ABSTRACT
DNA barcoding analysis, using tufA, revealed considerable differences between the expected and observed species inventory of Ulva sensu lato in the Baltic and North Sea areas of the German state of Schleswig-Holstein. Of 20 observed genetic entities, at least four (U. australis, U. californica, U. gigantea and Umbraulva dangeardii) had been introduced recently, whereas three others (one Ulva sp. and two Blidingia spp.) could not be identified at the species level and could also represent recently introduced species. In addition, the observed distributions of Kornmannia leptoderma and U. rigida were much more extensive than indicated by historical records, whereas Blidingia minima and Gayralia oxysperma were absent or much less common than expected. Barcoding analysis also revealed that both U. tenera (type material) and U. pseudocurvata (historical vouchers) from Helgoland, an off-shore island in the North Sea, actually belong to U. lactuca, a species that appears to be restricted to this island. Furthermore, past morphological descriptions of U. intestinalis and U. compressa have apparently been too restrictive and have been responsible for numerous misidentifications. The same is true for U. linza, which, in northern Germany, clusters into two genetically closely related but morphologically indistinguishable entities. One of these entities is present on Helgoland, while the second is present on North Sea and Baltic Sea mainland coasts.
Introduction
Macroalgae of the orders Ulvales and Ulotrichales are ubiquitous inhabitants of fully marine and brackish coastal waters. Several macroalgal species have recently increased, owing to opportunistic lifestyles and the capacity to benefit from eutrophication and other anthropogenic impacts, and the ability to accurately identify such taxa has become much more important (Charlier et al., Citation2007, Citation2008; Smetacek & Zingone, Citation2013). However, the identification of certain macroalgae, such as the genus Ulva, is notoriously difficult (Koeman & Van den Hoek, Citation1981, Citation1982a, Citation1982b, Citation1984; Hoeksema & Van den Hoek, Citation1983; Brodie et al., Citation2007), with the morphological instability of specific Ulva species being attributed to variation in salinity (Reed & Russell, Citation1978; Steinhagen et al., Citation2018b), nutrient concentrations (Blomster et al., Citation2002; Steinhagen et al., Citation2018b) and bacterial associations (Spoerner et al., Citation2012; Wichard, Citation2015), as well as to an elevated tendency for mutagenesis (Wichard, Citation2015). As a consequence, morphological plasticity (i.e. multiple morphotypes within species) or cryptic speciation may hinder identification and lead to taxonomic confusion. Such identification problems have been confirmed by DNA barcoding studies (e.g. Blomster et al., Citation1998, Citation2002; Tan et al., Citation1999; Hayden & Waaland, Citation2002; Hayden et al., Citation2003; Shimada et al., Citation2003; Brodie et al., Citation2007; Heesch et al., Citation2009; Wolf et al., Citation2012; Kirkendale et al., Citation2013), which have reported that the historical separation of Enteromorpha (for tubular ‘species’) and Ulva (for sheet-like taxa) is artificial and does not reflect phylogenetic relationships, as predicted by Linnaeus (Citation1753; Hayden et al., Citation2003). The genera Enteromorpha and Ulva were consequently synonymized and the currently accepted genus Ulva includes tubular, sheet-like and mixed-morphology taxa. Thus, allegedly unique morphological characteristics that were indicated in past species descriptions, and subsequently used in identification keys, are often uninformative, whereas molecular methods allow for reliable species differentiation (Blomster et al., Citation1998, Citation2002; Hayden et al., Citation2003; Brodie et al., Citation2007). In particular, tufA has been reported as a useful marker for identifying green algae (Saunders & Kucera, Citation2010). However, DNA-based species identification remains ambiguous when reference sequences of type material are missing, as is the case for most of the Ulvales and Ulotrichales. The DNA quality of historical voucher specimens is often low, thereby hampering sequencing efforts (Staats et al., Citation2011). Therefore, both molecular and morphological methods are still needed to link taxonomic concepts that were originally based on morphology with molecular traits (Hillis, Citation1987).
The documentation of seaweeds in northern Germany has been conducted since the mid-19th century, and seaweeds of the small island of Helgoland, located in the SE North Sea, have received much attention from marine botanists and phycologists, making Helgoland among the best-studied seaweed habitats in Europe (Bartsch & Kuhlenkamp, Citation2000). The solid rock pedestal of Helgoland provides a natural substratum for a macrophytobenthos in a fully marine environment and comprises a unique habitat in Germany (Reinke, Citation1889; Bartsch & Kuhlenkamp, Citation2000). However, even the most recent comprehensive descriptions of Helgoland’s macroalgae (Kornmann & Sahling, Citation1977, Citation1983, Citation1994) were exclusively based on morphological identification. Bartsch & Kuhlenkamp (Citation2000) also included rare and doubtful species and summarized taxonomic changes. Furthermore, the understanding of macroalgal species diversity on Helgoland is only transferable to a limited extent to Germany’s mainland coasts, which differ extensively in ecological conditions.
The tidal Wadden Sea is another fully marine ecosystem (salinity 30–33) within the North Sea, but it mainly consists of extended sand and mud flats, with relatively little hard substrate, and the German coast of the Baltic Sea is brackish, lacks tides, and is mainly composed of stones, gravel and sand (Rönnbäck et al., Citation2007). Furthermore, except for general identification keys (Rothmaler, Citation1984; Pankow, Citation1990), information about the identity and abundance of macroalgae in the Wadden and Baltic Sea areas of Germany is relatively sparse. Based on a summary of literature records, Schories et al. (Citation2009) described the distribution of macroalgae along the coast of Germany. However, the taxonomic concepts underlying historical records are often unclear and records based on molecular species identification are still sparse for the area.
Accordingly, the aim of the present study was to reassess the diversity of Ulva sensu lato at geographically separated areas along the coasts of the Baltic and North Seas in the German state of Schleswig-Holstein, as well as on Helgoland. The survey included both DNA barcoding and classical morphological identification approaches, and both field-collected and herbarium specimens were examined which allowed for the detection of several cryptic or newly introduced species and for the identification of several historical misinterpretations.
Materials and methods
Sample collection
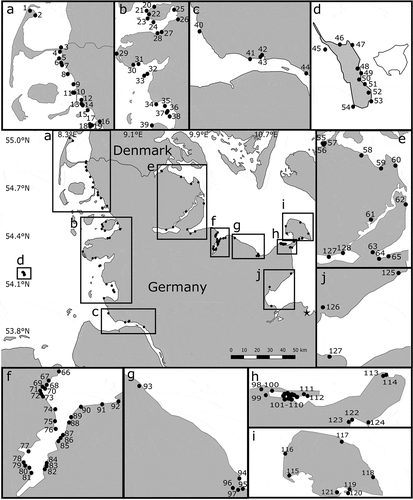
Samples of Ulva sensu lato were collected from 127 sites throughout the state of Schleswig-Holstein, Germany (), including sites in the Wadden Sea (n = 44), Baltic Sea (n = 73) and on Helgoland (n = 10). The sites represented a variety of habitats, such as estuaries, overflow basins and drainage channels, within each of the ecosystems. The sites were spread over 536 km along the coast of the Baltic Sea and over 466 km along the coast of the North Sea, with a maximum distance between sites of less than 25 km. Full data on the collection sites are available in Supplementary table S1. To ensure that seasonal species were sampled, collections were conducted during both summer (July–August 2014 and August–September 2015) and spring (April 2015 and March 2016). Single locations were also visited in 2017 and 2018, and only a limited number of sites were visited during winter (November 2014–early March 2015) owing to lower green algal growth. Sites along the coast of the North Sea (mainly groynes, bulwarks, rocks and mudflats) were sampled during low tide, whereas sites along the coast of the Baltic Sea were sampled when water levels were low using waders and an aquascope, which allowed for sampling to a depth of 1.2–1.5 m below mean sea level. Additional sampling (n = 3) was undertaken by divers in August 2014. Representative specimens were collected for each morphotype that was observed at each sample site, and epiphytes were also collected from host specimens. The collected thalli were stored in a cool box (~10°C) and transported to the laboratory.
Fig. 1. Map of sampling sites in northern Germany. Insets a–j provide higher resolution. Numbers 1–126 cross-reference to and Supplementary table S1, whereas numbers 127 and 128 indicate the sampling sites at Winning and Brodersby. The asterisk indicates a previously investigated site in Wohlenberg.
Morphological analysis
Pre-identification was based on typical morphological characters (e.g. overall thallus morphology, cell form, cell arrangement, number of pyrenoids per cell, etc.) using identification keys (Koeman & Van den Hoek, Citation1981, Citation1982a, Citation1982b, Citation1984; Hoeksema & Van den Hoek, Citation1983), and morphological characters were recorded separately at basal, middle and apical-thallus parts using light microscopy. Lugol’s solution (iodine-potassium iodide) was used to stain starch-containing compartments, such as pyrenoids. After morphological analysis, epiphyte-free pieces of remaining thallus tissue (1 cm2) or complete smaller thalli were either frozen and lyophilized or dried in silica gel for future molecular analysis.
Molecular analysis
Total DNA was extracted from lyophilized or silica-dried samples using the Invisorb Spin Plant Mini Kit (Stratec, Birkenfeld, Germany), according to the manufacturer’s protocol, and the plastid-encoded DNA barcoding marker tufA was PCR amplified using the primers tufGF4 (Saunders & Kucera, Citation2010) and tufAR (Famà et al., Citation2002). The following conditions for amplification were used: initial denaturation at 94°C for 4 min; 38 cycles of 94°C for 1 min, 55°C for 30 s and 72°C for 1 min; then a final extension of 72°C for 7 min. Both strands of the purified amplicons were directly sequenced by GATC Biotech (Konstanz, Germany) and both sequence alignment and reciprocal editing were performed using Sequencher (v. 4.1.4; Gene Codes Co., Ann Arbor, Michigan). The resulting sequences were uploaded to GenBank (Supplementary table 1). Sequence alignment was performed using MAFFT (Katoh et al., Citation2002), whereas editing was done visually with Sequencher (v. 4.1.4, Gene Codes Corporation, Ann Arbor, Michigan). The alignment represented a 777 bp portion of the tufA gene. An optimal substitution model was determined using MrModeltest software version v. 2.2. (Nylander, Citation2004) and found to be GTR+G+I. Subsequently, maximum likelihood analysis was performed using RAxML (v. 8; Stamatakis, Citation2014) with 1000 bootstrap iterations and the suggested substitution model, and Bayesian inference was performed using MrBayes (v. 3.2.2; Ronquist et al., Citation2012) with four simultaneously running Markov Chain Monte Carlo chains for 5 × 106 generations. The run was ended automatically when the standard deviation of split frequencies dropped below 0.01. Reference sequences from GenBank were also included in the analyses, with preference given to annotated sequences published in peer-reviewed articles. The trees were rooted by an outgroup that contained Urospora penicilliformis GenBank code HQ610440 and Urospora wormskioldii GenBank code HQ610441. Sequences used in the phylogenetic tree are listed in .
Comparison of recent species richness to historical findings
To assess the potential misidentification of historical specimens, historical vouchers of Ulvales taxa from the study area and neighbouring regions were obtained from several macroalgae collections and herbaria (Herbarium of the Alfred Wegener Institute for Polar and Marine Research, Bremerhaven, Germany (BRM), Herbarium of GEOMAR Helmholtz Centre for Ocean Research, Kiel, Germany (GEO); Herbarium of the Natural History Museum of Denmark, Copenhagen, Denmark (C)) and morphologically compared to specimens collected during the present study. The micro- and macromorphological characters of the vouchers were assessed using the above-mentioned criteria. When possible, small thallus pieces of the historical voucher specimens were sampled for molecular verification of species identity, as described in Steinhagen et al. (Citation2018a).
Results
A total of 370 Ulva sensu lato samples were processed genetically for species discrimination and identification, on the basis of tufA sequence data, and the full dataset was subject to phylogenetic analyses (see Supplementary table S1). In addition, an analysis with selected representative sequences was also performed (). The ML and BI analyses yielded congruent consensus trees. The species observed during the present study are described here, with a few particularly conspicuous species discussed in detail, and the majority are discussed in more depth in the Supplementary Information.
Fig. 2. Maximum likelihood phylogram of tufA sequences from taxa of Ulva sensu lato from northern Germany. Solid triangles indicate herbarium vouchers (see also ). The two shades of grey indicate clades that were present in the study area. Hatched boxes indicate species complexes and, thus, taxonomic entities that could not be clearly resolved phylogenetically. Numbers at nodes indicate bootstrap values (left) and Bayesian posterior probabilities (1000 replicates; right). Poorly supported nodes (< 70% bootstrap and < 0.70 Bayesian support) are not labelled. Branch lengths are proportional to sequence divergence.
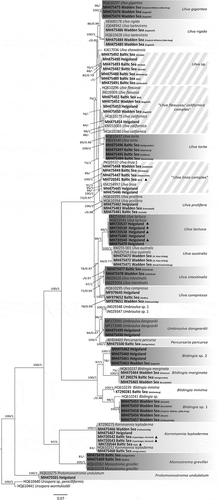
The phylogenetic analyses separated the investigated specimens into 20 taxonomic entities, nearly all of which could be resolved on the basis of peer-reviewed reference sequences provided by GenBank. More specifically, the taxa were identified as members of Ulva, Umbraulva, Percursaria, Blidingia, Kornmannia, Monostroma and Protomonostroma. One major branch within the consensus tree included members of Ulva, Umbraulva and Percursaria (i.e. the Ulvaceae) and was split into two subgroups, with the larger one containing Ulva taxa exclusively and the smaller subgroup containing several Ulva taxa (U. lactuca, U. australis, U. intestinalis and U. compressa), Umbraulva dangeardii and Percursaria percursa. However, this topology was only observed when P. percursa sequences were included; when P. percursa sequences were omitted, Umbraulva clustered as a sister group to Ulva (Supplementary fig. S1). Most of the species clades obtained full bootstrap and posterior probability support.
All U. gigantea sequences were identical to a sequence from New Brunswick, Canada (, ). The specimens of this species were always represented by distromatic blades and were only found in a limited area in the Wadden Sea, except for one specimen in the Baltic Sea (for details see Supplementary Information).
Table 1. List of green algal samples collected in 2014–2016 in northern Germany and used in the displayed phylogenetic tree
Table 2. List of genetically processed herbarium material
Ulva rigida was always distromatic, and attached specimens were found in all three investigated regions, whereas mats of drifting specimens were only observed in the Wadden Sea (for details see Supplementary Information). The cluster representing U. rigida in our phylogenetic tree () contained reference sequences for Ulva laetevirens Areschoug 1854 from Connecticut (JQ048942) and New Brunswick (HQ610428), as well as for U. rigida from the Italian Adriatic Sea (HE600178). All sequences were nearly identical, exhibiting divergences from the references of 0–0.26% and were placed in a well-delimited cluster.
Specimens that clustered most closely to U. shanxiensis type sequences were genetically more diverse than other taxa, with sequence distances ranging from 0 to 2.8% (). Furthermore, the specimens of Ulva sp. sampled during the present study were quite divergent (2.4–2.8%) from the U. shanxiensis type sequence. Specimens that belonged to this cluster were observed at all three main study areas and were typically found in areas of intense anthropogenic impact. The specimens were generally tubular but, nonetheless, of variable morphology (for details see Supplementary Information).
The clades delimiting the species U. flexuosa and U. californica received low to medium support. Both species were observed at all three main study areas. The more fragile and relatively rare species, U. flexuosa, exhibited only tubular morphologies and was generally observed unattached. The sequences of specimens in this clade differed from reference sequences from Canada (HQ610296) and South Korea (JN029309) by 0–0.17%. In contrast, the more robust of the two species, U. californica, exhibited more variable morphology, ranging from tubular to lanceolate or amorphous forms, and preferentially settled on artificial substrates. The sequences of specimens in this clade differed from reference sequences from California (KM255003) and Canada (HQ610279 and HQ610280) by 0–1.82% (, ).
Specimens that exhibited the relatively characteristic morphology of Ulva torta (long, narrow, with entangled unbranched tubular thalli and a central lumen of only 3–11 µm) were infrequently observed in either the Baltic or Wadden Seas and were not observed on Helgoland (for details see Supplementary Information). The specimens were clustered with reference sequences from southern Australia (JN029340) and British Columbia, Canada (HQ610437), but with only moderate support (85/0.96; ), even though the specimens exhibited relatively low genetic divergence from the reference sequences (0.12–0.3%).
Sequences of the U. linza specimens exhibited strong genetic divergence () and clustered in two strongly supported subgroups, U. linza 1 (100/1) and U. linza 2 (95/1). The U. linza 1 specimens were abundant over the whole study area (except Helgoland), formed a cluster with a genotype from Tasmania (JN029337) and exhibited very low sequence divergence (0–0.0014%). In contrast, the U. linza 2 specimens were exclusively collected from Helgoland, formed a cluster with a reference sequence from the North-east Pacific (KM254997) and exhibited slightly greater sequence divergence (0–0.49%). A historical herbarium voucher, originally identified as Enteromorpha ahlneriana Bliding nom. illeg., could be genetically assigned to Ulva linza sp. 1 (, ). However, the phylogenetic differentiation was not reflected morphologically, and both the U. linza 1 and U. linza 2 specimens exhibited a wide spectrum of tubular to lanceolate and partly distromatic morphologies (for details see Supplementary Information).
The U. prolifera reference sequences from Manitoba, Canada (HQ610395) and Labrador, Canada (HQ610394) clustered with specimens from all three main study areas. However, our samples were clearly more similar to one another (−/0.98; ), even though their genetic divergence from the reference ranged from 0 to 1.23%. Ulva prolifera was always attached when observed, was relatively abundant, exhibited a variety of tubular morphologies and frequently, but not always, possessed a characteristically twisted stipe-like base (for details see Supplementary Information).
Specimens that formed strongly supported clusters (99/1) with U. lactuca reference specimens from New Brunswick (HQ610341, 0–0.31% divergence) and California (KM255044, 0.12–0.47% divergence) were collected exclusively from Helgoland (). Even though Hughey et al. Citation2019 have suggested that the oldest available name for the European ‘U. lactuca’ is U. fenestrata, we will refer to the here mentioned genotype as U. lactuca for reasons of general understanding. They exhibited distromatic thalli of various shapes and were characterized by relatively strong attachment. The specimens were found abundantly within the intertidal zone and grew attached to hard substrata, such as naturally occurring rocks (), stones and mussel beds or artificial breakwaters and piers. Only a few drifting specimens were observed and such drifting thalli exhibited clear indications of recent ruptures in the rhizoidal zone, thereby suggesting that drifting is not tolerated for long time periods. Specimens of this clade were never found in rockpools that were subject to potential desiccation or influence by rainwater. All thalli were distromatic throughout, and their shape varied from rounded to lobed () or laciniate morphologies that could be straight, petiolate-like (, ) but also strongly curved (). Filled disc-like rhizoidal zones (, ) were frequently observed at the base of the blade. The margins of the thalli were never toothed (grazing traces were clearly distinguished) and were usually smooth, although the rounded individuals sometimes exhibited ruffled margins. Holes (2–6 mm diameter) were observed infrequently. The thalli reached lengths of 40 cm and widths of 35 cm, but smaller individuals (max. 5 cm length and 2 cm width) were also observed. Longitudinal ridges were observed in the basal regions of most of the investigated specimens but were often absent in young thalli. The thalli were attached by obconically shaped stipe-like structures that terminated in broad rhizoidal zones (, ). The cells of the middle and apical thalli were arranged in curved or short rows, whereas cells of the laciniate thalli were sometimes arranged in longitudinal rows. The cells were polygonal with rounded corners, 12–24 × 14–31 µm in surface view, and contained a parietal chloroplast (rarely filling the entire cell) and 1–2 (rarely up to 4) pyrenoids (, ). Cells of the rhizoidal zone contained up to seven pyrenoids, and both the stipe-like region and rhizoidal zone were filled by the elongated tails of the cell bodies, which became visible in microscopic transections. Thalli with these features were not only observed on Helgoland, but also along the mainland coasts, although they were always genetically assigned to U. compressa.
Figs 3–14. Morphology of Ulva lactuca specimens from Helgoland, Germany. Fig. 3. U. lactuca population growing on the northeast rocky tidal flats. Fig. 4. Typical lobular morphotype. Figs 5, 6. Petiolate-like morphotype. Fig. 7. Strongly curved morphotype, with (Fig. 8) a disk-like rhizoidal zone (cross section) and (Fig. 9) elongated club-shaped cells that extend to the centre of the rhizoidal disc. Fig. 10. Cells of the apical and middle thallus parts, with a hood-shaped chloroplast and one (sometimes two) central or marginal pyrenoids. Fig. 11. Marginal pyrenoid. Fig. 12. U. tenera syntypes collected from Helgoland in 1978 (Herbarium of the Alfred Wegener Institute, Bremerhaven; ID BRM007806). (I) However, by sequencing one individual (see also and ), its genetic affiliation to U. lactuca was confirmed. Fig. 14. U. pseudocurvata specimens collected from Helgoland in 1988 (Herbarium of the Alfred Wegener Institute, Bremerhaven; ID BRM007947); arrowhead indicates specimen that was genetically identified as U. lactuca (see also and ).

The sequence of one of over 100 syntypes of U. tenera Kornmann & Sahling – originating from Helgoland and stored at the BRM on Helgoland (voucher ID: BRM007806; , , ) – clearly clustered with U. lactuca (). Additionally, sequences from historical U. pseudocurvata vouchers (BRM001703 and BRM001700; ), which provided the first evidence of the species’ presence on Helgoland and were displayed in the publication of Kornmann & Sahling (Citation1994), were also placed, with full bootstrap support, in the clade representing U. lactuca and were identical to the sequences of recently collected U. lactuca samples from Helgoland ().
A few specimens from the west coast of Helgoland and an area around the peninsula of Eiderstedt in the Wadden Sea (i.e. fully marine environments) were clustered with an U. australis reference sample from Australia (JN029254, genetic dissimilarity 0–0.33%, , ). However, morphological comparison was impossible since only small thallus pieces conserved in silica gel could be obtained from our study area.
Sequences from the U. intestinalis specimens formed a fully supported clade (100/1) with reference specimens from Australia (JN029320) and California (KM255056; ) but consistently exhibited slight divergence from both reference sequences (0.12–0.45%). The species was abundantly present at all three main study areas and in salinities that ranged from fresh water to fully marine. All of the specimens investigated exhibited tubular morphology, and most individuals were inflated and unbranched, corresponding to the typical morphology of U. intestinalis. Furthermore, specimens that exhibited branched and unbranched morphologies could not be distinguished genetically (for details see Supplementary Information).
Ulva compressa was abundant in all three main study areas, although only tubular, usually branched individuals were observed on Helgoland, whereas only distromatic sheet-like specimens were found along the Baltic coast. Both morphologies were encountered along the Wadden coast, sometimes even at the same location. Furthermore, individuals exhibiting transition forms between the two morphologies were only rarely observed (for details see Supplementary Information). However, the morphotypes were not separated during the phylogenetic analyses and remained clustered with a U. compressa reference sample from Canada, New Brunswick (HQ610295, genetic dissimilarity 0–0.77%).
Sequences that were identical to those of Umbraulva dangeardii reference sequences from southern Italy (MF172090 and MF172091, genetic dissimilarity: 0–0.13%) were only recovered from specimens collected at Helgoland at a depth of 8 m. The specimens were distromatic sheets with a conspicuous dark olive colour and thin, soft texture, which corresponds to descriptions of specimens from the British Isles (Maggs et al., Citation2007a).
Percursaria percursa was only encountered once in a macroscopically visible state at Heiligenhafen (Baltic Sea), where it grew unattached in dense mats in the supralittoral zone. Microscopic examination confirmed the typical morphology of unbranched biseriate filaments (Maggs & Kelly, Citation2007) and our sequence was placed in a fully supported cluster with a P. percursa reference sequence (AY454403, genetic dissimilarity: 0.13%).
A well-delimited cluster that included Blidingia marginata, B. minima and Blidingia sp. reference sequences formed a sister clade to that including the Ulva, Umbraulva and Percursaria sequences and included four genetic entities (). Two of the four subgroups could not be resolved, since they did not match any references in GenBank, but were putatively identified as Blidingia specimens based on sequence similarity, overall morphology and growth habit.
Specimens that exhibited low genetic variability and clustered with a B. marginata reference sequence from New Brunswick, Canada (HQ610237, genetic dissimilarity: 0–0.28%) were abundant in all three main study areas. The specimens formed dense populations of variable tubular morphology in the upper intertidal and supralittoral zones and were often encountered as the only macroalgal settlers in microhabitats that are influenced by fresh water and that may fall dry for longer periods (for details see Supplementary Information).
Specimens that belonged to the unresolved entity Blidingia sp. 2 were morphologically indistinguishable from B. marginata but formed a separate and fully supported cluster with 8–8.2% divergence from the B. marginata reference sequence (HQ610237). These specimens were collected from Helgoland and one site in the Wadden Sea (Dagebüll).
Molecular analysis failed to confirm the presence of B. minima. However, in a previous project B. minima was found at Wohlenberg (, site marked by an asterisk in general map), a site only 30 km to the East in the neighbouring German state of Mecklenburg-Vorpommern. A sample from the previously reported population was included in the phylogenetic tree (KT290281, ) and formed a fully supported clade with a B. minima reference sequence from Canada (HQ610239).
Specimens that belonged to the unresolved entity Blidingia sp. 1 formed a fully supported cluster that was clearly delimited from other Blidingia species and genetically dissimilar (4.5–4.8%) from the B. minima reference sample (HQ610239). Specimens that belonged to this cluster had a broad distribution and were observed in all three main study areas, at remote and protected sites as well as in highly trafficked waters (see also Steinhagen et al., Citation2018b). The specimens grew as mats on various substrata in the supralittoral zone and were often found in the close vicinity of freshwater inflows, such as drainage pipes. The Blidingia sp. 1 specimens were relatively minute and they often exhibited a characteristic antler-like branched tubular morphology. However, macroscopically visible branches were rarely observed and appeared as spiralled or inflated.
The remaining three clades () only included specimens with monostromatic blades and formed fully supported clusters with reference sequences of Kornmannia leptoderma, Monostroma grevillei and Protomonostroma undulatum. Kornmannia leptoderma specimens from all three study areas clustered with reference sequences from Canada (HQ610252, dissimilarity: 0–0.38%) and northern Germany (KT290275) and exhibited little to no genetic diversity. Specimens that belonged to this cluster were found in the middle and lower intertidal zones and were typically attached to substrata. The specimens appeared to avoid exposure to direct sunlight and were frequently found on the shaded sides of stones () or jetties, or under piers. The 1–5 cm (rarely up to 8 cm) long thalli of the K. leptoderma specimens were nearly unrecognizable when the substrate became dry during low tide. The membranous and very soft thalli appeared funnel-shaped (), lanceolate or rosette-like (). Older thalli that had sporulated were amorphous in shape and deeply cut. The rhizoidal zone was not defined by a disc-like structure, and cells proceeded without tapering in a stipe-like region. Cells in the apical and middle thallus parts differed in shape from those in the basal thallus parts. The cells of the upper and middle thallus regions were either polygonal to round or with sharp and clearly defined angular edges, 9–15 × 11–16 µm in surface view, with a single centrally located pyrenoid and a chloroplast that was either marginal or filling the entire cell (, ). Meanwhile, the cells of the apical and middle thallus regions were thick-walled (), although progressively thinner and larger toward the basal region, and with 1–3 (rarely 4) pyrenoids per cell. In the lower mid-thallus parts, the cells were 11–21 × 11–27 (32) µm in surface view and sometimes appeared grainy, whereas others already resembled rhizoidal cells with long drawn-out tips (, ). Cells of the rhizoidal zone were up to 50 µm long, always grainy, with rhizoidal tips extending from the main body, and typically with 1–3 pyrenoids (rarely more; ).
Figs 15–21. Morphology of Kornmannia leptoderma specimens from northern Germany. Fig. 15. Typical sampling site along the coast of the Baltic Sea (Aschau lagoon), with K. leptoderma growing on the shaded side of a rock. Fig. 16. Rosette-shaped specimens from the Baltic Sea (Aschau lagoon). Fig. 17. Funnel-shaped specimen from the Baltic Sea (coastal inlet, Schlei, Lindaunis). Fig. 18. Cells in apical and middle thallus regions, in rows or pairs. Fig. 19. Marginal chloroplasts and central pyrenoids (one or rarely two). Figs 20, 21. Club-shaped cells of the rhizoidal zone.

All the M. grevillei specimens formed a cluster and were often identical to reference sequences from Maine, USA (HQ610262, dissimilarity: 0–0.51%) and New Brunswick, Canada (HQ610259, dissimilarity: 0–0.39%). The species was abundant in the Baltic Sea and also occurred on Helgoland. However, the species was only observed during spring (March to May), and in late spring drifting mats of M. grevillei frequently developed in sheltered bays, harbours and lagoons. The cells of the M. grevillei specimens were arranged in more distinct rows than those of the K. leptoderma specimens.
The only specimen that clustered with the P. undulatum reference sequence (dissimilarity: 0.13%) was found in the lower intertidal zone of Helgoland. As in M. grevillei, the cells were often arranged in rows. However, instead of a smooth transition from basal cells to rhizoidal cells, abrupt changes in cell shape were observed, and the rhizoidal cells were longer than those of the K. leptoderma specimens (60–90 µm, up to 110 µm; Supplementary fig. S2; for details see Supplementary Information).
An additional taxon that might represent Gayralia oxysperma was not detected at any of the study sites, even though Kützing (Citation1843) originally described its basionym, Ulva oxysperma, on the basis of material collected in Schleswig-Holstein at Winning (located at the inner Schlei, a narrow inlet of the Baltic Sea, site 127 in ). Unfortunately, the type material of U. oxysperma appears to be lost. However, historical G. oxysperma vouchers from Friedrichsort, Kiel, Germany, that were sampled in 1962 (MH720544) and from Copenhagen, Denmark, that were sampled in 2004 and 2007 (MH720542 and MH720543) were available for sequencing. Notably, all three voucher sequences clustered with K. leptoderma (, ). During subsequent visits (i.e. additional collections in 2017 and 2018), thalli exhibiting the described morphology of G. oxysperma were not detected at Winning (site 127 in , salinity 1) but were collected at the inner Schlei at Brodersby (site 128 in , salinity 7), which is 10 km from Winning, and at Lindaunis (site 61 in , Supplementary table S1), which is 30 km from Winning. However, sequences from these G. oxysperma-like specimens were also placed in the K. leptoderma cluster (MH720545–MH720547; Supplementary table S1).
Discussion
The 20 taxa of green algae that were detected in the present study () can be identified with variable degrees of certainty. Only one taxon could be assigned to a clade that included a reference sequence from type material, namely for Ulva tenera(Kornmann & Sahling, Citation1994). However, U. tenera was described relatively recently, and the corresponding cluster in our phylogenetic analysis also encompassed several reference sequences from specimens that were recognized elsewhere as U. lactuca L. Even though only one of the more than 100 U. tenera syntypes was examined, and given that the current concept of U. lactuca has been challenged (Butler, Citation2007), the phylogenetic analysis presented here strongly suggests that U. tenera is a synonym of U. lactuca. This view is further supported by the observation that young U. lactuca specimens from Helgoland exhibit the described morphology of U. tenera (Kornmann & Sahling, Citation1994). Other described characteristics of U. tenera are its restriction to the uppermost eulittoral and its exclusively vegetative propagation (i.e. with biflagellate spores; Kornmann & Sahling, Citation1994); apparently, the authors observed dwarfish forms of U. lactuca that were adapted to extended air exposure. Therefore, U. tenera is here reduced to synonymy with U. lactuca. Other homotypic synonyms of U. lactuca cited below are according to Guiry & Guiry (Citation2018).
Ulva lactuca
Linnaeus, C. 1753. Species plantarum, exhibentes plantas rite cognitas, ad gen era relatas, cum differentiis specificis, nominibus trivialibus, synonymis selectis, locis natalibus, secundum systema sexuale digestas. Vol. 2 pp. [i], 561–1200, [1–30, index], [i, err.]. Holmiae [Stockholm]: Impensis Laurentii Salvii.
Homotypic synonyms:
Phyllona lactuca (Linnaeus) F.H.Wiggers 1780
Monostroma lactuca (Linnaeus) J.Agardh 1883
Ulva tenera Kornmann & Sahling Citation1994
The genetic-based species identities of several other taxa corresponded to characteristic morphological traits. Indeed, this was the case for U. torta, which usually formed massively intertwined tubular thalli of small diameter; U. prolifera, which mostly exhibited characteristically twisted stipes; and Umbraulva dangeardii, which is characterized by its olive green pigmentation. Meanwhile, the genetic-based species identities of the three monostromatic taxa corresponded to known phenotypic traits. For example, M. grevillei was only observed during spring, which was not the case for any other entity; K. leptoderma exhibited a characteristic heteromorphic life cycle, as reported elsewhere (Weinberger et al., Citation2018); and P. undulatum, despite only being observed once, exhibited a typical morphology (see Supplementary Information). Furthermore, specimens that clustered with Ulva intestinalis mostly exhibited the tubular and unbranched morphology considered characteristic of the species (Kornmann & Sahling, Citation1977; Rothmaler, Citation1984; Pankow, Citation1990), but branched specimens were occasionally observed, as reported previously (Reed & Russell, Citation1978; Blomster et al., Citation1998), probably promoted by low salinity (Steinhagen et al., Citation2018b).
However, for most cases, genetic-based species identities failed to correspond to characteristic morphological traits. For example, specimens that exhibited the characteristic lanceolate and partly distromatic type morphology of U. linza (Kornmann & Sahling, Citation1977; Rothmaler, Citation1984; Pankow, Citation1990) clustered with U. linza reference sequences, but also with sequences from specimens that exhibited tubular and branched morphologies corresponding to descriptions of U. procera and U. ahlneriana(Kornmann & Sahling, Citation1977; Rothmaler, Citation1984; Pankow, Citation1990) and a sequence from a historical voucher of U. ahlneriana (). This observation supports the previous suggestions that U. procera (Maggs et al., Citation2007b) and U. ahlneriana(Guiry & Guiry, Citation2018) are synonyms of U. linza. It was interesting that the cluster representing U. linza in our phylogenetic tree included two lineages which were morphologically indistinct. One of the lineages was only detected on Helgoland, whereas the second was only detected on mainland coasts. However, more information is needed to determine whether the groups represent distinct species or simply unique genotypes that developed in response to geographic separation.
Meanwhile, the U. compressa specimens also exhibited multiple gross morphologies. One morphotype was only observed on North Sea coasts and corresponded to the morphology of the tubular and branched type material (Linnaeus, Citation1753). However, as already discussed elsewhere (Steinhagen et al., Citation2018a), genetically indistinguishable specimens from the Baltic and Wadden Seas exhibited a completely different morphology that was consistently distromatic and sheet-like. Evidently, the distromatic morphology of U. compressa strongly overlaps with the allegedly unique morphology of U. lactuca, thereby causing a considerable amount of historical taxonomic confusion (Steinhagen et al., Citation2018a). Based on the results of the present study, U. lactuca in northern Germany is only present on Helgoland (, Supplementary table 2), whereas historical records from the Baltic Sea (Schories et al., Citation2009) are misidentified U. compressa specimens (Steinhagen et al., Citation2018a). Notably, historical vouchers from Helgoland (Kornmann & Sahling, Citation1994) that exhibited the curved morphology of U. pseudocurvata (Hoeksema & Van den Hoek, Citation1983) yielded sequences that clustered with U. lactuca sequences, whereas specimens that were recently collected from mainland coasts of northern Germany (Steinhagen et al., Citation2018a) and elsewhere (Tan et al., Citation1999; Hayden & Waaland, Citation2004) exhibiting the same morphology yielded sequences that clustered with U. compressa sequences. This clearly challenges the validity of U. pseudocurvata as a taxonomic entity, because its description is based on morphological traits that are clearly not specific, and it also confirms the strong morphological plasticity of U. lactuca on Helgoland and U. compressa, in its distromatic form, on the mainland coasts of northern Germany.
In addition to U. lactuca and the distromatic form of U. compressa, three additional entities with consistently distromatic blades were also observed in the present study. These specimens clustered with U. gigantea, U. australis, U. rigida and U. laetevirens reference specimens (). In these cases, the observed morphologies generally paralleled the corresponding type morphologies. However, based on morphological observations the taxa were not reliably distinguishable from one another, U. lactuca, or the distromatic form of U. compressa. Furthermore, as recently demonstrated by ITS and rbcL analysis (Horta et al., Citation2018), U. rigida and U. laetevirens could not be distinguished using tufA gene sequences. Therefore, U. laetevirens Areschoug 1854 should be considered a synonym of U. rigida C. Agardh 1823.
Some clades with tubular morphologies could not be clearly resolved. As reported previously (Heesch et al., Citation2009; Kraft et al., Citation2010; Saunders & Kucera, Citation2010; Kirkendale et al., Citation2013), there was no clear species boundary between U. flexuosa and U. californica (). However, despite this observation, Hiraoka et al. (Citation2017) used hybridization experiments to confirm the biological separation of U. flexuosa and U. californica from Japan. Because cross-breeding experiments were not included in the present study, we have chosen to indicate the species’ lack of genetic resolution using the term ‘Ulva flexuosa/californica complex’.
A reference sequence for type material of the tubular species U. shanxiensis, which was recently described from a freshwater stream in northern China (Chen et al., Citation2015), was placed basal to a clade of tubular specimens in the phylogenetic analysis of the present study (). However, the clade encompassing U. shanxiensis and the tubular specimens was poorly supported, indicating relatively high sequence divergence (, note branch length). Therefore, the tubular specimens are unlikely to belong to U. shanxiensis, and the identity of the clade remains unidentified as a result.
Identities could also not be determined for two genetic entities in the genus Blidingia (Blidingia sp. 1 and Blidingia sp. 2), since they did not match any available reference sequences. Specimens of the Blidingia sp. 2 clade exhibited strong morphological overlap with a second clade encompassing a reference sequence of Blidingia marginata and could only be distinguished molecularly. The morphology of both clades was consistent with that of B. marginata but, perhaps, also with that of B. ramifera (Garbary & Barkhouse, Citation1987), a species that has not yet been reported from the area and which is, for formal reasons, invalid (Cormaci et al., Citation2014) and currently regarded as a synonym of B. marginata (Guiry & Guiry, Citation2018). In contrast, specimens of the relatively abundant Blidingia sp. 1 exhibited unique genetic and morphological traits that clearly distinguished them from other Blidingia taxa in northern Germany. In addition to B. marginata and B. minima, two other Blidingia species (B. chadefaudii and B. subsalsa) have also been reported from the German coasts of the North Sea (Kornmann & Sahling, Citation1978; Bartsch & Kuhlenkamp, Citation2000; Schories et al., Citation2009). However, no molecular reference data were available for B. chadefaudii and B. subsalsa, and morphological identification criteria for the species remain ambiguous and overlapping. Therefore, in order to identify Blidingia sp. 1 and Blidingia sp. 2 and to confirm the identities of B. marginata and B. minima, type material of different Blidingia species should be analysed by molecular markers and species life cycles should be documented using cultivated material. The same strategy might also facilitate the identification of ambiguous Ulva specimens in the future.
Notably, our phylogenetic analyses did not support the monophyly of the genus Ulva (). In our study the inclusion of U. lactuca, U. australis, U. intestinalis and U. compressa as a sister clade of Umbraulva species and Percursaria percursa was revealed (), in contrast to previous studies which used other marker genes (Hayden et al., Citation2003; Heesch et al., Citation2009; Kirkendale et al., Citation2013). This topology was not observed when P. percursa was excluded from the analysis (Supplementary fig. S1). However, the inclusion of more, rather than fewer, taxa is more likely to yield true phylogenetic relationships.
The species inventory of Ulva sensu lato of the present study diverged considerably from the expected inventory (Schories et al., Citation2009). Four species (U. australis, U. californica, U. gigantea and Umbraulva dangeardii) were observed in the area for the first time (). Ulva australis was first introduced to southern France and very recently reported from the Dutch Oosterschelde estuary (Fort et al., Citation2019). Now, the species is also present in the North Friesian Wadden Sea. The same is true for U. gigantea, which, in Europe, had only been reported from Britain and other westerly locations (Maggs et al., Citation2007b). Single individuals of U. californica were first observed in Germany in 2008 on the Wadden Sea island of Wangerooge in Lower Saxony (Lackschewitz et al., Citation2015) and, over the next six years, eventually reached the SW Baltic Sea. In the present study, Umbraulva dangeardii was only observed at one site on Helgoland (, Supplementary table S1). It is interesting that even though Helgoland is a phycological hotspot in Germany, U. dangeardii has never before been included in inventories (Kornmann & Sahling, Citation1977, Citation1983, Citation1994; Bartsch & Kuhlenkamp, Citation2000), suggesting recent introduction. Yet, the presence of U. dangeardii in Germany may have been ignored for some time, due to the preference of the species for subtidal habitats. In addition to the above-mentioned newly introduced species, three (Blidingia sp. 1, Blidingia sp. 2, Ulva sp.) or even four (if one of the two genetic entities within U. linza is included) additional taxa that were observed in our study probably represent cryptic and perhaps undescribed species that have so far not been recognized.
Fig. 22. Comparison of molecular (tufA)-based identification from the present study and the inventory list from Schories et al. (Citation2009). List of species predicted by Schories et al. (Citation2009) and detected in the present study (2019) from the Baltic Sea, Wadden Sea and Helgoland. X, species not observed; ✓, species observed; empty, unexpected by Schories et al. (Citation2009) and not observed in present study. Light grey shading indicates agreement between Schories et al. (Citation2009) and the present study, whereas dark grey shading indicates disagreement. For additional annotations or taxonomic notes by other authors see also Supplementary table S2.

Despite these new records, the morphology-based species inventories of all three main study areas were expected to be larger than the genetically validated ones (). Altogether, 14 of the species (members of Ulva, Blidingia, Monostroma, Gayralia and Ulvaria) that were listed by Schories et al. (Citation2009) and are currently accepted taxonomically (Guiry & Guiry, Citation2018) were not encountered genetically in the present study. This lack of detection could indicate their absence but might also be attributed to other factors, such as low abundance or lack of molecular reference material. Indeed, no tufA reference sequences are available for 11 of the 14 missing species, and the numerous historical records from the area may be the result of misidentification and taxonomic confusion.
As discussed above, records of Ulva pseudocurvata from northern Germany are often, and perhaps always, due to the misidentification of either U. compressa or U. lactuca. Also, the only record of U. splitiana from our area (as Enteromorpha jugoslavica; Kaminski, Citation1980) was due to the misidentification of U. linza, as demonstrated through sequencing of the ITS marker gene from the corresponding herbarium voucher (Gesche Bock, pers. comm.). Furthermore, analysis of historical Gayralia oxysperma vouchers from northern Germany and adjacent areas indicated that all the vouchers were genetically identical to Kornmannia leptoderma, which had until now been considered a relatively rare species that was only present on Helgoland (Kornmann & Sahling, Citation1983) and, therefore, has not been included in identification keys for other parts of Germany (Rothmaler, Citation1984; Pankow, Citation1990) or adjacent areas (Brodie et al., Citation2007). However, K. leptoderma was present in all three main areas of the present study (see also Weinberger et al., Citation2018). In striking contrast, G. oxysperma was not observed, even at the type locality of its basionym U. oxysperma Kützing (see the Supplementary Information for a description of the relatively complicated nomenclatural history of G. oxysperma). For further details see Doty (Citation1947), Gayral (Citation1965) and Womersley (Citation1984). This apparent absence or rarity of G. oxysperma is surprising because the species should be present across the entire Baltic Sea (Schories et al., Citation2009). Descriptions of G. oxysperma (Rothmaler, Citation1984; Pankow, Citation1990) are in complete agreement with the morphology of K. leptoderma in our area (–, see also Weinberger et al., Citation2018). The two species have very different life cycles (Vinogradova, Citation1969), but ontogenetic observations are time consuming, and for this reason most historical records of G. oxysperma are probably based on the morphological traits of field-collected material. As a consequence, it is likely that most records of G. oxysperma are due to the misidentification of K. leptoderma. Similarly, the molecular analysis of G. oxysperma-like specimens from the North-west Atlantic yielded two clusters attributed to Monostroma grevillei (Saunders & Kucera, Citation2010). Therefore, a thorough taxonomic reassessment of G. oxysperma and its populations is urgently needed.
Some species that were reported to occur only in parts of northern Germany were found to have broader distributions than expected. For instance, U. rigida, which was only expected to occur in the Wadden Sea and on Helgoland, was also observed in the Baltic Sea (, Supplementary table S1), and K. leptoderma, which had only been reported to occur on Helgoland, was observed in both the Baltic and Wadden Seas (, Supplementary table S1).
In summary, the current morphological concepts that are used for the identification of Ulva species and related taxa in northern Germany are neither in agreement with the species inventory of the area nor with the actual morphology of species that are present. Past morphological descriptions of U. linza, U. intestinalis and U. compressa have been too restrictive, thereby resulting in frequent misidentifications of these abundant taxa. Furthermore, several cryptic and/or newly introduced species, including K. leptoderma, U. australis, U. californica, U. gigantea and Protomonostroma undulatum, are now present in northern Germany, and several genetic entities, namely Ulva sp., Blidingia sp. 1 and Blidingia sp. 2, have yet to be identified. Meanwhile, B. minima and G. oxysperma were either absent or much rarer than expected, and certain other taxa that were expected in the area, namely U. tenera, are actually synonyms. The observations of the present study provide a basis for the development of improved identification keys, although it is unlikely that it will be possible to distinguish all species morphologically owing to the considerable overlap of traits. The DNA barcoding approach used in the present study clearly provides better resolution. However, U. californica and U. flexuosa cannot be clearly distinguished using the analysis of tufA alone and more sequences of type material will be needed to improve the identification of species in the future. Furthermore, additional genetic markers should be investigated and cultivation studies should be performed to resolve remaining issues, such as the taxonomic affiliation of the newly found Blidingia species or relations among the genera Ulva, Umbraulva and Percursaria.
Supplementary information
Supplementary Information. (1) Distribution and specific characteristics of observed species, (2) nomenclatural history of Gayralia oxysperma and (3) seasonal species variation.
Supplementary table S1. Full collection data.
Supplementary table S2. Comparison of molecular (tufA)-based identification from the present study and the inventory list from Schories et al. (2009).
Supplementary fig. S1. Maximum likelihood phylogram of tufA sequences from taxa of Ulva sensu lato from northern Germany.
Supplementary fig. S2. Morphology of Protomonostroma undulatum specimens from Helgoland, Germany.
Author contributions
S. Steinhagen: experimental design, fieldwork and algae collection, laboratory work, macro- and microscopic observation, phylogenetic analysis, drafting and editing manuscript; R. Karez: experimental design, algae collection, drafting and editing manuscript; F. Weinberger: original concept, collection of specimens, drafting and editing manuscript.
TEJP-2018-0117-File009.tif
Download TIFF Image (7.7 MB)TEJP-2018-0117-File008.tif
Download TIFF Image (3.5 MB)TEJP-2018-0117-File007.docx
Download MS Word (211.8 KB)Acknowledgements
We would like to express gratitude to T. Dolch, K. Reise and R. Kuhlenkamp for support in specimen collection; the herbaria and macroalgae collections for granting us access to their herbaria and vouchers; our colleague D. Afanasyev for translating the Russian publications cited in our article; and G. Bonthond for valuable comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bartsch, I. & Kuhlenkamp, R. (2000). The marine macroalgae of Helgoland (North Sea): an annotated list of records between 1845 and 1999. Helgoland Marine Research, 54: 160–189.
- Blomster, J., Maggs, C.A. & Stanhope, M.J. (1998). Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles. Journal of Phycology, 34: 319–340.
- Blomster, J., Back, S., Fewer, D.P., Kiirikki, M., Lehvo, A., Maggs, C.A. & Stanhope, M.J. (2002). Novel morphology in Enteromorpha (Ulvophyceae) forming green tides. American Journal of Botany, 89: 1756–1763.
- Brodie, J., Maggs, C.A., Society, B.P. & John, D.M. (2007). The Green Seaweeds of Britain and Ireland. British Phycological Society, London.
- Butler, D. (2007). Ancient algal mixup sorted. Nature. https://www.nature.com/news/2007/071220/full/news.2007.396.html.
- Charlier, R.H., Morand, P., Finkl, C.W. & Thys, A. (2007). Green tides on the Brittany coasts. Environmental Research, Engineering and Management, 3: 52–59.
- Charlier, R.H., Morand, P. & Finkl, C.W. (2008). How Brittany and Florida coasts cope with green tides. International Journal of Environmental Studies, 65: 191–208.
- Chen, L., Feng, J. & Xie, S.-L. (2015). Ulva shanxiensis (Ulvaceae), a new species from Shanxi, China. Novon, 23: 397–405.
- Cormaci, M., Furnari, G., & Alongi, G. (2014). Flora marina bentonica del Mediterraneo: Chlorophyta. Bollettino dell’Accademia Gioenia di Scienze Naturali di Catania, 47: 11–436
- Doty, M.S. (1947). The marine algae of Oregon. Part 1. Chlorophyta and Phaeophyta. Farlowia, 3: 1–65.
- Famà, P., Wysor, B., Kooistra, W.H.C.F. & Zuccarello, G.C. (2002). Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene. Journal of Phycology, 38: 1040–1050.
- Fort, A., Lebrault, M., Allaire, M., Esteves-Ferreira, A.A., McHale, M., Lopez, F., Fariñas-Franco, J.M., Alseekh, S., Fernie, A.R. & Sulpice, R. (2019). Extensive variations in diurnal growth patterns and metabolism among Ulva spp. strains. Plant Physiology, 180: 109–123.
- Garbary, D.J. & Barkhouse, L.B. (1987). Blidingia ramifera (Bliding) stat. nov. (Chlorophyta): a new marine alga for eastern North America. Nordic Journal of Botany, 7: 359–363.
- Gayral, P. (1965). Monostroma Thuret, Ulvaria Rupr. emend. Gayral, Ulvopsis Gayral. (Chlorophycées, Ulotrichales): structure, reproduction, cycles, position systématique. Revue générale de botanique, 72: 627–638.
- Guiry, M.D. & Guiry, G.M. (2018). AlgaeBase. World-wide electronic publication. National University of Ireland, Galway. http://www.algaebase.org.
- Hayden, H.S. & Waaland J.R. (2002). Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences. Journal of Phycology, 38: 1200–1212.
- Hayden, H.S. & Waaland J.R. (2004). A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific. Phycologia, 43: 364–382.
- Hayden, H.S., Blomster, J., Maggs, C.A., Silva, P.C., Stanhope, M.J. & Waaland, J.R. (2003). Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. European Journal of Phycology, 38: 277–294.
- Heesch, S., Broom, J.E., Neill, K.F., Farr, T.J., Dalen, J.L. & Nelson, W.A. (2009). Ulva, Umbraulva and Gemina: genetic survey of New Zealand taxa reveals diversity and introduced species. European Journal of Phycology, 44: 143–154.
- Hillis, D.M. (1987). Molecular versus morphological approaches to systematics. Annual Review of Ecology and Systematics, 18: 23–42.
- Hiraoka, M., Ichihara, K., Zhu, W., Shimada, S., Oka, N., Cui, J., Tsubaki, S. & He, P. (2017). Examination of species delimitation of ambiguous DNA-based Ulva (Ulvophyceae, Chlorophyta) clades by culturing and hybridisation. Phycologia, 56: 517–532.
- Hoeksema, B.W. & Van Den Hoek, C. (1983). The taxonomy of Ulva (Chlorophyceae) from the coastal region of Roscoff (Brittany, France). Botanica Marina, 26: 65.
- Horta, P., Bernardes Batista, M., Cunha, R.L. & Castilho, R. (2018). Sea lettuce systematics: lumping or splitting? In BioRxiv. https://doi.org/https://doi.org/10.1101/413450.
- Hughey, J.R., Maggs, C.A., Mineur, F., Jarvis, C., Miller, K.A., Shabaka, S.H. & Gabrielson, P.W. (2019). Genetic analysis of the Linnaean Ulva lactuca (Ulvales, Chlorophyta) holotype and related type specimens reveals name misapplications, unexpected origins, and new synonymies. Journal of Phycology, doi: https://doi.org/10.1111/jpy.12860.
- Kaminski, E. (1980). Über Funde von Enteromorpha jugoslavica Bliding in der Kieler Förde (west. Ostsee). Botanica Marina, 23: 309–312.
- Katoh, K., Misawa, K., Kuma, K.-I. & Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30: 3059–3066.
- Kirkendale, L., Saunders, G.W. & Winberg, P. (2013). A molecular survey of Ulva (Chlorophyta) in temperate Australia reveals enhanced levels of cosmopolitanism. Journal of Phycology, 49: 69–81.
- Koeman, R.P.T. & Van den Hoek, C. (1984). The taxonomy of Ulva (Chlorophyceae) in the Netherlands. British Phycological Journal, 16: 9–53.
- Koeman, R. & Van den Hoek, C. (1982a). The taxonomy of Enteromorpha Link, 1820, (Chlorophyceae) in the Netherlands I: the section Enteromorpha. Archiv für Hydrobiologie, 63: 279–330.
- Koeman, R. & Van den Hoek, C. (1982b). The taxonomy of Enteromorpha Link, 1820, (Chlorophyceae) in the Netherlands I. The section Proliferae. Cryptogamie, Algologie, 3: 37–70.
- Koeman, R. & Van den Hoek, C. (1984). The taxonomy of Enteromorpha Link, 1820, (Chlorophyceae) in the Netherlands III. The sections Flexuosae and Clathratae and an addition to the section Proliferae. Cryptogamie, Algologie, 5: 21–61.
- Kornmann, P. & Sahling, P.-H. (1977). Meeresalgen von Helgoland. Helgoländer wissenschaftliche Meeresunter-suchungen, 29: 1–289.
- Kornmann, P. & Sahling, P.-H. (1978). Die Blidingia-Arten von Helgoland (Ulvales, Chlorophyta). Helgoländer wissenschaftliche Meeresuntersuchungen, 31: 391–413.
- Kornmann, P. & Sahling, P.-H. (1983). Meeresalgen von Helgoland, Ergänzung. Helgoländer wissenschaftliche Meeresuntersuchungen, 36: 1–65.
- Kornmann, P. & Sahling, P.-H. (1994). Marine algae of Helgoland – 2nd Supplement. Helgoländer Meeresunter-suchungen, 48: 365–406.
- Kraft, L.G.K., Kraft, G.T. & Waller, R.F. (2010). Investigations into southern Australian Ulva (Ulvophyceae, Chlorophyta) taxonomy and molecular phylogeny indicate both cosmopolitanism and endemic cryptic species. Journal of Phycology, 46: 1257–1277.
- Kützing, F.T. (1843). Phycologia generalis: oder, Anatomie, Physiologie und Systemkunde der Tange. Brockhaus, Leipzig.
- Lackschewitz, D., Reise, K., Buschbaum, C. & Karez, R. (2015). Neobiota in deutschen Küstengewässern. Eingeschleppte und kryptogene Tier- und Pflanzenarten an der deutschen Nord- und Ostseeküste. Landesamt für Landwirtschaft, Umwelt und ländliche Räume des Landes Schleswig-Holstein (LLUR).
- Linnaeus, C. (1753). Species Plantarum, Exhibentes Plantas Rite Cognitas Ad Genera Relatas: Cum Differentiis Specificis, Nominibus Trivialibus, Synonymis Selectis, Locis Natalibus, Secundum Systema Sexuale Digestas. Trattner, Stockholm.
- Maggs, C.A. & Kelly, J. (2007). Percursaria. In The Green Seaweeds of Britain and Ireland (Brodie, J., Maggs, C.A. & John, D., editors). British Phycological Society, London.
- Maggs, C.A., Blomster, J. & Kelly, J. (2007a). Umbraulva. In The Green Seaweeds of Britain and Ireland (Brodie, J., Maggs, C.A. & John, D., editors). British Phycological Society, London.
- Maggs, C.A., Blomster, J., Mineur, F. & Kelly, J. (2007b). Ulva. In The Green Seaweeds of Britain and Ireland (Brodie, J., Maggs, C.A. & John, D., editors). British Phycological Society, London.
- Nylander, J.A.A. (2004). MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Pankow, H. (1990). Ostsee-Algenflora. Gustav Fischer Verlag, Jena.
- Reed, R.H. & Russell, G. (1978). Salinity fluctuations and their influence on bottle brush morphogenesis in Enteromorpha intestinalis (L) Link. British Phycological Journal, 13: 149–153.
- Reinke, J. (1889). Notiz über die Vegetationsverhältnisse in der deutschen Bucht der Nordsee. Berichte der Deutschen Botanischen Gesellschaft, 7: 367–369.
- Rönnbäck, P., Kautsky, N., Pihl, L., Troell, M., Soerqvist, T. & Wennhage, H. (2007). Ecosystem goods and services from Swedish coastal habitats: identification, valuation, and implications of ecosystem shifts. Ambio, 36: 534–544.
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61: 539–542.
- Rothmaler, W. (1984). Exkursionsflora für die Gebiete der DDR und BRD Band 1: Niedere Pflanzen. Volk und Wissen Volkseigener Verlag, Berlin.
- Saunders, G.W. & Kucera, H. (2010). An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie, Algologie, 31: 487–528.
- Schories, D., Selig, U. & Schubert, H. (2009). Species and synonym list of the German marine macroalgae based on historical and recent records. Meeresbiologische Beiträge, University of Rostock.
- Shimada, S., Hiraoka, M., Nabata, S., Iima, M. & Masuda, M. (2003). Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva. Phycological Research, 51: 99–108.
- Smetacek, V. & Zingone, A. (2013). Green and golden seaweed tides on the rise. Nature, 504: 84–88.
- Spoerner, M., Wichard, T., Bachhuber, T., Stratmann, J. & Oertel, W. (2012). Growth and thallus morphogenesis of Ulva mutabilis (Chlorophyta) depends on a combination of two bacterial species excreting regulatory factor. Journal of Phycology, 48: 1433–1447.
- Staats, M., Cuenca, A., Richardson, J.E., Vrielink-van Ginkel, R., Petersen, G., Seberg, O. & Bakker, F.T. (2011). DNA damage in plant herbarium tissue. PLoS ONE, 6: e28448.
- Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312–1313.
- Steinhagen, S., Karez, R. & Weinberger, F. (2018a). Molecular analysis of Ulva compressa (Chlorophyta, Ulvales) reveals its morphological plasticity, distribution and potential invasiveness on German North Sea and Baltic Sea coasts. European Journal of Phycology, 54: 102–114.
- Steinhagen, S., Karez, R. & Weinberger, F. (2018b). Surveying seaweeds from the Ulvales and Fucales in the world’s most frequently used artificial waterway, the Kiel Canal. Botanica Marina, 62: 51–61.
- Tan, I.H., Blomster, J., Hansen, G., Leskinen, E., Maggs, C.A., Mann, D.G., Sluimam, H.J. & Stanhope, M.J. (1999). Molecular phylogenetic evidence for a reversible morphogenetic switch controlling the gross morphology of two common genera of green seaweeds, Ulva and Enteromorpha. Molecular Biology and Evolution, 16: 1011–1018.
- Vinogradova, K.L. (1969). K. sistematike poryadka Ulvales (Chlorophyta) s.l. A contribution to the taxonomy of the order Ulvales. Botanicheskij Zhurnal SSSR, 54: 1347–1355.
- Weinberger, F., Steinhagen, S., Afanasyev, D.F. & Karez, R. (2018). New records from the southern North Sea and first records from the Baltic Sea of Kornmannia leptoderma. Botanica Marina, 62: 63–73.
- Wichard, T. (2015). Exploring bacteria-induced growth and morphogenesis in the green macroalga order Ulvales (Chlorophyta). Frontiers in Plant Science, 6.
- Wolf, M.A., Sciuto, K., Andreoli, C. & Moro, I. (2012). Ulva (Chlorophyta, Ulvales) biodiversity in the North Adriatic Sea (Mediterranean, Italy): cryptic species and new introductions. Journal of Phycology, 48: 1510–1521.
- Womersley, H.B.S. (1984). The Marine Benthic Flora of Southern Australia. Government Printer, Adelaide.
