ABSTRACT
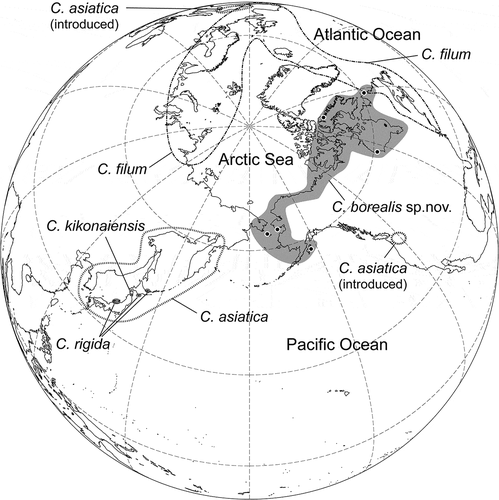
Chorda borealis sp. nov. is newly described from Alaska and northern Canada. Chorda borealis resembles C. filum and C. asiatica in gross morphology, but has a thinner cortical layer than those two taxa and is genetically distinct in the mitochondrial cox1 and cox3, plastid atpB, psbA and rbcL, and nuclear rDNA ITS1-5.8S-ITS2 region DNA sequences. The erect thalli are simple, cord-shaped, medium to light brown, and up to 90 cm in length and 2 mm in diameter. The mid-thallus cortex is composed of 2–4 cells and is 110–230 µm thick. When mature, unilocular zoidangia develop among unicellular clavate paraphyses and measure 30–48 µm long and 8–13 µm in diameter, containing presumably 16 zooids, each with an eyespot. The distributional pattern of C. borealis, which is distributed from the northern Pacific Ocean to the Labrador Sea and connected to the north-western Atlantic through the Arctic, suggests that the dispersal of Chorda species from the Pacific to the Atlantic could have occurred along the coasts of the north-western Arctic Sea.
Introduction
Chorda filum (L.) Stackhouse (Laminariales, Chordaceae) was described from the Atlantic Ocean, but considerably greater species diversity in the genus has been revealed in the Pacific Ocean. Based on morphological and genetic analyses, Kawai et al. (Citation2001) described a second species C. rigida H. Kawai & Arai in the Sea of Japan, followed by the description of C. kikonaiensis H. Sasaki & H. Kawai from the Tsugaru Strait (Sasaki & Kawai, Citation2007). Furthermore, Sasaki & Kawai (Citation2007) showed that the species identified as C. filum in the Pacific Ocean was distinct from true C. filum in the Atlantic Ocean and proposed a new name C. asiatica Sasaki & Kawai. In contrast, the population of Chorda in Washington, USA was shown to be an introduced population of C. asiatica, similar to Mediterranean populations (Kawai et al., Citation2015).
Among the known Chorda species, C. asiatica, C. kikonaiensis and C. rigida are distributed only in the North-west Pacific (Kawai et al., Citation2001, Citation2015), while C. filum is only distributed in the Atlantic. Furthermore, since genetic diversity within Japanese C. asiatica is greater than that within Atlantic C. filum, it was suggested that Chorda originated and diverged in the Pacific Ocean, and spread into the Atlantic through the Arctic (Kawai et al., Citation2016). These findings are consistent with the notion that the Laminariales originated in the Pacific Ocean, based on the rich diversity of laminarialean taxa in the Pacific, the occurrence of basal taxa only in the Pacific, and the distribution of the Pseudochordaceae (Kawai & Kurogi, Citation1985; Kawai & Nabata, Citation1990; Lüning & tom Dieck, Citation1990; Bolton, Citation2010) and Akkesiphycaceae (Kawai & Sasaki, Citation2000), limited to this area.
In previous studies an additional species was discovered in the Bering Sea (northernmost Pacific Ocean), but did not receive formal taxonomic treatment owing to the insufficient number of specimens (Sasaki & Kawai, Citation2007; Kawai et al., Citation2015). Around the same time an undescribed Chorda species was recorded in a barcoding survey of macroalgal species in Churchill, Hudson Bay, Canada, which was hypothesized to be a range extension from the north Pacific flora (Saunders & McDevit, Citation2013). Subsequently, collections from Nome, Alaska, USA and eastern Baffin Island and northern Labrador, Canada were acquired, greatly extending the range for the Churchill genetic group and establishing an Atlantic–Pacific link for this northern entity (Bringloe & Saunders, Citation2019). The possibility that the previous authors had encountered the same species warranted study. The aim of the present study was to resolve this uncertainty through comparative anatomical and genetic analyses, which resulted here in our description of C. borealis sp. nov.
Materials and methods
Materials
For morphological and genetic studies, fresh specimens of the new Chorda species were collected at Kodiak Island (Alaska, USA). Dried specimens collected from St. Lawrence Island (Alaska, USA), Newfoundland, Labrador and Manitoba (Canada) were also used (, Supplementary table S1). The type specimen of the new species (SAP115413) is housed in the Herbarium of the Graduate School of Hokkaido University (SAP), and additional silica gel-dried specimens used for the molecular analyses are deposited in the Kobe University Research Center for Inland Seas. In addition, unialgal gametophyte culture strains isolated from the type material (Kodiak, Alaska, USA) are housed in the Kobe University MacroAlgal Culture Collection (KU-MACC; KU-3361, -3364). Voucher specimens collected from the Arctic and Atlantic Canada are housed in the Connell Memorial Herbarium, Department of Biology, University of New Brunswick.
Morphological observations
For comparisons of the number of cells constituting cortical layers, cross-sections were made by hand using a razor blade and measured midway between the meristem and base. Photomicrographs were taken using a VB–7010 Digital Camera (Keyence, Tokyo, Japan) attached to an Axioplan microscope (Zeiss, Oberkochen, Germany).
Molecular phylogenetic analysis
For molecular phylogenetic studies, field-collected specimens rapidly desiccated in silica gel or fragments taken from the voucher specimens were used for DNA extractions (Supplementary table S1). Genomic DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Polymerase chain reaction (PCR) amplifications of the mitochondrial cox1 and cox3; plastid atpB, psbA and rbcL; and nuclear rDNA ITS1–5.8S–ITS2 region were carried out using the KOD FX (ToYoBo, Osaka, Japan) and the TaKaRa PCR Thermal Cycler Dice (Takara Bio, Kusatsu, Japan). Primers used for PCR and sequencing are listed in Supplementary table S2. The profiles of PCRs were as described by Kawai et al. (Citation2015) and Kojima et al. (Citation2015). After PEG purification (Lis, Citation1980), PCR products were sequenced by a DNA sequencing service (FASMAC, Atsugi, Japan). For molecular phylogenetic analyses, published and newly determined sequence data of the Chorda specimens were used (Supplementary table S1). Cox1 and cox3 sequences for 53 taxa, atpB and psbA sequences for 11 taxa, rbcL sequences for 29 taxa, and rDNA ITS1–5.8S–ITS2 region for 75 taxa were aligned using the program MAFFT v.6 (Katoh & Toh, Citation2008) and then manually adjusted prior to phylogenetic analyses. The ambiguous regions of the alignments were removed. Samples with identical nucleotide sequences were treated as a single operational taxonomic unit (OTU). Akkesiphycus lubricum Yamada & Tak.Tanaka, Pseudochorda gracilis H. Kawai & Nabata and P. nagaii (Tokida) Inagaki were selected as the outgroup for the analyses, except for the aligned ITS1–5.8S–ITS2 rDNA analysis. Phylogenetic analyses of the aligned sequences from each data set (data set 1: 42 OTUs, cox1+cox3 genes, total 1362 bp; data set 2: 17 OTUs, rbcL gene, 1356 bp; data set 3: 57 OTUs, ITS1+5.8S+ITS2 rDNA region, 542 bp; data set 4: 10 OTUs, atpB+psbA+rbcL genes, total 3450 bp; data set 5: 11 OTUs, cox1+cox3+atpB+psbA+rbcL genes, total 4812 bp) were subjected to maximum likelihood (ML) and Bayesian analysis (BI). For Maximum likelihood analysis, we used RAxML GUI v.1.31 (Silvestro & Michalak, Citation2012) run to conduct 10,000 Rapid Bootstrap searches followed by an ML search. The GTR + G model were implemented with partitioning by gene or region for the analyses. Bayesian analyses were run using MrBayes v.3.2.2 (Ronquist et al., Citation2012), as described previously (Kawai et al., Citation2015). The Bayesian analyses were initiated with a random starting tree and ran four chains of Markov chain Monte Carlo iterations simultaneously for 2 000 000 generations, keeping one tree every 500 generations. The first 5 000 trees sampled were discarded as ‘burn-in’, based on the stationarity of ln L as assessed using Tracer v.1.7.1 (Rambaut & Drummond, Citation2018). A consensus topology and posterior probability values were calculated from the remaining trees. The appropriate substitution models were selected on the basis of hierarchical likelihood ratio test using MrModeltest 2.3 (Nylander, Citation2004) with partitioning by gene or region for the analyses. The Bayesian analyses are summarized in Supplementary table S3.
Results
Habit and morphology
Sporophytes of Chorda borealis sp. nov. were collected from diverse habitats. At St. Lawrence Island, Alaska, they grew on pebbles in a shallow waterway connecting the open sea and a salt marsh near Gambell. At Kodiak Island, Alaska, the specimens were collected on an artificial substrate of a pier in Kodiak. In Canada, specimens were collected on pebble, cobble, solid rock or dead shells ranging from mid intertidal pools to subtidal at 10 m (as were subsequent specimens from Nome; Bringloe & Saunders, Citation2019).
The sporophytic thalli were simple, cord-shaped, medium to light brown, and up to ~90 cm in length and 2 mm in diameter (–, ). This species resembles Chorda filum and C. asiatica and they are virtually indistinguishable in gross morphology. Mid-thallus the cortex was composed of 2–4 cell layers and was 110–230 µm thick (, ). When mature, unilocular zoidangia were formed among clavate unicellular paraphyses (, ). Unilocular zoidangia measured 30–48 µm long and 8–13 µm in diameter, containing presumably 16 zooids, each with an eyespot ().
Table 1. Comparison of selected morphological features and distributional range of Chorda species
Figs 2–4. Habit of Chorda borealis sp. nov. Fig. 2. Type specimen; fertile sporophyte (Crescent Harbor, Kodiak, Kodiak Island, Alaska, USA on 11 July 2016 by H. Kawai. SAP115413). Fig. 3. Young immature sporophytes (St. Lawrence Island, Bering Sea, USA on 6 August 1996 by H. Kawai). Fig. 4. Young sporophyte (Gordon Pt., Churchill, Manitoba, Canada on 21 August 2006 by G.W. Saunders, B. Clarkston and D. McDevit; GWS005304).
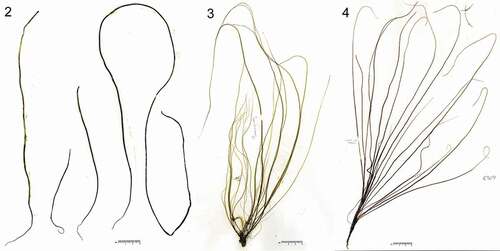
Figs 5–9. Morphology of sporophytes and gametophytes of Chorda borealis sp. nov. 5–7, Type specimen collected on 11 July 2016 by H. Kawai (sporophyte, SAP115413). 8, 9, Gametophytes. Fig. 5. Cross section in the middle portion showing paraphysis, cortical (asterisk) and medullary layers. Double arrowhead indicates trumpet-shaped hyphae. Fig. 6. Cross section showing paraphysis (arrow), unilocular zoidangium (arrowhead) and cortical cell (asterisk). Fig. 7. Surface view showing unilocular zoidangium with release structure at the tip (arrowhead), paraphysis (arrow) and phaeophycean hair (double-headed arrow); Fig. 8. Putative male gametophyte (KU-3367). Fig. 9. Putative female gametophyte (KU-3364).
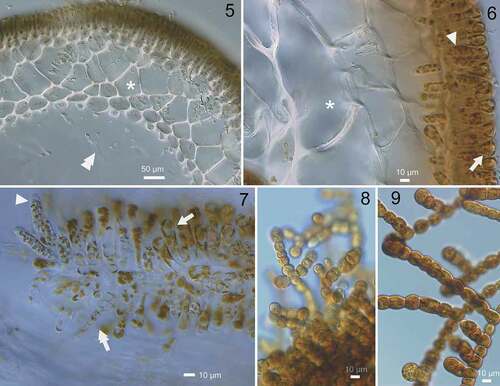
The gametophytes showed typical morphology as in other Chorda species: filaments of presumptive male gametophytes (; ~10–15 µm in diameter) were considerably thinner than those of presumptive female gametophytes (; ~15–20 µm in diameter).
Molecular phylogenetic analysis
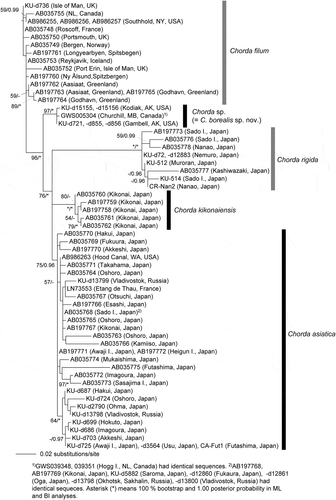
In the molecular phylogeny based on concatenated DNA sequences of mitochondrial cox1+cox3 genes (), the plastid rbcL genes () and the ITS1+5.8S+ITS2 rDNA region (), Chorda specimens comprised five independent clades corresponding to distinct species: C. asiatica, C. filum, C. kikonaiensis, C. rigida and the new species (= C. borealis sp. nov.). Tree topologies were somewhat different among the analyses, but the new species was either sister to C. filum or all of the other species of Chorda (–). In a rooted tree of mitochondrial cox1+cox3 genes, C. kikonaiensis branched first in the Chorda clade, and the clade of C. asiatica/C. rigida was sister to the C. filum/C. borealis clade (). Statistical support was generally high (86–100%) except for the branch connecting C. asiatica/C. rigida and C. filum/C. borealis (). The genetic divergences of cox1 and cox3 between Chorda borealis sp. nov. and C. filum were 6.3–6.8% and 6.5–6.6%, respectively. These values were greater than those between related laminarialean species (Neoagarum fimbriatum vs. N. oharaense: 3.7% (cox1; AB775225, LC148148, LC148149) and 2.4–2.5% (cox3; AB775242, LC148163, LC148164), Agarum clathratum vs. A. turneri: 2.9–3.1% (cox1; LC148146, LC148147, LC149897, LC148151) and 1.5–1.9% (cox3: LC148160, LC148161, LC149898, LCLC148166), Saccharina angustata vs. S. japonica: 3.8% (cox1; AP011493, AP011498) and 2.3% (cox3; AP011493, AP011498)).
Fig. 10. Maximum likelihood (ML) molecular phylogeny based on concatenated DNA sequences of mitochondrial cox1 and cox3 genes (1362 bp). Numbers below the branches indicate the bootstrap values (BP, right) and Bayesian posterior probabilities (PP, left). Only the BP (≥ 50%) and PP (≥ 0.95) are shown.
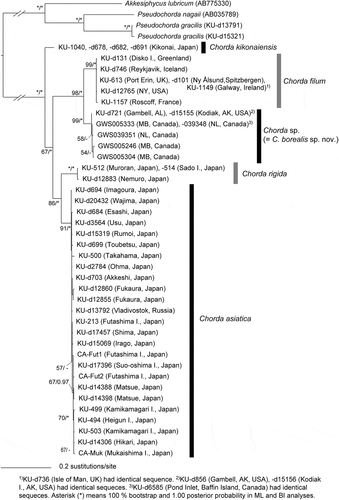
Fig. 11. Maximum likelihood (ML) molecular phylogeny based on DNA sequences of plastid rbcL gene (1356 bp). Numbers below the branches indicate the bootstrap values (BP, right) and Bayesian posterior probabilities (PP, left). Only the BP (≥ 50%) and PP (≥ 0.95) are shown.
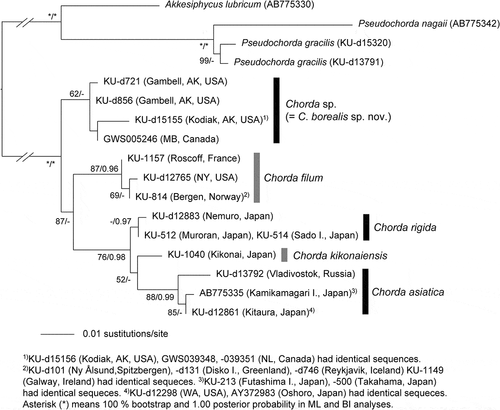
Fig. 12. Maximum likelihood (ML) molecular phylogeny based on concatenated DNA sequences of 5.8S, ITS1 and ITS2 rDNA (542 bp). Numbers below the branches indicate the bootstrap values (BP, right) and Bayesian posterior probabilities (PP, left). Only the BP (≥ 50%) and PP (≥ 0.95) are shown.
In the plastid rbcL gene tree, statistical support was not typically low and monophyly of C. borealis was weak (). However, in the tree of concatenated atpB + psbA + rbcL genes, monophyly of this species had high support (94%), but C. borealis branched first among species of Chorda and the branching order was different from other trees (Supplementary fig. S1). In the concatenated tree of five mitochondrial and plastid genes, Chorda species formed two clades of C. filum/C. borealis and C. kikonaiensis/C. rigida/C. asiatica (Supplementary fig. S2).
In the unrooted tree of nuclear ITS1+5.8S+ITS2 rDNA sequences, independence of the five species was supported, although C. asiatica was not monophyletic (). There was no sign of gene flow by hybrid formation among the species.
In addition, we performed an analysis for data sets 1, 2, 4 and 5 with partitioning by codon for cox1, cox3, atpB, psbA and rbcL genes. All phylogenetic trees generated with or without partitioning by codon showed almost the same topologies (data not shown).
Discussion
Chorda borealis resembles C. filum and C. asiatica in gross morphology and can scarcely be distinguished by external morphology and texture (). In contrast, C. rigida is distinctive in having rather tough thalli, whereas C. kikonaiensis has remarkably softer thalli reminiscent of Scytosiphon spp. The number of cells making up the cortical layer is considered to be a reliable taxonomic character with which to distinguish Chorda sporophytes, because it is predetermined in the meristem and shows considerable stability irrespective of environmental conditions (Kogame & Kawai, Citation1996; Kawai et al., Citation2001). Also, this morphological feature strongly influences the texture of the thallus: tough C. rigida has a thicker cortical layer, whereas soft C. kikonaiensis has a thin cortical layer. Chorda asiatica has a cortex of 4–8 cell layers (Sasaki & Kawai, Citation2007), whereas C. filum displays considerable variation, being composed of 4–10 cell layers (Kylin, Citation1918; South & Burrows, Citation1967; Russell, Citation1985; Kawai et al., Citation2001). Mid-thallus C. borealis has 3–4 cell layers and is thus morphologically distinguishable from C. asiatica and C. filum in having a thinner cortical layer.
The range of Chorda borealis in the north-eastern Arctic Sea is not clear because of limited sampling for that area, but its known distribution extends from the North Pacific to the North Atlantic through the Arctic Sea, overlapping with that of Chorda filum. Molecular phylogenetic analyses suggest that Chorda species originated in the Pacific and spread into the Atlantic. In almost all phylogenetic analyses, Chorda asiatica, C. kikonaiensis and C. rigida, which are distributed in north-eastern Asia, formed monophyletic clades and were basal in the Chorda clade suggesting their origin in the north-western Pacific. The distributional pattern of C. borealis, which is distributed from the North Pacific Ocean to the Labrador Sea connected to the north-western Atlantic through the Arctic, suggests that the dispersal of Chorda species from the Pacific to the Atlantic could have occurred along the coasts of the north-western Arctic Sea. Since the distributional ranges of C. filum and C. borealis appear to overlap in north-eastern Canada (Labrador and its vicinity), intensive sampling and genetic analyses including nuclear markers may reveal whether the two species interact ecologically or genetically.
Description and diagnosis
Chorda borealis H. Kawai, T. Hanyuda & G. W. Saunders sp. nov. (–).
Sporophyte macroscopic, epilithic, intertidal, solitary or sparse, simple, cord-shaped, up to 91 cm in length and up to 2.0 mm in diameter, medium to dark brown, parenchymatous, hollow, composed of hyphae, cortex, epidermis, hairs, unicellular paraphyses without mucilaginous cap, and disc-shaped rhizoidal holdfast. Cortex composed of 2–4 cells. Unilocular zoidangia sessile and narrowly ovate.
This species is similar to Chorda filum and C. asiatica in habit, but it differs in the lower number of cell layers composing the cortex. Nucleotide sequences of mitochondrial cox1 and cox3, plastid atpB, psbA and rbcL, and nuclear rDNA ITS1–5.8S–ITS2 region are also distinctive.
Holotype: SAP115413. 2016.7.11. Collected at Crescent Harbor, Kodiak, Kodiak Island, Alaska, USA by H. Kawai (, –).
Etymology: The specific epithet originates from the distributional range of the species.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at http://10.1080/09670262.2019.1610580
Supplementary table S1. Origin of specimens and sequence data used for molecular phylogenetic analysis, including their database accession numbers. Accession numbers in bold were newly determined for this study. Specimen codes in [KU-###] correspond to KU-MACC (Kobe University MacroAlgal Culture Collection) strain code, and [KU-d###] corresponds to silica gel-dried specimens housed at Kobe University Research Center for Inland Seas.
Supplementary table S2. List of primers used for PCR and sequencing.
Supplementary table S3. Summary for the Bayesian analyses based on cox1, cox3, atpB, psbA, rbcL and ITS1-5.8S-ITS2 rDNA data sets.
Supplementary fig. S1. Maximum likelihood (ML) molecular phylogeny based on concatenated DNA sequences of plastid atpB, psbA and rbcL genes (3450 bp). Numbers below the branches indicate the bootstrap values (BP, right) and Bayesian posterior probabilities (PP, left). Asterisk (*) indicates 100% BP and 1.00 PP in ML and BI analyses. Only the BP (≥ 50%) and PP (≥ 0.95) are shown.
Supplementary fig. S2. Maximum likelihood (ML) molecular phylogeny based on concatenated DNA sequences of mitochondrial cox1 and cox3; plastid atpB, psbA and rbcL genes (4812 bp). Numbers below the branches indicate the bootstrap values (BP, right) and Bayesian posterior probabilities (PP, left). Asterisk (*) indicates 100% BP and 1.00 PP in ML and BI analyses. Only the BP (≥ 50%) and PP (≥ 0.95) are shown.
Author contributions
H. Kawai: Original concept, collection of specimens, morphological studies, culture studies, drafting and editing manuscript; M. Suzuki: genetic analyses; G. Saunders: collection of specimens, genetic analyses, editing manuscript; T. Hanyuda: genetic analyses
TEJP-2018-0136-File010.tif
Download TIFF Image (1 MB)TEJP-2018-0136-File009.tif
Download TIFF Image (969.6 KB)TEJP-2018-0136-File008.doc
Download MS Word (218.5 KB)Acknowledgements
We are grateful to Dr Eric Henry for critically reading and improving the manuscript, and Dr Sandra Lindstrom for support in collecting specimens. GWS thanks the many collectors from the Saunders lab who have accompanied him to the Arctic and contributed to gathering some of the specimens used here.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolton, J. (2010). The biogeography of kelps (Laminariales, Phaeophyceae): a global analysis with new insights from recent advances in molecular phylogenies. Helgoland Marine Research, 64: 263–279.
- Bringloe, T.T. & Saunders, G.W. (2019). DNA barcoding of the marine macroalgae from Nome, Alaska (Northern Bering Sea) reveals many trans-Arctic species. Polar Biology, 5: 851–864.
- Katoh, K. & Toh, H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics, 9: 286–298.
- Kawai, H. & Kurogi, M. (1985). On the life history of Pseudochorda nagaii (Pseudochordaceae fam. nov.) and its transfer from the Chordariales to the Laminariales (Phaeophyta). Phycologia, 24: 289–296.
- Kawai, H. & Nabata, S. (1990). Life history and systematic position of Pseudochorda gracilis sp. nov. (Laminariales, Phaeophyceae). Journal of Phycology, 26: 721–727.
- Kawai, H. & Sasaki, H. (2000). Molecular phylogeny of Akkesiphycus and Halosiphon (Laminariales), resulting in the circumscription of the new families Akkesiphycaceae and Halosiphonaceae. Phycologia, 39: 416–428.
- Kawai, H., Sasaki, H., Maeda, Y. & Arai, S. (2001). Morphology, life history and molecular phylogeny of Chorda rigida sp. nov. (Laminariales, Phaeophyceae) from the Sea of Japan and the genetic diversity of Chorda filum. Journal of Phycology, 37: 130–142.
- Kawai, H., Hanyuda, T., Mumford, T. & Waaland, J.R. (2015). An introduced population of Chorda asiatica (Chordaceae, Laminariales) in Puget Sound, Pacific coast of North America. Phycological Research, 63: 154–158.
- Kawai, H., Hanyuda, T. & Uwai, S. (2016). Evolution and biogeography of laminarialean kelps. In Seaweed Phylogeography (Hu, Z. & Fraser, C., editors), 227–249. Springer, Berlin.
- Kogame, Y. & Kawai, H. (1996). Development of the intercalary meristem in Chorda filum (Laminariales, Phaeophyceae) and other primitive Laminariales. Phycological Research, 44: 247–260.
- Kojima, R., Hanyuda, T. & Kawai, H. (2015). Taxonomic re-examination of Japanese Halimeda species using genetic markers, and proposal of a new species Halimeda ryukyuensis (Bryopsidales, Chlorophyta). Phycological Research, 63: 178–188.
- Kylin H. (1918). Studien über die Entwicklungsgeschichte der Pheophyceen. Svensk Botanisk Tidskrift, 12: 1–64.
- Lis, J.T. (1980). Fractionation of DNA fragments by polyethylene glycol induced precipitation. Methods in Enzymology, 65: 347–353.
- Lüning, K. & tom Dieck, I. (1990). The distribution and evolution of the Laminariales: North Pacific–Atlantic relationships. In NATO ASI Series G22. Evolutionary Biogeography of the Marine Algae of the North Atlantic (Garbary, D.J. & South, G.R., editors), 187–204. Springer Verlag, Berlin.
- Nylander, J.A.A. (2004). MrModeltest 2.3. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- Oltmanns, F. (1922). Morphologie und Biologie der Algen, ed. 2, II. Fischer, Jena.
- Rambaut, A. & Drummond, A. J. (2018). Tracer v.1.7.1. http://tree.bio.ed.ac.uk/software/tracer/.
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L, Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M. A. & Huelsenbeck, J.P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61: 539–542.
- Russell, G. (1985). Some anatomical and physiological differences in Chorda filum from coastal waters of Finland and Great Britain. Journal of the Marine Biological Association of the United Kingdom, 65: 343–349.
- Sasaki, H. & Kawai, H. (2007). Taxonomic revision of the genus Chorda (Chordaceae, Laminariales) based on sporophyte anatomy and molecular phylogeny. Phycologia, 46: 10–21.
- Saunders, G.W. & McDevit, D.C. (2013). DNA barcoding unmasks overlooked diversity improving knowledge on the composition and origins of the Churchill algal flora. BMC Ecology, 13: 9. doi:https://doi.org/10.1186/1472-6785-13-9.
- Silvestro, D. & Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution, 12: 335–337.
- South, G.R. & Burrows, E.M. (1967). Studies on marine algae of the British Isles. 5. Chorda filum (L.) Stackh. British Phycological Bulletin, 3: 379–402.