ABSTRACT
The gelatinous, calciferous red alga Renouxia antillana was described in 1995 based on material from Guadeloupe, French West Indies, and accommodated in a new family and order (Rhodogorgonaceae, Rhodogorgonales) along with the genus Rhodogorgon from Belize and Caribbean Panama. For more than 20 years, Renouxia has remained monotypic, with rare reports in the Caribbean and the Indo-Pacific (from Réunion Island to French Polynesia). DNA-based analyses of recently collected Renouxia specimens from Egypt showed that they are not conspecific with the Caribbean R. antillana and are described as R. marerubra sp. nov. Uncorrected p-distances between the Red Sea specimens and the generitype were 8.0% for COI, 6.5-7.3% for rbcL and 3.1% for UPA. Morphological and anatomical features are also presented for the newly described species and compared to its congener, with the first documented report of monoecism in the Rhodogorgonales. Besides the new record of Renouxia from the Red Sea, the geographic distribution of the genus is here extended with additional records from Sri Lanka, Indonesia, as well as the islands of Guam and Kosrae in the Western Pacific. The UPA phylogeny suggests that these new distribution records may also represent undescribed species, with representatives in two distinct genetic groups.
INTRODUCTION
Littler et al. (Citation1989) reported an unknown red alga from the Caribbean as ‘quite an oddity’, with the unusual presence of calcium carbonate within its highly gelatinous fronds, to which no species name, genus, order or even family could be assigned.
Six years later, the genus Renouxia Fredericq & J.N.Norris was erected to accommodate the species R. antillana Fredericq & J.N.Norris, a taxon that was placed together with Rhodogorgon J.N.Norris & K.E.Bucher in the new family Rhodogorgonaceae, order Rhodogorgonales (Fredericq & Norris, Citation1995). The order currently encompasses four species: Renouxia antillana; Rhodenigma contortum J.A.West, Verbruggen & Loiseaux; Rhodogorgon ramosissima J.N. Norris & Bucher; and Rhodogorgon flagellifera Huisman (Guiry & Guiry, Citation2019). Earlier, Ogden (Citation1992) synonymized Rhodogorgon carriebowensis J.N. Norris & Bucher from Belize with R. ramosissima, suggesting that habit differences resulted from differences in hydrodynamic conditions.
The genus Renouxia remained monotypic with new records of the alga reported from the Caribbean (Jamaica, Freshwater et al., Citation1994; Puerto Rico, Ballantine et al., Citation2004), and from various tropical locations in the Indo-Pacific (, with ecoregions based on Spalding et al., Citation2007): Tanzania and Rodrigues (de Clerck et al., Citation2004), Réunion Island (Zubia et al., Citation2018), Thailand (Liao & Aungtonya, Citation2000), Philippines (Kraft et al., Citation1999), Papua New Guinea (Millar et al., Citation1999; Coppejans & Millar, Citation2000), French Polynesia (Payri et al., Citation2000), New Caledonia (Bittner et al., Citation2011), American Samoa (Littler & Littler, Citation2003), and Fiji (N’Yeurt, Citation2001). Besides the original 1995 description, little is known about the morphological and reproductive diversity of these Renouxia collections, with only a few studies scarcely reporting on internal anatomy (e.g. N’Yeurt, Citation2001; Lewmanomont & Noiraksa, Citation2010). Although DNA sequences of Renouxia taxa have been included in various studies (e.g. Freshwater et al., Citation1994; Le Gall & Saunders, Citation2007; Sauvage et al., Citation2016; West et al., Citation2016; Lee et al., Citation2018), they were obtained from a limited number of specimens (two from the Caribbean and one from the Red Sea, the latter obtained in the present study) and therefore represent a limited perspective on the genetic diversity of the genus. The Rhodogorgonales, along with the Corallinales, Hapalidiales and Sporolithales, represent four well-supported, monophyletic orders in the subclass Corallinophycidae of the class Florideophyceae (Yang et al., Citation2016; Lee et al., Citation2018).
Fig. 1. Geographic distribution of the genus Renouxia. Ecoregions (Spalding et al., Citation2007) with previous reports are shaded in dark grey, and those with new reports in light grey. Collection sites of specimens included in the present study are colour-coded according to the different putative species in (see online version for colours). The type locality of R. antillana (the type species of the genus) is shown by a black star.
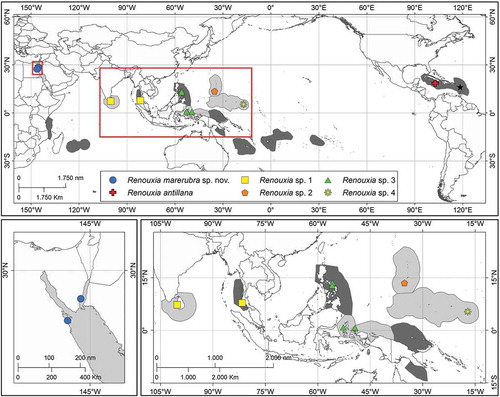
Recent fieldwork conducted in the Red Sea, in the Indo-Pacific and in some Pacific Islands revealed new records of Renouxia antillana. The present work aims to (1) use a multigene approach to assess the phylogenetic position of the new specimens and their relationship to other taxa in the Rhodogorgonales, and (2) describe the morphological and reproductive features of Renouxia from Egypt.
Materials and methods
Specimens resembling the typical habit of Renouxia antillana were recently collected () along the Red Sea coast of Egypt (), in the Indo-Pacific basin (–) and the Pacific Ocean (). Collection data are provided in Supplementary table S1. Newly collected specimens were preserved and characterized following Gabriel et al. (Citation2017), with microphotographs and measurements (presented as length × width) taken using an optical microscope (Leica DM2500, Wetzlar, Germany) with a microphotography system (Leica LAS V3.8). DNA was extracted and three genetic markers (mitochondrial COI, cytochrome c oxidase subunit 1, 5’-prime end; plastid rbcL, ribulose-1,5- bisphosphate carboxylase/oxygenase large subunit; and the Universal Plastid Amplicon (UPA, domain V of the plastid large subunit 23S, Sherwood et al., Citation2010) were sequenced following the protocols in Gabriel et al. (Citation2017). Additional sequences were retrieved from GenBank (Supplementary table S1) and members of Sporolithales were used as an outgroup based on Yang et al. (Citation2016) and Lee et al. (Citation2018).
Figs 2–6. In situ pictures of Renouxia spp. from different locations. Fig. 2. Renouxia marerubra sp. nov. (LAF6170) from Hurghada, Egypt; Fig. 3. Renouxia sp. 1 (SGAD1801001) from Phuket, Thailand; Fig. 4. Renouxia sp. 3 (SGAD0911025) from Ternate, Indonesia; Fig. 5. Renouxia sp. 3 (SGAD1606052) from Catanduanes, Philippines; Fig. 6. Renouxia sp. 4 (GH13674) from Kosrae, Federated States of Micronesia.

Sequence divergences were estimated in MEGA v.7.0.18 (Kumar et al., Citation2016), using the uncorrected p-distance method (Nei & Kumar, Citation2000), i.e. the proportion of nucleotide sites at which the sequences being compared are different without any correction for multiple substitutions at the same site, substitution rate biases, or differences in evolutionary rates among sites. Maximum likelihood (ML) and Bayesian inference (BI) phylogenetic analyses were implemented on the CIPRES Science Gateway (https://www.phylo.org/; Miller et al., Citation2010). ML analyses were conducted in RAxML v.8 (Stamatakis, Citation2014) using the GTR+I+G model and the default rapid hill-climbing algorithm. Nodal support was estimated using a non-parametric bootstrap estimate with 1000 replicates and a random starting tree. Bayesian inference reconstructions were performed in MrBayes v.3.2.6 (Ronquist et al., Citation2012), using 10 million generations with two independent runs of four Monte Carlo Markov chains (MCMC), sampled every 1000 generations. After discarding the initial 20% generations as burn-in, the remaining trees were used to construct the 50% majority-rule consensus tree to estimate the Bayesian posterior probabilities (PP). Tree nodes with ML bootstrap value (BS) greater than 80% and BI posterior probability (PP) greater than 0.95 were considered as strongly supported (Erixon et al., Citation2003).
Results
The level of genetic divergence of the different markers was consistent with previous studies (e.g. Gabriel et al., Citation2017), with COI being the most variable and UPA the most conservative marker (, Supplementary table S2). Likewise, the success of DNA sequencing varied among markers, with UPA being the easiest and COI the most difficult to amplify (Gabriel et al., Citation2017). Uncorrected p-distances between the Red Sea specimens and the generitype were 8.0% for COI, 6.5–7.3% for rbcL and 3.1% for UPA. Although COI exhibited higher divergence values within the genus, rbcL presented higher values between the genera of Rhodogorgonaceae probably because of increased genetic saturation of COI at the genus level (Gabriel et al., Citation2017).
Table 1. COI (lower left) and rbcL (upper right) uncorrected p-distances between members of the Rhodogorgonales. Note: COI distance values are higher than rbcL within the genus, and lower between genera. p-distances are colour-coded from low values in yellow to high values in green (see online version for colours).
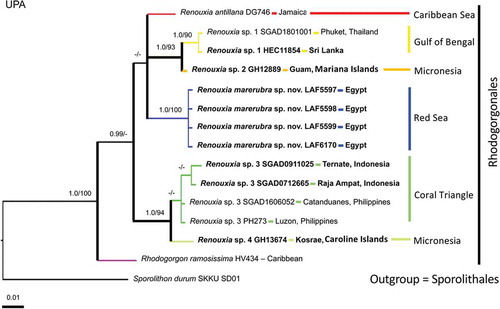
Phylogenetic analyses of the two most informative genes resulted in similar topologies (), therefore COI and rbcL were concatenated into a single dataset (), while UPA sequences were only used to report new records of the genus (). The UPA phylogeny suggests the existence of additional undescribed species within two main groups of the genus Renouxia (). One group, without strong BS and PP support, has representatives from the Caribbean Sea (R. antillana), the Red Sea (R. marerubra), the Bay of Bengal (Renouxia sp. 1), and the Mariana Islands (Renouxia sp. 2), while a second group includes representatives from the Coral Triangle (Renouxia sp. 3) and the Caroline Islands (Renouxia sp. 4).
Fig. 7. Phylogeny of the genus Renouxia, including R. marerubra sp. nov. (indicated in bold) based on Bayesian inference analyses of COI sequences (left) and rbcL sequences (right). Bayesian inference posterior probabilities (PP) and Maximum likelihood bootstrap (BS) presented as ‘PP/BS’ near branches ('-' indicates BS below 80). Scale bar indicates number of substitutions per site. The other ingroup taxa are members of the Rhodogorgonales and the outgroup taxa are members of the Sporolithales.
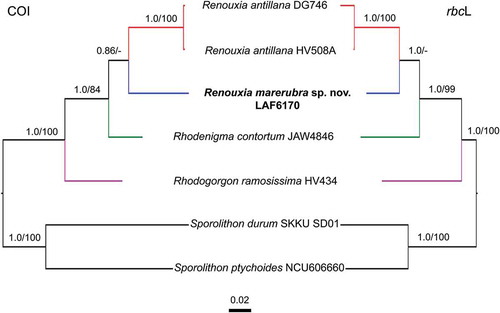
Fig. 8. Phylogenetic reconstruction of Renouxia, including R. marerubra sp. nov. (indicated in bold) based on Bayesian inference of a concatenated dataset of COI and rbcL sequences. Bayesian inference posterior probabilities (PP) and Maximum likelihood bootstrap (BS) presented as ‘PP/BS’ near branches ('-' indicates BS below 80). Scale bar indicates number of substitutions per site. Additional ingroup taxa represent members of the Rhodogorgonales while the outgroup taxa are members of the Sporolithales.
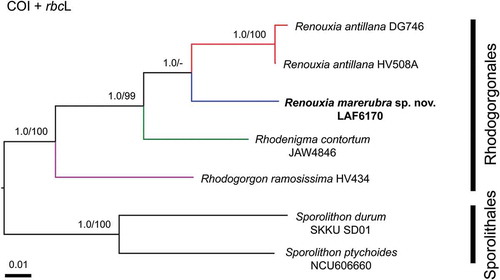
Fig. 9. Phylogenetic reconstruction of Renouxia based on Bayesian inference analysis of UPA sequences. Bayesian inference posterior probabilities (PP) and Maximum likelihood bootstrap (BS) presented as ‘PP/BS’ near branches ('-' indicates PP below 0.95 and BS below 80). Scale bar indicates number of substitutions per site. New Renouxia spp. records are indicated in bold (Red Sea, Sri Lanka, Indonesia, Guam and Kosrae). The additional ingroup taxon is a member of the Rhodogorgonales and the outgroup taxon is a member of the Sporolithales.
Figs 10–12. In vivo pictures of Renouxia marerubra sp. nov. Fig. 10. Holotype, monoecious gametophyte, LAF5597. Fig. 11. Monoecious gametophyte, LAF5598. Note the sub-dichotomous habit of the branches. Fig. 12. Sterile specimen, LAF6170. Note: skeleton-like refractive axis formed by the shade of calciferous cells borne in the inner cortex. Scale bars = 1.0 cm.
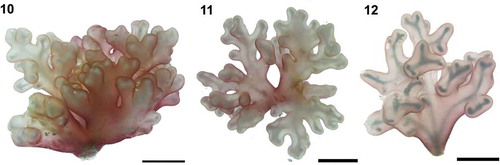
Figs 13–20. Renouxia marerubra sp. nov. Fig. 13. Holotype (herbarium-pressed), monoecious gametophyte, LAF5597, Dahab, Egypt. Fig. 14. Pseudo-dichotomously branched cortical fascicles with numerous rhizoids (arrows). Fig. 15. Adventitious rhizoids (arrows) borne at or close to cortical dichotomies, growing inward to form the medulla. Fig. 16. Clavulate calciferous cells (arrows) in inner cortex (calcite husks not visible in stained material). Note: remnants of disrupted cell walls (arrowheads). Fig. 17. Partially dissolved calcite husks giving calciferous cells a hairy appearance (arrows) in non-stained material. Fig. 18. Pseudo-dichotomous cortical fascicles bearing carpogonial branch initials (arrows). Fig. 19. Mature two-celled carpogonial branch (arrow) in outer cortex. Fig. 20. Supporting cell (sc) bearing a carpogonial branch comprised of a hypogynous cell (hy), carpogonium (cp) and elongated trichogyne (t). Note: darkly staining carpogonium and tip of trichogyne. Scale bars: Fig. 13 = 1.0 cm, Fig. 14 = 200 μm, Figs 15, 18 = 100 μm, Figs 16, 19 = 50 μm, Figs 17, 20 = 20 μm.
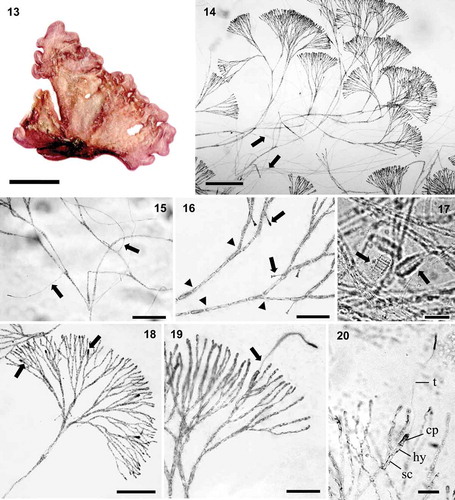
Figs 21–29. Renouxia marerubra sp. nov. Fig. 21. First gonimoblast initials (arrows). Fig. 22. Numerous gonimoblast filaments (arrows). Fig. 23. Gonimoblast filaments (arrows) dividing and growing outwardly reaching the cortex surface. Fig. 24. Gonimoblast filaments with swollen apical cells (arrows). Fig. 25. Remnant terminal carposporangial cell walls (arrows) borne on swollen gonimoblast cells (arrowheads). Fig. 26. Remnant terminal and lateral carposporangial cell walls (arrows) borne on swollen gonimoblast cells (arrowhead), from which secondary gonimoblast initials are issued (double arrowheads). Fig. 27. Spermatangial parent cells (arrows) on the outer cortex. Fig. 28. Fan-like spermatangia bearing numerous spermatia (arrows). Fig. 29. Old clusters of spermatangial parent cells (arrows) located in mid-cortex. Scale bars: Figs 21–24, 27, 29 = 50 μm, Fig. 25 = 100 μm, Figs 26, 28 = 20 μm.
Based on molecular and morphological data, the new species from the Red Sea is here described as follows:
Renouxia marerubra D.Gabriel, J.N.Norris & Fredericq, sp. nov. (, –)
Holotype: LAF 5597 (monoecious gametophyte), collected by D. Gabriel, T. Sauvage & W.E. Schmidt on 5 May 2012 at 17 m depth. Deposited in the Herbarium of the University of Louisiana at Lafayette (LAF, USA).
Type locality: The Island, Dahab, Gulf of Aqaba, Egypt (28°28'38.3"N, 34°30'45.6"E).
Etymology: The name marerubra refers to its collection site in the Red Sea.
Description
Gelatinous pink thallus, from 1.9 to 3.6 cm in height and 2.3 to 4.3 cm in width (, –), sub-dichotomously branched growing from a short firm stalk, attached to the substratum by a discoid holdfast (0.2–0.5 cm in diameter). A refractive axis reminiscent of a ‘skeleton’ (, , ) can be observed using strong lighting, such as under a camera flash or over drawing light box. This axis is not formed by a solid structure, but by the overlap of calcite husks occurring at the same position on cortical fascicles throughout the thallus. Cortical fascicles are pseudo-dichotomously branched () with adventitious rhizoidal filaments growing inward towards the medulla (, ). Inner cortex contains 1–2 clavulate calciferous cells (12–22 × 4–6 μm) per cortical branch (), though calcite husks become dissolved when stained. In unstained material, calciferous cells look hairy () due to partial dissolution of the husk structures under prolonged preservation in diluted formalin/seawater. Remnants of disrupted cell walls are found in inner cortical filaments, usually before a dichotomy ().
Gametophyte monoecious. Numerous carpogonial branch initials observed close to the outer cortex (), although mature carpogonial branches are usually found in the inner cortex (, ). Carpogonial branch two-celled, sessile () or borne on a supporting cell (), composed of a subhypogynous cell (8–19 × 4–5 μm), and a carpogonium longer (14–28 μm) and thicker (4–6 μm) than cortical cells in the same position, bearing a straight, terminally enlarged trichogyne (123–195 μm). Carpogonium and the terminal, swollen portion of trichogyne stain densely with aniline blue (, ). After presumed fertilization, the fertilized carpogonium bifurcates () and continues to branch, resulting in numerous gonimoblast filaments (, ), which grow outwardly. At the thallus surface, the tips of the gonimoblast thallus are enlarged and produce carposporangia (). Mature carposporangia were not observed, though structures composed of a series of empty oval sporangial walls (24–34 × 17–27 μm) were observed in the outer cortex (, ), grown from enlarged gonimoblast filaments, from which secondary gonimoblast filaments are issued (). It is possible that carpospores are released sequentially as they mature from gonimoblast filaments. Outer cortical cells are enlarged and darkly stained as they become spermatangial parent cells (). A mature fan-like spermatangial cluster is formed (), with 2–3 spermatangial parent cells (4.2–5.1 × 1.9–2.6 μm), each bearing 1–3 spermatia (1.9–2.7 μm in diameter). After the release of spermatia, cortical branches continue to grow beyond the clusters of spermatangial parent cells, whose remnants remain enlarged (13–22 × 5 μm) and darkly stained in a mid-cortical zone (). Tetrasporangia were not observed.
Additional specimens examined
LAF5598 (monoecious gametophyte), LAF5599 (monoecious gametophyte), Dahab; LAF6170 (sterile), Hurghada – Northern Red Sea, Egypt.
Representative sequences deposited in GenBank
MK174358 (COI), MK174342 (rbcL) and MK174346 (UPA), generated from specimen LAF6170. KU362135 (partial elongation factor Tu; Sauvage et al., Citation2016), MH281629 (complete plastid genome) and MH281622 (complete mitochondrion genome), MK091141 (partial 5.8S ribosomal RNA, internal transcribed spacer 2, and partial large subunit ribosomal RNA), MK091140 (partial small subunit ribosomal RNA; Lee et al., Citation2018) are also available for this specimen.
Remarks
Renouxia is not a monotypic genus as is currently reported in Algaebase (Guiry & Guiry Citation2019). The new species Renouxia marerubra differs from the generitype R. antillana on the basis of its simpler, shorter and more compact habit with a distinguishing refractive axis, lower abundance of calciferous cells, monoecious gametophyte, simpler development of gonimoblasts and spermatangia, and less numerous spermatia (). Intermediate carposporophyte formations were not observed and therefore cannot be compared with the generitype. The new species is possibly restricted to the Red Sea or the western Indian Ocean.
Table 2. Comparison of vegetative and reproductive characters between Renouxia spp.
Renouxia Fredericq & J.N.Norris (1995, Cryptogamic Botany 5: 329) emend. mut. char. D.Gabriel & Fredericq
Male and female reproductive structures on separate (R. antillana) or on the same thalli (R. marerubra).
Remarks
Renouxia marerubra is the first taxon in the order where monoecism was observed. Renouxia antillana and Rhodogorgon ramosissima are dioecious. Only monosporangia have been reported for R. flagelifera while no reproductive structure is known for Rhodenigma contortum. Additional emendation might occur as more specimens are collected and microscopically investigated.
Discussion
New records of the genus Renouxia are presented for Egypt (Red Sea), Sri Lanka, Indonesia, Guam and Kosrae, extending the previously known geographic limits of the genus further north in the Indian and Pacific Oceans (). The expanded distribution range suggests that Renouxia may be a Tethyan relict (Leliaert et al., Citation2018), though this pattern may disappear when more collections are added. The present study proves that Renouxia is not monotypic and might occur in many other tropical locations, where it could have been overlooked due to its resemblance to sea anemones, soft corals or other gelatinous algae (Littler & Littler, Citation2003). Herbivore pressure may have led to the selection of different traits to improve species fitness, but mimicry is rarely reported for macroalgae (Richards & Huisman, Citation2014). Mimicry, however, has also been observed in Renouxia’s sister genus Rhodogorgon, which was named after the striking resemblance of these algae to gorgonian corals (Norris & Bucher, Citation1989; Richards & Huisman, Citation2014).
A few records of Renouxia were previously thought to possibly represent unknown species within the genus (Littler & Littler, Citation2003; De Clerck et al., Citation2004; Le Gall & Saunders, Citation2007; Bittner et al., Citation2011; West et al., Citation2016; Zubia et al., Citation2018), but no further information was provided. Although rbcL and COI sequences could not be generated in the present work for all specimens because of failed amplification reactions, sequencing of these genetic markers is necessary to confirm the existence of new putative species and their respective genetic groups. Moreover, the description of new species besides Renouxia marerubra will depend on adequate collections of additional material for detailed morphological studies, particularly to document reproductive characters.
Uncorrected p-distances between the Red Sea specimens and the generitype were 8.0% for COI, 6.5-7.3% for rbcL and 3.1% for UPA. COI distance values are higher than those of rbcL within the genus, and lower between genera. Infrageneric sequence divergence values of Renouxia were similar to divergence values between species of Renouxia and Rhodenigma contortum, suggesting that Rhodenigma could be an alternate phase of an unknown Renouxia species, as also previously suggested by West et al. (Citation2016). Monoecism is firstly reported for the Rhodogorgonales, indicating the current limited knowledge of this order. Molecular and morphological studies of new collections are required to further unveil the taxonomic diversity within the genus Renouxia and the order Rhodogorgonales (West et al., Citation2016). The order seems to be more diverse in the Indo-Pacific, but expanded sampling and sequencing of Atlantic specimens are necessary to confirm this pattern.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article's online page at https://doi.org/10.1080/09670262.2019.1670362
Supplementary table S1. List of taxa with collection data and GenBank accession numbers. Newly generated sequences shown in bold.
Supplementary table S2. UPA uncorrected p-distances between members of the Rhodogorgonales. Values between conspecifics are limited by rectangles.
Author contributions
D. Gabriel and S. Fredericq: original concept; D. Gabriel, S.G.A. Draisma, T. Schils, W.E. Schmidt and T. Sauvage: sample collection and production of molecular data; D. Gabriel, J. Norris and S. Fredericq: description of new species; D. Gabriel: drafting; D. Gabriel, S.G.A. Draisma, T. Schils, W.E. Schmidt, T. Sauvage, D.J. Harris and S. Fredericq: analysis of molecular data and editing of manuscript.
TEJP-2019-0059-File011.docx
Download MS Word (24.8 KB)TEJP-2019-0059-File010.docx
Download MS Word (23.3 KB)Acknowledgements
We thank Eric Coppejans and Olivier de Clerk (Herbarium Ghent) for providing specimens, and António Medeiros for providing the maps. We also thank John Huisman and an anonymous reviewer for their constructive comments.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ballantine, D.L., Ruiz, H. & Nilda, E. (2004). Notes on the benthic marine algae of Puerto Rico VIII. Additions to the flora. Botanica Marina, 47: 335–340.
- Bittner, L., Payri, C.E., Maneveldt, G.W., Couloux, A., Cruaud, C., de Reviers, B. & Le Gall, L. (2011). Evolutionary history of the Corallinales (Corallinophycidae, Rhodophyta) inferred from nuclear, plastidial and mitochondrial genomes. Molecular Phylogenetics and Evolution, 61: 697–713.
- Coppejans, E. & Millar, A.J.K. (2000). Marine Red Algae from the North Coast of Papua New Guinea. Botanica Marina, 43: 315–346.
- De Clerck, O., Coppejans, E., Schils, T., Verbruggen, H., Leliaert, F., de Vriese, T. & Marie, D. (2004). The marine red algae of Rodrigues (Mauritius, Indian Ocean). Journal of Natural History, 38: 3021–3057.
- Erixon, P., Svennblad, B., Britton, T. & Oxelman, B. (2003). Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Systematic Biology, 52: 665–673.
- Fredericq, S. & Norris, J.N. (1995). New order (Rhodogorgonales) and family (Rhodogorgonaceae) of red algae composed of two tropical calciferous genera, Renouxia gen. nov. and Rhodogorgon. Cryptogamic Botany, 31: 616–632.
- Freshwater, D.W., Fredericq, S., Hommersand, M.H. & Chase, M.W. (1994). A gene phylogeny of the red algae (Rhodophyta) based on plastid rbcL. Proceedings of the National Academy of Sciences USA, 91: 7281–7285.
- Gabriel, D., Draisma, S.G.A., Schmidt, W.E., Schils, T., Sauvage, T., Maridakis, C., Gurgel, C.F.D., Harris, D.J. & Fredericq, S. (2017). Beneath the hairy look: the hidden reproductive diversity of the Gibsmithia hawaiiensis complex (Dumontiaceae, Rhodophyta). Journal of Phycology, 53: 1171–1192.
- Guiry, M.D. & Guiry, G.M. (2019). AlgaeBase. World-wide electronic publication. National University of Ireland, Galway. http://www.algaebase.org.
- Kraft, G.T., Millar, A.J.K., Coppejans, E.G.G., Hommersand, M.H., Liao, L. & Freshwater, D.W. (1999). Marine benthic red algae (Rhodophyta) from Bulusan, Sorsogon Province, Southern Luzon, Philippines. Philippine Scientist, 36: 1–50.
- Kumar, S., Stecher, G. & Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33: 1870–1874.
- Le Gall, L. & Saunders, G.W. (2007). A nuclear phylogeny of the Florideophyceae (Rhodophyta) inferred from combined EF2, small subunit and large subunit ribosomal DNA: establishing the new red algal subclass Corallinophycidae. Molecular Phylogenetics and Evolution, 43: 1118–1130.
- Lee, J.M., Song, H.J., Park, S.I., Lee, Y.M., Jeong, S.Y., Cho, T.O., Kim, J.H., Choi, H.-G., Choi, C.G., Nelson, W.A., Fredericq, S., Bhattacharya, D. & Yoon, H.S. (2018). Mitochondrial and plastid genomes from coralline red algae provide insights into the incongruent evolutionary histories of organelles. Genome Biology and Evolution, 10: 2961–2972.
- Leliaert, F., Payo, D.A., Gurgel, C.F.D., Schils, T., Draisma, S.G.A., Saunders, G.W., Kamiya, M., Sherwood, A.R., Lin, S.M., Huisman, J.M., Le Gall, L., Anderson, R.J., Bolton, J.J., Mattio, L., Zubia, M., Spokes, T., Vieira, C., Payri, C.E., Coppejans, E., D’hondt, S., Verbruggen, H. & De Clerck, O. (2018). Patterns and drivers of species diversity in the Indo-Pacific red seaweed Portieria. Journal of Biogeography, 45: 2299–2313.
- Lewmanomont, K. & Noiraksa, T. (2010). Rhodogorgon and Renouxia (Rhodogorgonales, Rhodophyta) the gorgonian-like marine red algae from Thailand. Algal Resources, 3: 73–80.
- Liao, L.M. & Aungtonya, C. (2000). Note on Renouxia antillana (Rhodophyta, Rhodogorgonales), a new addition to the marine flora of Thailand. Phuket Marine Biological Center Research Bulletin, 77: 77–80.
- Littler, D.S. & Littler, M.M. (2003). South Pacific Reef Plants: A Divers’ Guide to the Plant Life of South Pacific Coral Reefs. Offshore Graphics, Inc., Washington, D.C.
- Littler, D.S., Littler, M.M., Bucher, K.E. & Norris, J.N. (1989). Marine Plants of the Caribbean; A Field Guide from Florida to Brazil. Smithsonian Institution Press, Washington, D.C.
- Millar, A.J., Clerck, O. De, Coppejans, E. & Liao, L.M. (1999). Annotated and illustrated survey of the marine macroalgae from Motupore Island and vicinity (Port Moresby area, Papua New Guinea). III. Rhodophyta. Australian Systematic Botany, 12: 549–591.
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), 1–8. 14 November 2010, New Orleans, LA.
- N’Yeurt, A.D.R. (2001). Marine algae from the Suva Lagoon and Reef, Fiji. Australian Systematic Botany, 14: 689–869.
- Nei, M., & Kumar, S. (2000). Molecular Evolution and Phylogenetics. Oxford University Press, New York.
- Norris, J.N. & Bucher, K.E. (1989). Rhodogorgon, an anamolous new red algal genus from the Caribbean Sea. Proceedings of the Biological Society of Washington, 102: 1050–1066.
- Ogden, N.B. (1992). Morphology, reproduction and range extension of Rhodogorgon (? Nemaliales, Rhodophyta). Phycologia, 31: 470–477.
- Payri, C., N’Yeurt, A.D.R. & Orempuller, J. (2000). Algae of French Polynesia. Au Vent des Iles Editions, Tahiti.
- Richards, Z.T. & Huisman, J.M. (2014). Coral-mimicking alga Eucheuma arnoldii found at Ashmore Reef, north-western Australia. Coral Reefs, 33: 441.
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61: 539–542.
- Sauvage, T., Schmidt, W.E., Suda, S. & Fredericq, S. (2016). A metabarcoding framework for facilitated survey of endolithic phototrophs with tufA. BioMed Central Ecology, 16: 8.
- Sherwood, A.R., Sauvage, T., Kurihara, A., Conklin, K.Y. & Presting, G.G. (2010). A comparative analysis of COI, LSU and UPA marker data for the Hawaiian florideophyte Rhodophyta: implications for DNA barcoding of red algae. Cryptogamie, Algologie, 31: 451–465.
- Spalding, M.D., Fox, H.E., Allen, G.R., Davidson, N., Ferdaña, Z.A., Finlayson, M.A.X., Halpern, B.S., Jorge, M.A., Lombana, A.L., Lourie, S.A. & Martin, K.D. (2007). Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. BioScience, 57: 573–583.
- Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312–1313.
- West, J.A., Zuccarello, G.C., de Goër, S.L., Stavrias, L.A. & Verbruggen, H. (2016). Rhodenigma contortum, an obscure new genus and species of Rhodogorgonales (Rhodophyta) from Western Australia. Journal of Phycology, 52: 397–403.
- Yang, E., Boo, S., Bhattacharya, D., Saunders, G., Knoll, A., Fredericq, S., Graf, L. & Yoon, H. (2016). Divergence time estimates and evolution of major lineages in the florideophyte red algae. Scientific Reports 6, 1–11.
- Zubia, M., De Clerck, O., Leliaert, F., Payri, C., Mattio, L., Vieira, C., Cambert, H., Quod, J.P., Loiseau, N., Golubic, S., Lin, S.M., Liu, S.L. & Pinault, M. (2018). Diversity and assemblage structure of tropical marine flora on lava flows of different ages. Aquatic Botany, 144: 20–30.
