ABSTRACT
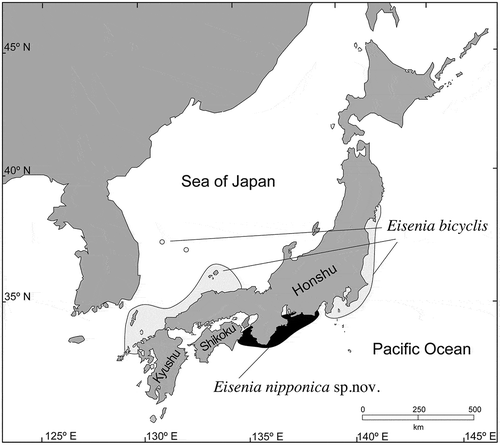
Recently, synonymizing Eisenia with Ecklonia was suggested based on a molecular phylogeny using the rbcL gene and ITS1–5.8S rDNA sequences. However, in a multigene molecular phylogeny based on mitochondrial cox1, cox3 genes and the atp8–16S rDNA region, as well as the plastid atpB, psaA, psbA and rbcL genes, Ecklonia spp. formed a monophyletic clade supported by high statistical values, and Eisenia spp. showed monophyly depending on analytical methods. The Japanese Eisenia species that used to be identified as Eisenia arborea (E. arborea sensu Arasaki) was shown to be genetically distant from E. arborea from the NE Pacific and E. bicyclis from Japan. Eisenia spp. were morphologically distinct in having a split meristematic zone in the mature thallus forming a dorsi-ventral blade with false branches. Therefore, we propose reinstatement of Eisenia as an independent genus and describe a new species, Eisenia nipponica (= E. arborea sensu Arasaki) from Japan. E. nipponica is distributed on the Pacific Coast of central Honshu, having a separate geographic range from E. bicyclis, which is distributed on the Pacific coast of eastern Honshu and the Sea of Japan coast of north-western Honshu and northern Kyushu.
Introduction
The laminarialean genus Eisenia was described by Areschoug (Citation1876) based on specimens collected by Dr G. Eisen from Santa Catalina Island, California, proposing the new species Eisenia arborea Areschoug. Ecklonia and Eisenia are virtually indistinguishable as young sporophytes. However, in Eisenia the distal portion of the primary blade becomes eroded as the thallus grows and the region becomes divided longitudinally, establishing two meristematic regions and eventually two twisted stipe-like portions (bifurcate false branches). Yendo (Citation1902) suggested transfer of Ecklonia bicyclis Kjellman described from Japan, which also had this morphological feature, to Eisenia as Eisenia arborea f. bicyclis (Kjellman) Yendo, and later Setchell (Citation1905) suggested treating the taxon as an independent species, Eisenia bicyclis (Kjellman) Setchell. Since then, five new species have been described in the genus (i.e. E. cokeri M.Howe, E. desmarestioides Setchell & N.L.Gardner, E. galapagensis W.R.Taylor, E. gracilis E.Y.Dawson, Acleto & Foldvik, and E. masonii Setchell & N.L.Gardner), although little taxonomic study has been applied to those taxa since their original descriptions.
In Japan, a new taxon of Eisenia that has a distinctive distributional range () and different blade shapes was noted, and identified as E. arborea due to morphological similarity (Arasaki, Citation1953, Citation1964). E. bicyclis blades have side branches, whereas E. arborea lacks such side branches. The genus Ecklonia was described by Hornemann (Citation1828) based on Fucus buccinalis Linnaeus, and since then more than 10 species have been described. Recently, Rothman et al. (Citation2015) analysed the molecular phylogeny of Ecklonia/Eisenia species, and because two Eisenia species (E. arborea and E. bicyclis) were nested in the Ecklonia clade in the phylogenetic tree based on rbcL and ITS1–5.8S rDNA, they proposed synonymizing Eisenia to Ecklonia. However, in our preliminary analyses Japanese Eisenia arborea specimens appeared to be genetically distinct from those of the north-eastern Pacific (Washington and California, USA) including the type locality. Therefore, in the present study we have re-examined the molecular phylogeny of Ecklonia/Eisenia species, especially Eisenia species, using multiple gene sequences.
Materials and methods
Materials
For morphological and genetic studies, fresh specimens of Eisenia arborea sensu Arasaki (Citation1964) were collected at Mugisaki, Mie, Japan. Specimens of E. arborea collected from various localities, housed in the University Herbarium, University of California, Berkeley (UC) were also examined.
Morphological observations
For comparisons of the number of cells constituting cortical layers, cross-sections were made by hand using a razor blade and measured midway between the meristem and base. Photomicrographs were taken using a VB–7010 Digital Camera (Keyence, Tokyo, Japan) attached to an Axioplan microscope (Zeiss, Oberkochen, Germany).
Molecular phylogenetic analysis
Field-collected specimens rapidly desiccated in silica gel, or fragments taken from herbarium specimens, were used for DNA extractions (Supplementary table S1). Genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Polymerase chain reaction (PCR) amplifications of the mitochondrial cox1, cox3 and atp8–16S rDNA region including atp8, rps10, rpl31, and 16S rDNA and their intergenic spacers, as well as plastid atpB, psaA, psbA and rbcL were conducted using the KOD FX (ToYoBo, Osaka, Japan) and the TaKaRa PCR Thermal Cycler Dice (Takara Bio, Kusatsu, Japan). Primers used for PCR and sequencing are listed in Supplementary table S2 (Yoon et al., Citation2002; Hanyuda et al., Citation2004; Kawai et al., Citation2007, Citation2012, Citation2013, Citation2019; Lane et al., Citation2007; Ni-Ni-Win et al., Citation2008; Le Corguillé et al., Citation2009; Cock et al., Citation2010; Silberfeld et al., Citation2010). The profiles of the PCRs were as described by Kawai et al. (Citation2015) and Kojima et al. (Citation2015). After PEG purification (Lis, Citation1980), PCR products were sequenced by a DNA sequencing service (FASMAC, Atsugi, Japan). For molecular phylogenetic analyses, published and newly determined sequence data of the Ecklonia and Eisenia specimens were used (Supplementary table S1; Kawai et al., Citation2013; Wang et al., Citation2013; Chen et al., Citation2019; Starko et al., Citation2019). The obtained sequences were aligned using the program MAFFT v.6 (Katoh & Toh, Citation2008) and then manually adjusted prior to phylogenetic analyses. Samples with identical nucleotide sequences were treated as a single operational taxonomic unit (OTU). Egregia menziesii (Turner) Areschoug, Macrocystis pyrifera (Linnaeus) C.Agardh (=M. integrifolia Bory) and Saccharina japonica (Areschoug) C.E.Lane, C.Mayes, Druehl & G.W.Saunders were selected as the outgroup based on a recent phylogenetic study of Laminariales (Starko et al., Citation2019). The pair-end reads of Egregia menziesii were trimmed, quality filtered and assembled using Trim Galore 0.6.0 (available at http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), FASTX-Toolkit 0.014 (available here: http://hannonlab.cshl.edu/fastx_toolkit/download.html), and ABySS 2.0 (Jackman et al., Citation2017), respectively, and the above-mentioned 6 genes and 1 region of sequence data were detected by BLAST 2.2.30 (Camacho et al., Citation2009). Phylogenetic analyses of the aligned sequences (21 OTUs, 8223 bp) were subjected to Maximum likelihood (ML) and Bayesian (BI) and Neighbour-joining (NJ) analyses. For Maximum likelihood (ML) analysis, we used RAxML GUI v.1.31 (Silvestro & Michalak, Citation2012) run to conduct 10 000 Rapid Bootstrap searches followed by an ML search. The GTR + G model were implemented with partitioning by genes for the analyses. Bayesian analyses were run using MrBayes v.3.2.2 (Ronquist et al., Citation2012). The best-fit evolutionary model was determined using Kakusan4 (Tanabe, Citation2011). The Bayesian analyses were initiated with a random starting tree and ran four chains of Markov chain Monte Carlo iterations simultaneously for 10 000 000 generations, keeping one tree every 100 generations. The first 10 000 trees sampled were discarded as burn-in, based on the stationarity of log-likelihood as assessed using Tracer v.1.7.1 (Rambaut & Drummond, Citation2018). A consensus topology and posterior probability values were calculated from the remaining trees. The Neighbour-joining (NJ) tree was constructed with maximum composite likelihood + G model using MEGA X (Kumar et al., Citation2018). The tree branching pattern was evaluated by 10 000 bootstrap analyses.
Results
Habit and morphology
Eisenia arborea sensu Arasaki (= Eisenia nipponica sp. nov.) was distributed along the Pacific coast of central Honshu, growing on lower intertidal and subtidal rocks to ~10 m depth (). The sporophytic thalli were composed of blades, stipe and holdfast of dichotomously branched haptera (). Stipes were nearly terete near the base and became somewhat compressed in upper portions. Juvenile sporophytic thalli were at first lanceolate (), then continuously formed new side branches at the transition zone (meristematic zone) between stipe and blade (, ). The continuous development of the lateral meristematic zone formed new lateral branches, and the upper part of the thallus became in-rolled to one side, creating a dorsi-ventral morphology (). The primary blade deteriorated from the tip and eventually was lost, probably at the end of the first growing season (–). There was no epidermal meristematic activity at the tip of the blade after the deterioration there, so that no epidermis covered the cortical and medullary layers of the blade, as if the side had been cut by a knife (). Then, as the second-year thallus developed, each side of the meristematic zone continued to form new blades from one side, so that by a remarkable thickening of the tissue associated with this elongation, new dichotomously branched stems were formed (, ). As a result of this developmental process, one side of the stem lacked meristoderm and looked as if the surface were cut by a knife (, ).
Figs 2–13. Habit and morphology of sporophytes of Eisenia nipponica sp. nov. Fig. 2. Underwater habit of sporophyte. Fig. 3. Upper part of a mature thallus. Fig. 4. Holdfast of dichotomously branched haptera. Figs 5, 6. Juvenile sporophytes. Asterisk shows distal part of primary central blade. Fig. 7. Young (first-year) thallus with developed lateral blades and retaining primary central blade. Asterisk shows eroding apical portion of primary blade. Figs 8–11. Early stage of mature thallus development. Apical portion of the primary blade (asterisk) is lost and the bare side of the cortical and medullary layer of the blade is observed (arrowheads). Upper part of thallus becomes in-rolled to one side, creating a dorsi-ventral morphology. Arrow shows initials of lateral blades. Double arrowhead shows regenerated meristoderm midway along the split blade. Figs 12, 13. Mature sporophyte with developed false branches, showing dorsal side (Fig. 12) and ventral side (Fig. 13) with bare medulla and cortex (arrowheads).

Molecular phylogenetic analysis
In the Maximum likelihood (ML) molecular phylogeny based on concatenated DNA sequences of mitochondrial cox1, cox3 genes and atp8–16S rDNA region, and chloroplast psaA, psbA, atpB, rbcL gene sequences, Ecklonia cava, Ec. kurome, Ec. radicosa and Ec. stolonifera formed a clade supported by full statistical support (100% bootstrap value in ML/1.0 posterior probability in BI), although the support for elucidating their species-level relationships were low to moderate (full support for E. radicosa in ML/BI, but < 86% in ML for others) (). The clade comprised of Ecklonia cava/Ec. kurome/Ec. radiata/Ec. stolonifera was sister to the clade comprised of Ec. maxima and Ec. radiata, supported by full statistical values. Although only single specimens represented Ec. maxima (from South Africa) and Ec. radiata (from Victoria, Australia), published sequence data (Rothman et al., Citation2015) confirmed their phylogenetic positions in the tree.
Fig. 14. Maximum likelihood (ML) molecular phylogeny based on concatenated DNA sequences of mitochondrial cox1, cox3 genes, atp8–16S rDNA region, and chloroplast psaA, psbA, atpB, rbcL gene sequences (Total 8223 bp). Numbers below the branches indicate the bootstrap values (BP, right) and Bayesian posterior probabilities (PP, left). Asterisk (*) indicates 100% BP and 1.00 PP in ML and BI analyses. Only the BP ≥ 70% and PP ≥ 0.95 are shown.
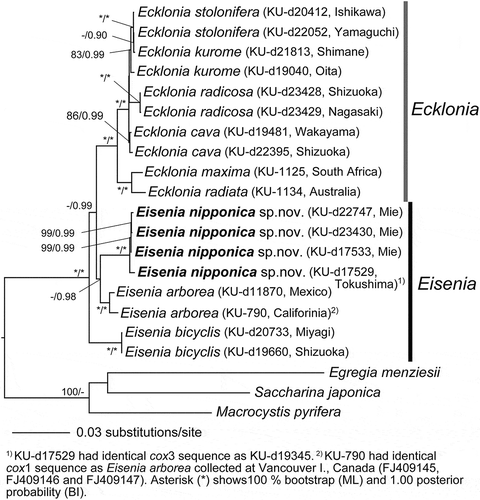
In contrast, Eisenia arborea from the north-eastern Pacific (Canada to Mexico) and E. arborea from Japan (E. arborea seusu Arasaki) formed independent clades supported by high statistical values, showing a sister relationship with the clade comprised of Ecklonia cava/Ec. kurome/Ec. radiata/Ec. stolonifera and Ec. maxima/Ec. radiata. In the tree, E. bicyclis was basal to these clades. In contrast, in the Neighbour-joining (NJ) phylogenetic tree, E. arborea, E. arborea sensu Arasaki and E. bicyclis formed a monophyletic clade, although the statistical support was low (Supplementary fig. S2, 51%).
Discussion
Eisenia species show a peculiar type of development in forming second year thalli which is unique in Laminariales. Setchell (Citation1896) described this in detail: ‘As the growth proceeds, the involutions become more and more pronounced and are accompanied by a thickening of the margins along the in-rolled edges. Consequently, one surface of the blade becomes somewhat rounded and convex at this point, while the other becomes indented with a broad and rather shallow longitudinal furrow. This gives to the blade a certain dorsi-ventral character, i.e., gives to the two faces different characteristics.’ Furthermore, following this process, Setchell continued: ‘the meristematic region in Eisenia becomes elongated and soon separated into two definite regions, an upper and a lower one. The lower meristematic region seems certainly to persist, but the stipe continues to increase in length as well as in thickness, even to a very marked degree. Thereby, first a transverse splitting in which the portion belonging to the stipe is separated from that belonging to the blade, which is unique in members of Laminariales. In contrast, in most laminarialean species, the stipe and blade possess a common meristem at the transition-plane.’
However, because one side of the upper stem (upper branch of the Y-shaped thallus) lacks meristoderm, showing bare medulla as in the tip of the first-year blade at the late stage, we consider that the branch does not represent a stipe but rather thickened and elongated blades. Setchell (Citation1896) interpreted this structure as a branched stipe formed by the meristematic activity of the lower meristem, after the separation of the meristem into two portions. However this structure is interpreted, this type of development of the sporophyte is unique among Laminariales and distinct from Ecklonia, in which the meristematic zone does not split into two portions, even in the perennial thallus.
Although the sporophyte morphology of Eisenia arborea, E. bicyclis and E. arborea sensu Arasaki is rather plastic in thallus size, length of the stipe, shape of the blade and presence or absence of marginal dentations, E. bicyclis is distinguishable from the other two taxa by the frequent occurrence of secondary side branches. In contrast, E. arborea and E. arborea sensu Arasaki are very similar in their gross morphology, and distinctive differences have not been noticed. Nevertheless, they are genetically remarkably distant (2.21–3.06% in cox3 and 0.50–0.57% in rbcL DNA sequences). These values are similar to or higher than the distances between Ec. maxima (KU-1125) and Ec. radiata (KU-1134) (1.39% in cox3 and 0.78% in rbcL). Therefore, also considering their disjunct geographic distribution and genetic similarity within each taxon, we concluded that they are distinct species.
Rothman et al. (Citation2015), in their molecular phylogeny based on the concatenated rbcL gene and rDNA ITS region sequences, found that Eisenia bicyclis and E. arborea did not form monophyletic clades but nested within the clade of Ecklonia spp. However, the statistical values supporting major nodes in the tree were generally low. This is perhaps because of the low phylogenetic resolution of the rbcL and rDNA ITS sequences. Rothman et al. (Citation2015) also used atp8–trnWI region sequences, which are considered to have higher phylogenetic resolution, but Eisenia species were not included in the analysis. In our molecular phylogeny using concatenated DNA sequences of mitochondrial cox1, cox3 genes and the atp8–16S rDNA region, and chloroplast psaA, psbA, atpB, rbcL genes (8223 bp), Ecklonia species excluding Eisenia species (i.e. Ec. cava, Ec. kurome, Ec. maxima, Ec. radiata, Ec. radicosa and Ec. stolonifera) formed a clade supported by full statistical values. Furthermore, in the Neighbour-joining (NJ) analyses Eisenia arborea, E. bicyclis and E. arborea sensu Arasaki formed a clade, although the statistical support was low (51%). Morphologically, the clade (Ecklonia spp.) shares the capability of regeneration of secondary blades (second year thallus) from the meristematic zone with laminariacean species, whereas Eisenia spp. share the formation of a dorsi-ventral thallus with false branches by splitting of the meristematic zone. Therefore, in conclusion, we propose reinstatement of Eisenia as a distinct genus and describe a new species Eisenia nipponica (= E. arborea sensu Arasaki) from Japan.
Although no genetic data are available for other Ecklonia/Eisenia species, Eisenia desmarestioides and Eiseina masonii were shown to be originally described based on fragments of the same specimens, and hence Eisenia masonii was suggested to be a synonym of E. desmarestioides (Silva, Citation2008). E. desmarestioides was also reported to have characteristic false stipes bearing dimorphic blades resembling the E. arborea type of morphology (Silva, Citation2008; fig. 7). Therefore, E. desmarestioides possibly belongs to Eisenia rather than Ecklonia.
Description and diagnosis
Eisenia nipponica H.Kawai, S.Akita, K.Hashimoto & T.Hanyuda sp. nov. –, Supplementary fig S1
Sporophyte perennial, up to ~1.5 m high, epilithic, tightly attached by a dichotomously branched hapteroid holdfast. Stipe exceeding 1 m long and 2.5 cm thick, rounded near the base and compressed in the upper portion. In the mature thallus, stipe becoming bifurcate by forming two compressed false branches with thickened lower margins of the original and subsequently eroded blade. Blades of mature thallus more or less longitudinally grooved, lanceolate to linear with denticulations along margins, up to 60 cm long and 6 cm broad. This species is similar to E. arborea and E. bicyclis, but genetically distinct in mitochondrial cox1, cox3 genes, atp8–16S rDNA region, and chloroplast psaA, psbA, atpB, rbcL gene sequences, and morphologically distinct from E. bicyclis in lacking side branches of the blade.
Holotype: SAP115478. 2019.4.2. Collected at Mugisaki, Shima, Mie, Japan by H. Kawai (Supplementary fig. S1).
Etymology: The specific epithet refers to the distributional range of the species.
Supplementary Information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at http://10.1080/09670262.2019.1692911
Supplementary table S1. Origins of samples and sequence data used for molecular analyses, including their database accession numbers.
Supplementary table S2. List of primers used for polymerase chain reaction (PCR) and sequencing.
Supplementary fig. S1. Holotype specimen of Eisenia nipponica sp. nov. (SAP115478), collected at Mugisaki, Shima, Mie, Japan on 2 April 2019 by H. Kawai.
Supplementary fig. S2. Neighbour-joining (NJ) phylogenetic tree based on concatenated DNA sequences of mitochondrial cox1, cox3 genes, atp8–16S rDNA region, and chloroplast psaA, psbA, atpB, rbcL gene sequences (Total 8223 bp). Numbers below the branches indicate the bootstrap values. Only the BP (≥50%) are shown.
Author contributions
H. Kawai: Original concept, collection of specimens, morphological studies, drafting and editing manuscript; Shingo Akita: genetic analyses; Kazuki Hashimoto: genetic analyses; Takeaki Hanyuda: collection of specimens, genetic analyses.
TEJP-2019-0093-File008.tif
Download TIFF Image (2 MB)TEJP-2019-0093-File007.tif
Download TIFF Image (23.3 MB)TEJP-2019-0093-File006.docx
Download MS Word (17.7 KB)TEJP-2019-0093-File005.docx
Download MS Word (19.8 KB)Acknowledgements
We are grateful to Dr Eric Henry for critically reading and improving the manuscript, Dr Kathy Ann Miller for her help in examining specimens in UB, Dr Jessie Alstatt for providing field data of E. arborea, Drs Akira Kurashima, Tatsuya Ishikawa, You Kato and Taku Yoshimura for their support in collecting specimens. Part of the study was supported by a JSPS Grant-in-Aid for Scientific Research (No. 16H04832) to H.K.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Arasaki, S. (1953). On Eisenia bicyclis Setchell. Bulletin of Japanese Society of Phycology, 1: 49–53. (In Japanese)
- Arasaki, S. (1964). How to know the seaweeds of Japan and its vicinity fully illustrated in colours. Hokuryukan, Tokyo. 217 pp. (In Japanese)
- Areschoug, J.E. (1876). De tribus Laminarieis (Egregia Aresch., Eisenia Aresch., Nereocystis) et de Stephanocystide osmundacea (Turn.). Trevis. observationes praecursorias offert. Botaniska Notiser, 1876: 65–73.
- Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K. & Madden, T. (2009). BLAST+: architecture and applications. BMC Bioinformatics, 10: 421.
- Chen, J., Zang, Y., Shang, S. & Tang, X. (2019). The complete mitochondrial genome of the brown alga Macrocystis integrifolia (Laminariales, Phaeophyceae). Mitochondrial DNA Part B, 4: 635–636.
- Cock, J.M., Sterck, L., Rouzé, P., Scornet, D., Allen, A.E., Amoutzias, G., … & Beszteri, B. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature, 465: 617–621.
- Hanyuda, T., Suzawa, Y., Suzawa, T., Arai, S., Sato, H., Ueda, K. & Kumano, S. (2004). Biogeography and taxonomy of Batrachospermum helminthosum Bory (Batrachospermales, Rhodophyta) in Japan inferred from rbcL gene sequences. Journal of Phycology, 40: 581–588.
- Hornemann, J.W. (1828). Om Fucus buccinalis Linnaei. Kongelige Danske Videnskabernes Selskabs Naturvidenskabelige og Mathematiske Afhandlinger, 3: 379–390, 1 folded plate.
- Jackman, S.D., Vandervalk, B.P., Mohamadi, H., Chu, J., Yeo, S., Hammond, S.A., Jahesh, G., Khan, H., Coombe, L., Warren, R.L. & Birol, I. (2017). ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Research, 27: 768–777.
- Katoh, K. & Toh, H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics, 9: 286–298.
- Kawai, H., Hanyuda, T., Draisma, S.G.A. & Müller, D.G. (2007). Molecular phylogeny of Discosporangium mesarthrocarpum (Phaeophyceae) with a reassessment of the Discosporangiales. Journal of Phycology, 43: 186–194.
- Kawai, H., Kogishi, K., Hanyuda, T. & Kitayama, T. (2012). Taxonomic revision of the genus Cutleria proposing a new genus Mutimo to accommodate M. cylindrica (Cutleriaceae, Phaeophyceae). Phycological Research, 60: 241–248.
- Kawai, H., Hanyuda, T., Ridgway, L.M. & Holser, K. (2013). Ancestral reproductive structure in basal kelp Aureophycus aleuticus. Scientific Reports, 3: 2491.
- Kawai, H., Hanyuda, T., Yamagishi, T. & Kai, A. (2015). Reproductive morphology and DNA sequences of the brown alga Platysiphon verticillatus support the new combination Platysiphon glacialis. Journal of Phycology, 51: 910–917.
- Kawai, H., Hanyuda, T., Shibata, K., Kamiya, M. & Peters, A.F. (2019). Proposal of a new brown algal species, Mesogloia japonica sp. nov. (Chordariaceae, Phaeophyceae), and transfer of Sauvageaugloia ikomae to Mesogloia. Phycologia, 58: 63–69.
- Kojima, R., Hanyuda, T. & Kawai, H. (2015). Taxonomic re-examination of Japanese Halimeda species using genetic markers, and proposal of a new species Halimeda ryukyuensis (Bryopsidales, Chlorophyta). Phycological Research, 63: 178–188.
- Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35: 1547–1549.
- Lane, C.E., Lindstrom, S.C. & Saunders, G.W. (2007). A molecular assessment of northeast Pacific Alaria species (Laminariales, Phaeophyceae) with reference to the utility of DNA barcoding. Molecular Phylogenetics and Evolution, 44: 634–648.
- Le Corguillé, G., Pearson, G., Valente, M., Viegas, C., Gschloessl, B., Corre, E., Bailly, X., Peters, A.F., Jubin, C., Vacherie, B. & Cock, J.M. (2009). Plastid genomes of two brown algae, Ectocarpus siliculosus and Fucus vesiculosus: further insights on the evolution of red-algal derived plastids. BMC Evolutionary Biology, 9: 253–266.
- Lis, J.T. (1980). Fractionation of DNA fragments by polyethylene glycol induced precipitation. Methods in Enzymology, 65: 347–353.
- Ni-Ni-Win, Hanyuda, T., Arai, S., Uchimura, M., Abbott, I.A. & Kawai, H. (2008). New records of Padina species from the western coast of the Pacific Ocean. Phycological Research, 56: 288–300.
- Rambaut, A. & Drummond, A.J. (2018). Tracer v.1.7.1. [cited 11 October 2018]. Available from: http://tree.bio.ed.ac.uk/software/tracer/.
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61: 539–542.
- Rothman, M.D., Mattio, L., Wernberg, T., Anderson, R.J., Uwai, S., Mohring, M.B. & Bolton, J.J. (2015). A molecular investigation of the genus Ecklonia (Phaeophyceae, Laminariales) with special focus on the southern hemisphere. Journal of Phycology, 51: 236–246.
- Setchell, W.A. (1896). Eisenia arborea Aresch. Erythea, 4: 129–133, 155–162.
- Setchell, W.A. (1905). Post-embryonal stages of the Laminariaceae. University of California Publications in Botany, 2: 115–138.
- Silberfeld, T., Leigh, J.W., Verbruggen, H., Cruaud, C., de Reviers, B. & Rousseau, F. (2010). A multi-locus time-calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): investigating the evolutionary nature of the “brown algal crown radiation”. Molecular Phylogenetics and Evolution, 56: 659–674.
- Silva, P.C. (2008). Conspecificity of Eisenia desmarestioides and E. masonii (Laminariales, Phaeophyceae) from Isla Guadalupe, Baja California, Mexico. Hidrobiológica, 18: 155–159.
- Silvestro, D. & Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution, 12: 335–337.
- Starko, S., Gomez, M.S., Darby, H., Demes, K.W., Kawai, H., Yotsukura, N., Lindstrom, S.C., Keeling, P.J. & Graham, S.W. (2019). A comprehensive kelp phylogeny sheds light on the evolution of an ecosystem. Molecular Phylogenetics and Evolution, 136: 138–150.
- Tanabe, A.S. (2011). Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Molecular Ecology Resources, 11: 914–921.
- Wang, X., Shao, Z., Fu, W., Yao, J., Hu, Q. & Duan, D. (2013). Chloroplast genome of one brown seaweed, Saccharina japonica (Laminariales, Phaeophyta): its structural features and phylogenetic analyses with other photosynthetic plastids. Marine Genomics, 10: 1–9.
- Yendo, K. (1902). On Eisenia and Ecklonia. Botanical Magazine, Tokyo, 16: 203–206.
- Yoon, H.S., Hackett, J.D. & Bhattacharya, D. (2002). A single origin of the peridinin- and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proceedings of the National Academy of Sciences USA, 99: 11724–11729.