophyta, Spain, MUB-ALGAS (Diatomeas) 2785. Coll. Belando, M.D., in the Mar Menor lagoon (SE Spain), July 2012.
Genbank Sequences: KY679465 (SSU rDNA), KY679464 (rbcL).
Type Strain: HyaLan-MMen. Cultures are available upon request from the Laboratory of Algology of Murcia University.
Type Locality: El Ciervo Island (37°39.595′N, 00°44.435′W) from the Mar Menor lagoon (SE Spain), at a depth of 0.4 m.
Ecology: Epiphytic on Cymodocea nodosa and macroalgae, such as Cladophora dalmatica in hypersaline coastal waters. It is abundant in summer (25–28°C) in locations far from the influence of metal and nutrient contamination. In a more general study, the physico-chemical characteristics of the type locality were characterized by: salinity 44.2, pH = 8.1, dissolved O2 = 8.22 mg l–1, NO3− and PO43– = 2.9±0.39 and <1.05 µmol l–1, respectively, and Zn and Pb = < 0.45 and 3.01±2.80 µg l–1, respectively. For further details, see Belando et al. (Citation2017).
Etymology: From Latin lanceolatus, referring to the lanceolate form of the sternum.
Extended description: The two long lobed chloroplasts lie against the valves along the whole length of the cell (–), though they may split up in unhealthy material, giving the impression of numerous discoid chromatophores (–). Under LM, valves appear hyaline, except for the lanceolate sternum that appears as a white line which becomes fainter in apices (–). The wide range of cell sizes found in the field (20.7–266 µm) is persistent in cultures, but cultured cells are wider in the centre (4.0–5.5 µm). Large cells tend to be linear, while smaller ones are linear-lanceolate (–). Cells tend to decrease to the minimum size when they are in cultures for a long time, according to the asexual reproduction process. Recovery of the largest size () occurred as a fast and synchronized process in which most cultured cells reached the largest size for one or two days and the small ones died. However, the auxospores were not directly observed.
The lanceolate sternum can reach up to one-third of the valve width in the centre (). The striae are biseriate externally (, ), containing tiny areolae arranged alternately, which appear to be elongated and slightly oblique to the transapical axis. Internally the biseriate arrangement is invisible since each stria is alveolate, opening internally by a single foramen close to the valve face–mantle junction (, ; compare , ). The rounded external apertures of rimoportulae are located at the end of the striation (, ), and the internal opening is in a depressed area with a beak-like appearance in most cases (), but sometimes with two almost parallel lips (). The ocellulimbus is formed by three rows of pores, larger than pores of areolae, and three or four small spines overhang it at each apex (). The cingulum consists of a series of alternating, incomplete and ligulate bands (–). It is formed by a plain valvocopula, a copula (similar in structure but shorter), and by a much narrower pleura with a single row of poroids (–).
Evaluation and comparison with other Hyalosynedra species: The morphological and diagnostic characters of the studied material and other Hyalosynedra species are summarized in . Hyalosynedra lanceolata morphologically matches the description of Hyalosynedra, except for the lanceolate sternum. The valve shape of the small specimens is similar to H. laevigata, but the latter is broader, and has a less dense striation. The striation has been described as uniseriate where each areola is occluded by a small closing plate with struts orientated on the transapical plane. Under LM, small specimens may also be confused with H. sublaevigata (26.5–34.1 μm in length, 2.3–3.0 μm wide), but H. sublaevigata has no biseriate striation, no lanceolate sternum, nor non-occluded poroids on the valve apex present in H. lanceolata. The valve of H. hyalina is strongly lanceolate, with capitately constricted ends.
Table 1. Diagnostic characters of Hyalosynedra lanceolata compared with other species of the genus.
Synedra toxoneides Castracane
Synonym: Hyalosynedra toxoneides (Castracane) Coca, Chang, Wang & Wang (Citation2017).
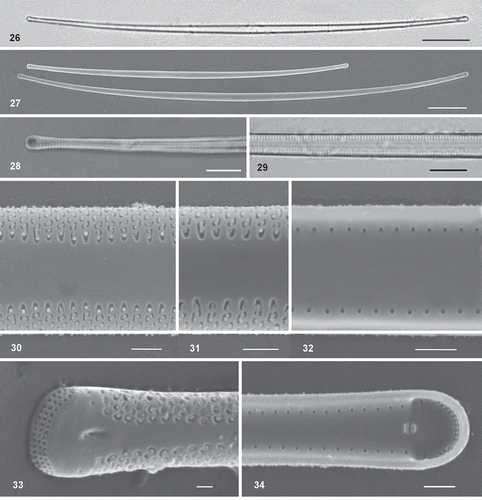
Description: Solitary cells or forming radiate colonies. Cells long, narrow, slightly curved towards the ends in the valve view (, ). Valves linear-lanceolate, slightly inflated both at the centre and close to the slightly capitate poles (–). Under LM sternum appears wide, delimited by the visible striation (). Cells are 221–310 µm long, 2.3–4.0 (4.2) µm wide in the centre and 2.3–2.9 µm at the poles, with 23–25 transapical striae in 10 µm. Externally, the striae are biseriate and parallel, with elongated and curved areolae; the terminal areola (closest to the sternum) is crescent-shaped (, , ). The biseriate striae are not interrupted at the junction of the valve face and mantle. Internally the striae can be seen to be alveolate since the valve appears structureless, except for the small rounded foramina located between the valve face and mantle (, ). The sternum is very wide (one-third, or more, of the valve width in the centre), tapering slightly towards the apices (, ). One rimoportula is present at each pole of the valve, opening to the outside by a small pore located in a depressed area () and internally between two parallel small lips (). One to three single pores is/are present on each side of the unornamented extremities of valves, opening also inside. Apical pore fields are present, consisting of a rectangular grid of pores organized in rows and columns that occupy the whole margin at the ends of the valve (); the pore fields are not sunken into the valve, in contrast to H. lanceolata (compare and ).
Figs 26–34. Light and scanning electron micrographs of Synedra toxoneides from the Mar Menor lagoon. Fig. 26. Hyaline thin and curved cells under LM. Fig. 27. SEM micrographs of large curved cell with capitate poles. Figs 28–29. Detail of the visible striation under LM with immersion oil. Figs 30–31. External view of the wide sternum, striae and areolae. Fig. 32. Internal view of valves with rounded pores. Fig. 33. External view of rimoportula in a depressed area and a row of pores occupying the whole apex. Fig. 34. Internal opening of rimoportulae showing two parallel lips. Scale: Figs 26–27 = 20 µm, Figs 28–29 = 5 µm, Figs 30–34 = 1 µm.
Ecology: Synedra toxoneides occurred as an epiphyte on C. nodosa and macroalgae, with low abundance in summer at the hypersaline Mar Menor lagoon, 0.4 m depth.
Comparison with other taxa and remarks on its inclusion in the genus Hyalosynedra: Our material morphologically resembles strain S. toxoneides WK57 included in other phylogenetic analysis (personal communication, W.H.C.F. Kooistra), but it does not match the description of Hyalosynedra. It has no sunken apical pore plate (i.e. the apical pore field is not of the ocellulimbus type), nor are spines present at the apices, as described for the genus. Furthermore, the valves of Hyalosynedra are straight and linear-lanceolate, and the striae are composed of rows of small pores. The areolae of S. toxoneides are similar to Tabularia affinis (Kützing) Snoeijs, but Tabularia has the ocellulimbus type of an apical pore field, and does not have chambered valves. Because of the long thin shape of S. toxoneides, it could be confused with specimens of the Thalassionemataceae (Thalassionema, Thalassiothrix and Trichotoxon), but these genera have no apical pore field. Moreover, Thalassionema has areolae with a bar across them externally, and spines in the apices, and Thalassiothix possesses heteropolar valves and spines all along the valve. The curved shape of S. toxoneides coincides with Trichotoxon, but the latter is longer (800–3500 µm) and broader (5–8 µm, measured in the middle section), only has a small pore in the apices (which is the rimoportule aperture), has areolae occluded by cribra and the striae are located principally on the valve face, only extending slightly onto the mantle. Internally the valve surface is slightly ribbed, especially in the centre.
Molecular phylogeny
A concatenated alignment of the nuclear SSU and plastid rbcL regions from 61 taxa yielded 3069 nucleotide sites (1593 SSU rDNA after masking with SSUalign, 1476 rbcL), of which 2196 were constant, 197 variable, but parsimony-uninformative, and 677 were parsimony-informative. Aligned sequences have been deposited in TreeBase (study accession number 22090, www.treebase.org/treebase/). Both the ML and Bayesian inference searches resulted in trees with a similar topology. Therefore, the Bayesian posterior probability/ML bootstrap values (Bpp/MLb) are provided in the same tree for all the analyses (). The outgroup taxa have been pruned away from the unconstrained tree, but are shown in the Supplementary figs S1, S2.
3.3. Maximum likelihood tree inferred from a concatenated alignment of SSU rRNA and rbcL markers of 61 araphid diatoms. The numbers on the nodes are Bayesian posterior probability/maximum likelihood bootstraps (Bpp/MLb). The newly described species is shown in bold. Any support values lower than 50% were omitted.
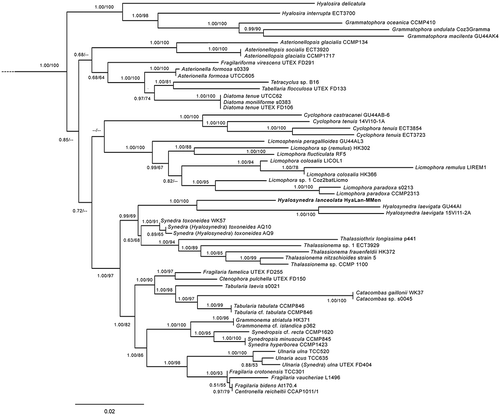
The phylogenetic tree () indicated that the Hyalosynedra strains were clustered in a strongly supported clade in the BI analysis, and were only moderately supported in the ML analysis. This group was sister to a weakly supported clade comprising S. toxoneides, Thalassionema and Thalassiothrix. In turn, these assemblages formed a well-supported monophyletic group that was sister to a well-supported clade which contained the seven genera Tabularia, Ctenophora, Catacombas, Centronella, Ulnaria, Grammonema and Synedropsis, and some species of Fragilaria.
The most likely tree topologies, inferred from the two gene data sets by RAxML and used in the AU test, obtained similar log likelihood values for the unconstrained tree and trees in which the strains of S. toxoneides were constrained as monophyletic with Thalasionema or Thalasiothrix (). The AU test results revealed that the unconstrained tree was the best (P>0.05), and did not allow the hypothesis that S. toxoneides forms a monophyletic group with Thalassionema or Thalassiothrix to be rejected (P>0.05). The monophyly of S. toxoneides or with Hyalosynedra was rejected (P<0.05).
Table 2. The AU test for the topologically unconstrained () and the four constrained trees in the monophyly analysis.
The divergence analysis results (Supplementary table S2) showed that the lowest percentage of sequence divergence corresponded to the group of S. toxoneides and Thalassionema (0.43–0.64%). The p-distance values among the strains of S. toxoneides and Hyalosynedra fell within the 0.86–1.72% range, and within 1.72–1.93% for S. toxoneides and Thalassiothrix. The highest divergence percentage was found among the strains of Hyalosynedra with Thalasiothrix (2.58–3.22%).
Discussion
In this study, we describe the new species Hyalosynedra lanceolata, based on the general morphological features shared by Hyalosynedra, e.g. valve shape, apical field of pores ocellulimbus-type and striae composed of rows of small pores, and molecular data that clearly separate it from two H. laevigata strains with which it forms a monophyletic clade. This is the first study to contribute new distinctive characters of live cells of this genus, and the description of Hyalosynedra needs to be extended to include radiate colonies, biseriate striation, lanceolate sternum, and two long laminar lobed plastids found in H. lanceolata. The original diagnosis of the genus was based on the uniseriate striation of the type species H. laevigata, although Round et al. (Citation1990) had already mentioned that areolae might be smaller and form biseriate rows in the mantle in some (undescribed) Hyalosynedra species, which is in agreement with our proposal. Moreover, Clavero (Citation2004) also reported a Hyalosynedra sp. with a biseriate striation and suggested the need to modify the diagnosis of the genus. At the end of the Discussion, we therefore present an amended description of Hyalosynedra.
Based on the morphological revision of both the described taxa and some strains included in the phylogenetic tree, we highlight that further molecular and SEM studies could provide the genus with wider interspecific variability. We observed that H. lanceolata sp. nov. coincided in various morphological features with Hyalosynedra sp., described by Clavero (Citation2004) from hypersaline waters in Baja California, South. It had a wide range of length (22.2–116.1 µm) and width (1.9–4.0 µm), and 45(48)–53 striae in 10 µm, but this author did not show a lanceolate form of the sternum. Therefore, we cannot be absolutely sure that these specimens correspond to H. lanceolata. Clavero (Citation2004) also mentioned the possible confusion of this Hyalosynedra sp. with H. laevigata var. angustata Grunow because, according to Hustedt (Citation1959), the differential character of the variety was a smaller valve (3 µm wide). The structureless hyaline characters of the H. lanceolata valves seen under LM could easily be confused with the aforementioned variety, but no detailed study of its morphology has been published to date as far as we know. Notably, Tomás (Citation1988) identified var. angustata with a similar size range to our material (10.2–204.5 µm long, 2.0–3.1 µm wide) and with an indiscernible striation under LM (more than 40 striae in 10 µm). It is not easy to differentiate the sternum shape (lanceolate or not) in the LM images shown by Tomás due to narrowness of cells, but these individuals probably belonged to H. lanceolata.
Overall our findings also indicate that Hyalosynedra sp. GU44AI should be named H. laevigata. The phylogenetic trees grouped it with H. laevigata (strain 15VI11-2A, Bpp/MLb: 1.00/100, 0.21% divergence), and it matched morphologically the description of this species (e.g. 37 striae/10 µm), see http://www.protistcentral.org/Taxa/get/taxa_id/586040 (Jordan et al., Citation2009–2017). Hyalosynedra cf. laevigata (WK52), which was one of the few strains used in previous phylogenetic studies, grouped with the monophyletic clade of H. lanceolata and H. laevigata in our preliminary trees. However, we did not include this sequence in our final trees as only SSU rDNA data were available, and its inclusion diminished the resolving power of the analysis. Further research into this strain is needed.
As far as we know, the present study (with material from the Mar Menor lagoon) provides the first detailed description of S. toxoneides, supported by the LM and SEM micrographs. Previously, the ultrastructure of this taxon has only been studied in detail for two deformed strains (AQ9, AQ10), from which the new H. toxoneides combination has been proposed (Coca et al., Citation2017). The morphological characteristics of our specimens agree with the description of S. toxoneides in Hustedt (Citation1959), and also with the material of the S. toxoneides strain WK57 included in some phylogenetic trees (provided by W.H.C.F. Kooistra). The morphological characters do not match the general features of Hyalosynedra; for example, the large, thin curved valve shape, the rectangular grid of pores (not sunken on the end of valves) and the coarse striation (visible under LM). Since the monophyly of S. toxoneides with Hyalosynedra was rejected, overall our findings suggest that S. toxoneides does not belong to Hyalosynedra. Moreover, the phylogenetic tree showed a closer relationship with Thalassionema, as Kooistra et al. (Citation2009) and Medlin et al. (Citation2008) have suggested, and with Thalassiothrix.
Synedra toxoneides does not match the Thalassionemataceae (Thalassionema, Thalassiothrix and Trichotoxon) either in habitat (it is epiphytic, while they are planktonic) or morphology. The most notable morphological difference is that Thalassionemataceae have no apical pore field; the only special pores at the poles are the rimoportulae (one at each apex). Furthermore, Thalassionema has areolae with an external bar crossing them and spines at the apices, and Thalassiothix possesses heteropolar valves and spines along the entire valve – these characters are not present in S. toxoneides. Synedra toxoneides shares some features of the genus Trichotoxon, e.g. long and thin valves, curved and inflated at the centre and apices, but the valves of S. toxoneides are much smaller, 221–310 µm long vs. 800–3500 µm long (Reid & Round, Citation1987; Round et al., Citation1990). In addition, the structure of the alveolate striae differs, as Trichotoxon striae open to the outside by elliptical to quadrate areolae with external cribra, which are restricted to the valve face (figs 6–8 in Reid & Round, Citation1987; fig. d in Round et al., Citation1990), whereas the biseriate rows of areolae in S. toxoneides extend onto the mantle. Externally, the valves of Trichotoxon sometimes have a few areolae scattered on the valve face, and internally they are slightly ribbed, especially in the central region, whilst they are not in S. toxoneides. The numerous small plastids of Trichotoxon contrast with the large plate of S. toxoneides (Coca et al., Citation2017).
The phylogenetic relationships between S. toxoneides and Thalassionema or Thalassiothrix remain unclear. Lack of resolution in the presented phylogenetic analyses probably derived from data being scarce (e.g. the strains of S. toxoneides only had SSU rDNA sequences). Hence further sequences of these strains and related genera, such as Trichotoxon, could help to elucidate the phylogenetic position of S. toxoneides.
Over the several years duration of this study, H. lanceolata was observed as being epiphytic on C. nodosa, at least from January to September. In summer, H. lanceolata is one of the dominant species in benthic diatom communities from relatively unaltered sites at the Mar Menor lagoon, while H. laevigata occurred exclusively at the historically contaminated site (Belando et al., Citation2017). Synedra toxoneides was not abundant in any sample. The present record is the first of this species at the hypersaline Mar Menor lagoon (Murcia). Tomás (Citation1988) had also reported finding it at another Spanish hypersaline lagoon (Almería, SE Spain), and it appears to be widely distributed in the Mediterranean Sea (Hustedt, Citation1959; Kooistra et al., Citation2009).
New extended description of the genus Hyalosynedra D.M. Williams & F.E. Round (Citation1986)
Radiate colonies, two long chloroplasts lying against the valves. Valves araphid, linear or linear-lanceolate, with rounded or capitate poles. Living epiphyte or colonizing hard substrata. Sternum narrow connecting both poles or wider lanceolate tapering gradually to apices. Striae very dense with small areolae extending from the mantle to the sternum, parallel, alveolate, opening to the outside by one (sometimes can appear paired due to a bar across pores) or two rows of small pores, and internally by a single round foramen at the angle between the valve face and mantle.
Valves each end with plain areas, and rimoportulae opening externally by a relatively large aperture, round or transversely elongate and close to the end of the areolae rows. Internally, it may appear single to lips or forming a structure like a parrot’s beak. An apical pore plate is present, type ocellulimbus, sunken and pores are larger than the areolae. A row of spines may overhang the ocellulimbus. Valves tend to be thick due to the chambering wall. Girdle consisting or three bands: a non-areolae valvocopula, copula and single pleura, which can have a row of pores.
Type species: Synedra laevigata Grunow (Citation1877) p.166, pl. 193, fig. 3.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at https://doi.org/10.1080/09670262.2018.1426787
Supplementary table. S1. List of the taxa and the GenBank accession numbers for the SSU rDNA and rbcL sequences used in the phylogenetic analyses.
Supplementary table. S2. Estimates of the evolutionary divergence of SSU rDNA gene sequences.
Supplementary fig. 1. Maximum likelihood tree inferred from a concatenated alignment of SSU rRNA and rbcL markers of 61 araphid diatoms with constrained S. toxoneides and Thalassionema as the monophyletic clade.
Supplementary fig. 2. Maximum likelihood tree inferred from a concatenated alignment of SSU rRNA and rbcL markers of 61 araphid diatoms with constrained S. toxoneides and Thalassiothrix as the monophyletic clade.
Supplementary_material.zip
Download Zip (490.9 KB)Acknowledgements
We would like to thank Helen Warburton for revising the English. We also thank Wiebe H.C.F. Kooistra for providing us with images of some Hyalosynedra strains (WK57 and WK52), which have been used for the morphological revision. Thanks also go to the anonymous reviewers for their helpful and valuable comments.
Additional information
Notes on contributors
Ma Dolores Belando
MD. Belando and M. Aboal original concept, drafting and editing manuscript; MD Belando sampling, culturing and electron microscopy; JFJiménez and MD Belando DNA extraction and molecular analysis; A. Marín sampling and finantial support.
References
- Álvarez-Blanco, I. & Blanco, S. (2014). Benthic diatoms from Mediterranean coasts. In Bibliotheca Diatomologica (Kociolek, P., editor), vol. 60. J. Cramer, Gebrüder Borntraeger Verlagsbuchhandlung, Stuttgart, Germany.
- Alverson, A.J., Jansen, R.K. & Theriot, E.C. (2007). Bridging the Rubicon: Phylogenetic analysis reveals repeated colonizations of marine and fresh waters by thalassiosiroid diatoms. Molecular Phylogenetics and Evolution, 45: 193–210.
- Belando, M.D., Aboal, M., Jiménez, J.F. & Marín, A. (2016). Licmophora colosalis sp. nov. (Licmophoraceae, Bacillariophyta), a large epiphytic diatom from coastal waters. Phycologia, 55: 393–402.
- Belando, M.D., Marín, A., Aboal, M., García-Fernández, A.J. & Marín-Guirao, L. (2017). Combined in situ effects of metals and nutrients on marine biofilms: shifts in the diatom assemblage structure and biological traits. Science of the Total Environment, 574: 381–389.
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y.S., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., Pande, N., Shang, Z.D., Yu, N. & Gutell, R.R. (2002). The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics, 3: 2.
- Car, A., Witkowski, A., Dobosz, S., Burfeind, D.D., Meinesz, A., Jasprica, N., Ruppel, M., Kurzydbowski, K.J. & Plocinski, T. (2012). Description of a new marine diatom, Cocconeis caulerpacola sp. nov. (Bacillariophyceae), epiphytic on invasive Caulerpa species. European Journal of Phycology, 47: 433–448.
- Carballeira, R., Trobajo, R., Leira, M., Benito, X., Sato, S. & Mann, D.G. (2017). A combined morphological and molecular approach to Nitzschia varelae sp. nov., with discussion of symmetry in Bacillariaceae. European Journal of Phycology, 53: 1–18.
- Clavero, E. (2004). Diatomees d’ambients hipersalins costaners. Taxonomia, distribució i emprentes en el registre sedimentari. PhD Thesis, University of Barcelona, Spain.
- Coca, J.J.P., Chang, Y.T., Lai, S.Y., Wang, W.L. & Wang, M.Y. (2017). Hyalosynedra toxoneides, in artificial culture conditions induce irreversible deformations. Nova Hedwigia, 103: 1–21.
- Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9: 772.
- Doyle, J.J. & Doyle, J.L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19: 11–15.
- Grunow, A. (1877). New diatoms from Honduras. The Monthly Microscopical Journal, 18: 165–186.
- Guindon, S. & Gascuel, O. (2003). A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology, 52: 696–704.
- Hall, T.A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95–98.
- Hustedt, F. (1959). Bemerkungen über die Diatomeenflora des Neusiedler Sees und des Salzlackengebietes. In Landschaft Neusiedler See (23) (Burgenland, W.A., editor), 180–240.
- Jordan, R.W., Lobban, C.S. & Theriot, E.C. (2009–2017). Western Pacific Diatoms Project. ProtistCentral. http://www.protistcentral.org. Revised in 2017.
- Kooistra, W., Forlani, G. & De Stefano, M. (2009). Adaptations of araphid pennate diatoms to a planktonic existence. Marine Ecology – An Evolutionary Perspective, 30: 1–15.
- Kumar, S., Stecher, G. & Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33: 1870–1874.
- Li, C.L., Ashworth, M.P., Witkowski, A., Dąbek, P., Medlin, L.K., Kooistra, W.H.C.F., Sato, S., Zgłobicka, I., Kurzydłowski, K.J., Theriot, E.C., Sabir, J.S.M., Khiyami, M.A., Mutwakil, M.H.Z., Sabir, M.J., Alharbi, N.S., Hajarah, N.H., Qing, S. & Jansen, R.K. (2015). New insights into Plagiogrammaceae (Bacillariophyta) based on multigene phylogenies and morphological characteristics with the description of a new genus and three new species. PLoS ONE, 10: e0139300.
- Li, C.L., Ashworth, M.P., Witkowski, A., Lobban, C.S., Zgłobicka, I., Kurzydłowski, K.J. & Qin, S. (2016). Ultrastructural and molecular characterization of diversity among small araphid diatoms all lacking rimoportulae. I. Five new genera, eight new species. Journal of Phycology, 52: 1018–1036.
- Lobban, C.S. & Ashworth, M.P. (2014). Hanicella moenia, gen. et sp. nov., a ribbon forming diatom (Bacillariophyta) with complex girdle bands, compared to Microtabella interrupta and Rhabdonema cf. adriaticum: implications for Striatellales, Rhabdonematales, and Grammatophoraceae, fam. nov. Journal of Phycology, 50: 860–884.
- Lobban, C.S., Ashworth, M.P., Car, A., Herwig, W. & Ulanova, A. (2015). Licmosphenia revisited: transfer to Licmophora, redescription of L. clevei Mereschkowsky and descriptions of three new species. Diatom Research, 30: 227–236.
- Marín-Guirao, L., Lloret, J., Marín, A., García, G. & García Fernández, A.J. (2007). Pulse-discharges of mining wastes into a coastal lagoon: water chemistry and toxicity. Chemistry and Ecology, 23: 217–231.
- Medlin, L., Jung, I., Bahulikar, R., Mendgen, K., Kroth, P., & Kooistra, W.H.C.F. (2008). Evolution of the Diatoms. VI. Assessment of the new genera in the araphids using molecular data. Nova Hedwigia, 133: 81–100.
- Moreno-González, R., Rodríguez-Mozaz, S., Gros, M., Pérez-Cánovas, E., Barceló, D. & León, V.M. (2014). Input of pharmaceuticals through coastal surface watercourses into a Mediterranean lagoon (Mar Menor, SE Spain): sources and seasonal variations. Science of the Total Environment, 490: 59–72.
- Nawrocki, E.P. (2009). Structural RNA homology search and alignment using covariance models, Washington University, St. Louis, MO.
- Nawrocki, E.P., Kolbe, D.L. & Eddy, S.R. (2009). Infernal 1.0: inference of RNA alignments. Bioinformatics, 25: 1335–1337.
- Reid, F.M. & Round, F.E. (1987). The Antarctic diatom Synedra reinboldii: taxonomy, ecology and transference to a new genus, Trichotoxon. Diatom Research, 2: 219–227.
- Ronquist, F. & Huelsenbeck, J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572–1574.
- Round, F.E., Crawford, R.M. & Mann, D.G. (1990). The Diatoms. Biology and Morphology of the Genera. Cambridge University Press, Cambridge.
- Shimodaira, H. & Hasegawa, M. (2001). CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics, 17: 1246–1247.
- Silvestro, D. & Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution, 12: 335–337.
- Theriot, E.C., Cannone, J.J., Gutell, R.R. & Alverson, A.J. (2009). The limits of nuclear-encoded SSU rDNA for resolving the diatom phylogeny. European Journal of Phycology, 44: 277–290.
- Theriot, E.C., Ashworth, M.P., Nakov, T., Ruck, E. & Jansen, R.K. (2015). Dissecting signal and noise in diatom chloroplast protein encoding genes with phylogenetic information profiling. Molecular Phylogenetics and Evolution, 89: 28–36.
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. & Higgins, D.G. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 24: 4876–4882.
- Tomás, X. (1988). Diatomeas de las aguas epicontinentales saladas del litoral mediterráneo de la Península Ibérica. University of Barcelona, Spain.
- Williams, D.M. & Round, F.E. (1986). Revision of the genus Synedra Ehrenb. Diatom Research, 1: 313–339.
