ABSTRACT
In the North-east (NE) Atlantic, most intertidal fucoids and warm-temperate kelps show unique low-latitude gene pools matching long-term climatic refugia. For cold-temperate kelps data are scarcer despite their unique cultural, ecological and economic significance. Here we test whether the amphi-Atlantic range of Laminaria digitata is derived from past glacial survival (and vicariance) in both NE and North-west (NW) Atlantic refugia (as suggested by niche modelling), or post-glacial (re)colonization (as suggested by low mtDNA divergence). We screened 14 populations from across the species range for 12 microsatellite loci to identify and map major gene pools and refugia. We assessed if NW Atlantic survival was supported by unique endemic variation, and if genetic diversity and structure were, as predicted from larger hindcasted glacial ranges, higher in the NE Atlantic. Microsatellite data subdivided L. digitata into three main genetic groups matching Brittany, northern Europe and the NW Atlantic, with finer-scale sub-structuring within European clusters. The relatively diverse NE Atlantic lineages probably survived the Last Glacial Maximum along unglaciated periglacial shorelines of the Armorican and Celtic Seas (Brittany cluster) and Ireland (northern European cluster), and remain well differentiated despite their relative proximity. The unique Brittany gene pool, at the contemporary European rear edge, is projected to disappear in the near future under high greenhouse gas emission scenarios. Low allelic diversity and low endemism in the NW Atlantic are consistent with recent post-glacial colonization from Europe, challenging the long-standing hypothesis of in situ glacial survival. Confusion with Hedophyllum nigripes may have led to underestimation of regional diversity of L. digitata, but also to overestimation of its presence along putative trans-Atlantic migration routes. Partial incongruence between modelling and genetic-based biogeographic inferences highlights the benefits of comparing both approaches to understand how shifting climatic conditions affect marine species distributions and explain large-scale patterns of spatial genetic structure.
Introduction
The extreme environmental shifts associated with the Quaternary glacial cycles (2.5 Ma ago to present) are among the most important factors shaping present-day bio- and phylogeographic patterns across the northern hemisphere. During glacial stages, such as the Last Glacial Maximum (LGM, 26–19 ka BP), the expansion of the cryosphere buried the Arctic and adjacent continents and shorelines under kilometre-thick ice sheets, ice shelves and perennial sea ice, causing the extirpation of coastal assemblages from much of their modern cold-temperate distributions. In the North-east (NE) Atlantic, the Eurasian ice sheet spread from the Severnaya Zemlya archipelago (Russia’s Arctic) to the British Isles across the emerged Kara, Barents and North seas, and reached as far south as Ireland (Brochmann et al., Citation2003). Iceland had a smaller, separate ice cap, but ice margins extended into the shelf break (Hubbard et al., Citation2006). In the North-west (NW) Atlantic, the Laurentide ice sheet spread across the modern Hudson Bay and the Canadian Arctic Archipelago to connect with the Greenland ice sheet (Carlson & Winsor, Citation2012). It reached as far south as Cape Cod, although some coastal regions such as Grand Banks off Newfoundland remained relatively ice-free (Carlson & Winsor, Citation2012). As a direct result of glacial ice and the southward position of isotherms, marine species’ ranges reached lower latitudes and were in general more latitudinally compressed. Amphi-Atlantic species were also presumably disconnected, because there are no island chains at mid-latitudes that could have provided habitat stepping stones, a role that Iceland and southern Greenland currently play.
Until the 2000s, the prevailing ‘tabula rasa’ hypothesis posited that most NW Atlantic temperate rocky-shore biota had been completely extirpated during the LGM and that present-day assemblages had (re)colonized the area from the NE Atlantic (or the Pacific) only post-glacially, during the last 13 ka or so (Vermeij, Citation1991; Ingólfsson, 1992). This view was based on the comparatively low species pool of the NW Atlantic, and the dominance of amphi-Atlantic (i.e. present also in Europe) and, to a lesser extent, amphi-boreal (i.e. Pacific immigrants) species, mirrored by few true NW Atlantic endemics. This apparent asymmetry was attributed to the equatorward compression of isotherms and apparent scarcity of suitable rocky substrate south of the Laurentide ice sheet. Upgraded species lists, and particularly the advent of molecular data and niche-modelling approaches, have allowed much stronger inferences regarding putative locations and extension of climatic refugia, as well as levels and timing of trans-Atlantic divergence, and helped resolve this dichotomy of glacial survival versus post-glacial colonization. The current paradigm is much more nuanced and acknowledges that, at least among invertebrates and macroalgae, both scenarios were common (Wares & Cunningham, Citation2001; Waltari & Hickerson, Citation2013; Li et al., Citation2016; Assis et al., Citation2018b; Bringloe & Saunders, Citation2018).
Post-glacial colonization versus long-term persistence (and isolation) are expected to leave distinct genetic signatures (Wares & Cunningham, Citation2001; Maggs et al., Citation2008; Li et al., Citation2015; Bringloe & Saunders, Citation2018). Under the scenario of post-glacial colonization, higher diversity (and structure) is expected in the source population, because successive founder events, gene surfing and priority effects would prevent most genetic diversity from progressing along expansion areas. Genetic differentiation among NW and NE Atlantic basins would also be expected to be low, because alleles should be shared with (or derived from) NE Atlantic populations, at least higher latitude ones. Conversely, in a scenario of survival in NW Atlantic refugia, some trans-Atlantic phylogeographic divergence and unique alleles are expected, and levels of genetic diversity are predicted to be greater. These predictions are, of course, contingent on additional factors, including life-history traits such as dispersal ability, habitat structure, and also the long-term size and location of refugial areas across the Atlantic. For instance, if glacial ranges were very small and/or largely non-overlapping with modern ranges, persisting genetic diversity may be extremely low and resemble the bottleneck effects experienced along expanding fronts. Conversely, secondary contact of distinct gene pools can lead to unexpected diversity throughout post-glacially colonized areas (Petit et al., Citation2003; Neiva et al., Citation2018).
Canopy-forming brown seaweeds have been pivotal to understanding these biogeographic and genetic consequences of past climatic shifts in the N Atlantic, including, in a few studies, patterns of trans-Atlantic colonization/persistence (Olsen et al., Citation2010; Assis et al., Citation2016a, Citation2016b, Citation2018a, Citation2018b; Lourenço et al., Citation2016; Neiva et al., Citation2016, Citation2018; Bermejo et al., Citation2018; Wilson et al., Citation2019). Fucoids (e.g. Fucus ceranoides), warm-temperate kelps (e.g. L. ochroleuca) and red seaweeds (Provan & Maggs, Citation2012; Li et al., Citation2016) frequently feature high geographic structuration and high genetic diversity and/or endemism in low-latitude refugial areas, often corresponding to modern species rear edges, when compared with more northern regions which were colonized post-glaciation. Surprisingly, and notwithstanding their cultural, ecological and economic importance, there is much less information regarding spatial genetic structure for cold-temperate kelps (e.g. Laminaria digitata, L. hyperborea, Alaria esculenta, but see Luttikhuizen et al. (Citation2018) and Neiva et al. (Citation2018) for Saccharina latissima).
A recent study modelling the dynamic geographic distributions of Atlantic kelps across the last glacial-interglacial transition found that the amphi-Atlantic kelp L. digitata had suitable climatic conditions during the LGM to subsist in both the NE and NW Atlantic, with wider ranges in the NE (Assis et al., Citation2018b, their fig. S6). Nonetheless, some environmental features can be important but difficult to model. Survival of coastal organisms in Iceland, for instance, remains contentious, as sea ice and/or ice shelves seem to have persisted year-round (Hubbard et al., Citation2006; Ingólfsson, Citation2009). Biotic interactions can also control distributions: L. digitata is outcompeted at >1.5 m below Lowest Astronomical Tide (LAT) by L. hyperborea (Kain & Jones, Citation1975; Lüning, Citation1979), and in disturbed habitats (e.g. after storms) by the annual Saccorhiza polyschides (Engelen et al., Citation2011). This species may thus be less able to colonize deeper or less pristine waters, including thermally buffered offshore banks (Assis et al., Citation2016a), becoming more vulnerable to unfavourable intertidal (e.g. air temperature) climates. Seaweeds may also exhibit multiple gene pools within a single refugial area (Neiva et al., Citation2012), for example resulting from independent colonizations or local barriers, that cannot be directly distinguished using modelling approaches. Finally, predictions may be compromised to some extent by species misidentifications, as reported particularly in the NW Atlantic (Longtin & Saunders, Citation2015).
The actual extent of LGM distributions and, more relevant here, of long-term climatic refugia (the overlap of glacial-interglacial ranges) of L. digitata in the NW and NE Atlantic remains somewhat uncertain, raising the question of how important climatic refugia have been (e.g. as opposed to intra- and trans-Atlantic expansions) to the contemporary distribution and genetic architecture of this important seaweed. Assessing the population structure of this species is also important because future climate change is expected to produce important regional contractions at low-latitude ranges where higher levels of diversity and endemism are predicted to occur (Assis et al., Citation2018b). This study describes the large-scale genetic architecture of L. digitata, and investigates whether L. digitata persisted, as suggested by environmental niche models (ENMs), during the LGM on both sides of the Atlantic. Using polymorphic microsatellite markers, we compared genetic compositions across the Atlantic to (1) identify (and map) major gene-pools and unique refugia, (2) assess if NW Atlantic survival is supported by unique endemic variation, and if so, (3) determine whether genetic diversity and structure are, as predicted from larger hindcasted glacial ranges, higher in the NE versus NW Atlantic.
Materials and methods
Focal species
Laminaria digitata (Hudson) J.V. Lamouroux (Laminariaceae, Phaeophyceae, Ochrophyta) is a boreal perennial kelp with a cold-temperate to Arctic distribution in the North Atlantic. It is distributed from southern Brittany (France) and Long Island (NY, USA) to Greenland and the Barents Sea, and has also been reported from Svalbard and Novaya Zemlya archipelagos well within the Arctic Circle (Filbee-Dexter et al., Citation2019, and references therein). In many regions, L. digitata forests occur along a relatively narrow bathymetric range (−5 to 0 m LAT), but they have been reported to occur at depths of at least −15 m at higher latitudes (Adey & Hayek, Citation2011; Hop et al., Citation2012), where confusion with Hedophyllum nigripes (J.Agardh) Starko, S. C. Lindstrom & Martone (=Saccharina nigripes (J.Agardh) Lontin & G. W. Saunders) is a possibility. In the NW Atlantic, L. digitata has been genetically confirmed from −30 to −10 m in multiple locations (e.g. Bonne Bay in Newfoundland, Gary Saunders, pers. comm.). This kelp has a typical heteromorphic haploid-diploid life cycle, with large diploid sporophytes (up to 2 m) alternating with microscopic male and female haploid gametophytes. Short-range dispersal and local population dynamics are assumed to be largely mediated by short-lived (<72 h before settlement (Billot et al., Citation2003)) planktonic zoospores. Using hierarchical sampling from 45 m to >100 km, effective dispersal distance in this species is estimated at less than 10 km (Billot et al., Citation2003; Robuchon et al., Citation2014). However, there may be occasional long-distance migration events (e.g. via rafting of spore-bearing blades or epiphytic microscopic gametophytes) that can play an essential role for long-distance colonization (e.g. offshore islands, or newly available habitat encountered during range expansions), but they are not sufficient to prevent differentiation even at relatively small regional scales (Robuchon et al., Citation2014).
Sampling, DNA extraction and genotyping
Populations of L. digitata were sampled between 2004 and 2016 from six temperate sites in the NW Atlantic (43–47º Lat.) and 11 sites in the NE Atlantic, between southern Brittany, France (~47° Lat.) and Finnmark in northern Norway (~70.5° Lat.), covering both putative refugial and post-glacially colonized areas (, ). Minimum marine distances between populations ranged from 5 to over 3500 km. At each site, blade tissue was collected from 20–48 individuals sampled randomly or along linear transects, but always ensuring at least 1 m distance between individuals. Tissue samples were wiped cleaned from epiphytes and excess water when required and stored dehydrated in silica-gel crystals until DNA extraction.
Table 1. Geographic origin and genetic diversity of populations of Laminaria digitata sampled in this study
Fig. 1. Sampling of Laminaria digitata and genetic structure inferred from multi-locus microsatellite genotypes. (a) Sampling locations coloured according to genetic structure (see below). Black dots (NW Atlantic) indicate misidentified collections of Hedophyllum nigripes. Modern shorelines (black solid lines) are superimposed over emerged unglaciated (grey areas) and glaciated (ice sheets, white patterns) landmasses at the time of the LGM. The dotted line is an approximation of perennial sea-ice during the LGM. (b) Hierarchical structure plots assuming K=3 (top), K=5 (middle) and K=9 (bottom) genotypic clusters. Selected regional diversity indices are shown at the bottom. Different colours represent percentage ancestry of each genotyped individual (vertical bars). (c) Discriminant Analysis of Principal Components (DAPC) scatter plot (K=3) based on the same individual multi-locus genotypes, coloured according to the regions defined in (b). Note the congruence between analyses
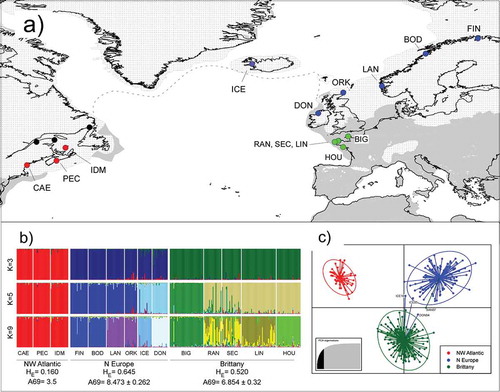
All individuals were genotyped for six microsatellites developed for L. digitata (Billot et al., Citation1998) and for six microsatellites developed for related L. ochroleuca (Coelho et al., Citation2014; see Supplementary table S1 for a summary of primer names, sequences, and amplification details). Four Brittany populations (BIG, SAN, LIN and HOU, see population names and locations in and ) had been previously genotyped (in Robuchon et al., Citation2014). Six other populations (CAE, PEC, FIN, LAN, ICE and DON) were genotyped at Station Biologique de Roscoff (SBR). Genomic DNA was extracted from 8–12 mg of dried tissue using the NucleoSpin 96 Plant II kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) following the manufacturer’s instructions. The lysis step was modified and performed at room temperature for 1–3 h instead of 65°C for 30 min. Microsatellite amplification and scoring was performed as detailed in Robuchon et al. (Citation2014). Alleles were sized using the SM594 size standard (Mauger et al., Citation2012) and scored manually using the software GeneMapper 4.0 (Applied Biosystems, Foster City, USA).
The four remaining populations (IDM, BOD, ORK and RAN) were genotyped at Centro de Ciências do Mar (CCMAR). PCR reactions were performed in a total volume of 15 μl containing 125 μM of each dNTP (Bioline, UK), 0.2–0.5 μM forward fluorescent-labelled primer FAM, NED, HEX, ROX and 0.5 µM reverse primer (MWG, France), 1.5–2.0 mM MgCl2, 1× GoTaq® Flexibuffer, 0.5–1 U GoTaq® FlexiDNA polymerase (Promega Corp., Madison, USA), and 1 μl of 1:10 or 1:100 diluted DNA template. Cycling conditions included an initial denaturation step at 94°C for 5 min, followed by 30 cycles at 94°C for 30 s, a primer-specific annealing temperature (Ta) for 30 s, and 72°C for 45 s, and a final extension at 72°C for 10 min. Amplified fragments were separated using an ABI PRISM capillary sequencer 3130XL Genetic Analyzer (Applied Biosystems, CCMAR, Portugal). Alleles were manually scored in STRand 2.4.110 (Toonen & Hughes, Citation2001) using the 500 LIZ™ size standard (Applied Biosystems, California, USA).
The two datasets were not directly comparable. Data were merged by transforming the called alleles (not raw allele sizes) in the CCMAR dataset into corresponding called alleles in the Roscoff dataset. The correspondence between called alleles was performed by genotyping in both laboratories a panel of 32 individuals from 11 locations covering our sampling area. Called alleles were then regressed and linear relationships were used to extrapolate the sizes of alleles sampled in only one of the datasets (Supplementary table S2). This calibration allowed correcting for systematic differences in allele size scoring attributable to different sequencers, labelled primers, size standards and scoring software and criteria (Supplementary fig. S1).
Genetic analyses
Summary statistics of genetic diversity within populations, including allele frequencies, mean (Am) and standardized (Astd) allelic richness, Nei’s gene diversity or expected heterozygosity (HE), observed heterozygosity (HO), multi-locus inbreeding coefficients (FIS) and number of private alleles were calculated with GENETIX 4.05 (Laboratoire Génome, Populations, Interactions, Université de Montpellier II; http://kimura.univ-montp2.fr/genetix) and the R package standArich v1.0 (available at http://alberto-lab.blogspot.pt/p/code.html). The same statistics were also computed for three selected regions, corresponding broadly to the NW Atlantic (npop=3, three other sampled populations were found to be Hedophyllum nigripes, see below), northern Europe (npop=6) and Brittany (npop=5).
Pairwise differentiation (Jost’s D, Weir and Cockerham Θ) of populations was calculated with the R package diveRsity 1.9 (Keenan et al., Citation2013). Genetic structure was analysed at multiple spatial scales using STRUCTURE 2.3 (Pritchard Lab, Stanford University; http://pritchardlab.stanford.edu/structure.html) without any prior population assignments. A range of assumed populations (K, set sequentially from 1 to 13) was run 10 times using a burn-in of 5 × 105 iterations and a run length of 1 × 106 iterations. Structure Harvester web v0.6.94 (http://taylor0.biology.ucla.edu/structureHarvester/) was used to summarize assignment results across independent runs. Structure analyses were complemented with a discriminant analysis of principal components (DAPC) implemented in the R package adegenet 2.1 (Jombart et al., Citation2008). The ‘best’ number of higher-level genetic clusters was assessed after checking the ΔK criterion (Evanno et al., Citation2005), the Bayesian information criterion (BIC), exploratory principal component analysis (PCA, adegenet) and factorial correspondence analysis (FCA, in GENETIX v. 4.05.2). Isolation-by-distance was assessed plotting geographic (minimum coastal routes in metres, as measured in Google Earth Pro v. 7.3.2.5776) and genetic (FST) distances and significance was assessed using Mantel tests using the R package vegan. Signatures of recent genetic bottlenecks in the NW Atlantic were assessed under a two-phase mutation model using the software BOTTLENECK v1.2.02 (Piry et al., Citation1999).
Results
Three populations of Laminaria digitata from the NW Atlantic could not be amplified for most loci, and a barcode marker (cox1) revealed that they were a distinct species (Hedophyllum nigripes; data not shown). The remaining 14 populations produced a total of 374 multi-locus genotypes with less than 25% missing data (>8 loci), that were used in the analyses. Microsatellite polymorphism within the 12 loci set varied from 2 (Lo454-27) to 24 (Ld371) alleles per locus, with a mean of 11.75 alleles per locus. Allelic richness was inflated by the high number of low-frequency alleles, dropping to 8.17 when excluding alleles with a global frequency ≤0.0067 (less than 5 alleles in total). Gene diversity (HE) within populations ranged from 0.061 (Cape Elizabeth, Maine, USA) to 0.600 (Donegal, Ireland), with high-latitude European populations consistently showing significant heterozygote deficiency ().
At the biogeographic scale, L. digitata was subdivided into three main genetic groups corresponding to Brittany, northern Europe and the NW Atlantic (), that could be easily recovered in exploratory PCAs (Supplementary fig. S2a) and FCAs (not shown). Structure clustering analyses revealed two or three well-supported (ΔK > 1000, Supplementary fig. S2b) and stable genotypic clusters, matching PCA-defined clusters (). The two European clusters were further subdivided into smaller (K=5, 6, 9) genotypic clusters with weaker support (Supplementary fig. S2b), but still showed stable compositions and a clear geographic signal. The ‘elbow’ in DAPC BIC criteria (Supplementary fig. S2c) supported with some ambiguity K=11 clusters, corresponding roughly to 10 NE genetic clusters plus a single NW Atlantic cluster. These genetic clusters matched to a large degree individual populations, but with a high proportion of mismatched/admixed individuals (data not shown). In STRUCTURE, K=11 produced variable sample groupings between runs, and thus this K value was not considered further. When three clusters were imposed in the DAPC analysis, they recovered the same STRUCTURE groupings, with minimal mismatch/admixed individuals between regions (). In this analysis, the most discriminant function separated NW from NE clusters (Supplementary fig. S3a).
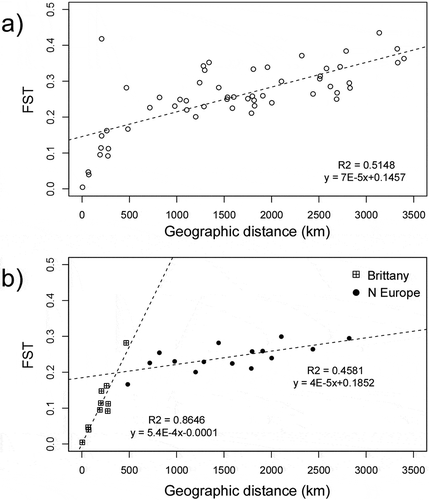
The prediction of higher genetic diversity and geographic subdivision of L. digitata in the NE than in the NW Atlantic was fully supported (, , bottom). NW Atlantic showed the lowest allelic and genotypic diversity (Astd=3.5; HE=0.160) at the population and regional levels (), with one or two alleles dominating across all loci (0.64<fa<1). BOTTLENECK results failed to support a recent bottleneck in the NW Atlantic (or the other regions), because no significant heterozygote excess was detected (one-tailed Wilcoxon test, P < 0.05) and allele frequency classes displayed a normal L-shaped distribution (Supplementary table S3). Within the NE Atlantic, genetic diversity was of the same order of magnitude as in Brittany (Astd=6.854; HE=0.520) and northern Europe (Astd=8.473; HE=0.645). Within the latter, there was a clear latitudinal decrease in diversity (). Pairwise FST ranged between 0.0045 (intra-Brittany) and 0.6799 (Cape Elizabeth versus Finnmark) (Supplementary fig. S4a). Absolute population differentiation, as measured by Jost’s D, ranged from 0.002 and 0.6129, corresponding to the same comparisons as FST(Supplementary fig. S4b). Within regions, pairwise population differentiation was higher within northern Europe than within Brittany, and lowest in the NW Atlantic (). The significant signal of differentiation-by-distance in the whole NE Atlantic (P = 0.001) was lost when the populations of Brittany were removed (P = 0.251) (), but Brittany alone (the only region in this study with population pairs separated by distances below 500 km) showed a significant increase in genetic differentiation with distance (P = 0.008).
Fig. 2. Genetic diversity and differentiation of populations of Laminaria digitata within selected regions. Above Nei’s gene diversity (HE) at population (box plot) and regional (open stars) levels. Below Pairwise differentiation of populations (Jost’s D) within regions. Box plots depict the median (horizontal line), the 25th and 75th percentiles (bottom and top of the box) and the minimum/maximum values (horizontal lines). The box plot’s width is proportional to sample size (number of regions or pairwise comparisons)
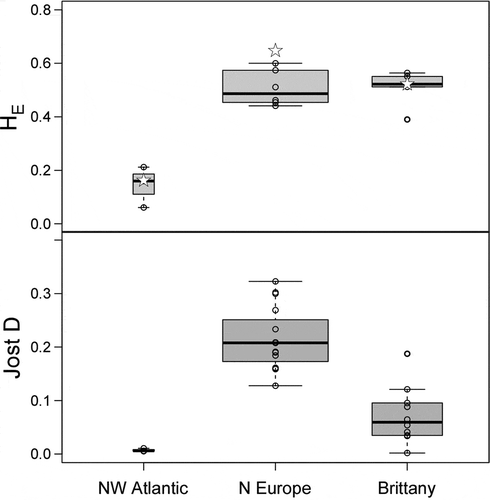
Fig. 3. Latitudinal trend in genetic diversity within the N Europe cluster. Black dots (and straight line) depict standardized allelic richness (A17) and open dots (and dotted line) unbiased gene diversity (HE)
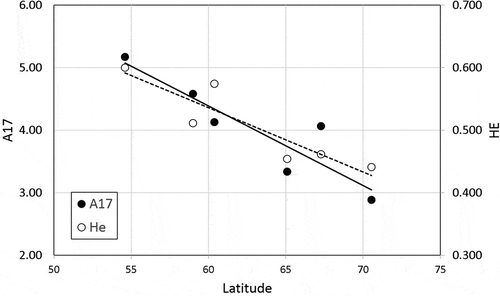
Fig. 4. Isolation-by-distance in Laminaria digitata. Estimates of pairwise differentiation (FST) are plotted against minimum marine distances (km) for (a) all NE Atlantic populations (Mantel r = 0.814, P = 0.001) and (b) separately for Brittany (Mantel r = 0.930, P =0.008) and N Europe (Mantel r = 0.677, P = 0.006)
Discussion
This study showed that Laminaria digitata is composed of three major genetic clusters, two in the NE (Brittany and northern Europe) and one in the NW Atlantic. Brittany, the present-day European rear edge (known range limit at HOU; Lüning, Citation1990; Oppliger et al., Citation2014), is a biogeographic transition zone (Spalding et al., Citation2007) long-identified as a genetic hotspot for a diverse range of coastal species (Provan et al., Citation2005; Hoarau et al., Citation2007; Neiva et al., Citation2014). Its diversity probably reflects its proximity to the LGM Armorican and Celtic shorelines, where distribution models have identified vast areas of suitable unglaciated shallow-water habitat (Waltari & Hickerson, Citation2013; Assis et al., Citation2018b; see also Assis et al., Citation2014, Citation2016b; Neiva et al., Citation2014; Bermejo et al., Citation2018).
The northern Europe cluster, sampled across more distant locations, spread from former periglacial (Ireland) to heavily glaciated (Iceland, N Norway) regions. Ireland and possibly western Scotland are the most likely sources of the extensive post-glacial colonization of northern Europe (see also Gómez et al., Citation2007; Hoarau et al., Citation2007; Neiva et al., Citation2016). Irish and Scottish populations showed the highest allelic diversity, and a clear decrease in allelic and genotypic diversity towards higher latitudes was observed, with minima in Iceland and Finnmark. Iceland has been identified as a possible glacial refugium for cold-tolerant kelps (Assis et al., Citation2016b) and invertebrates (Wares & Cunningham, Citation2001). Palaeo-environmental reconstructions of the Icelandic ice sheet and ice shelves (Hubbard et al., Citation2006), however, show that these coastal areas may have been completely glaciated (and hence inhospitable) during the LGM (Ingólfsson, Citation2009; Coyer et al., Citation2011; Neiva et al., Citation2018). This hypothesis is supported by the low diversities of our Iceland and Finnmark samples which could also be explained by their distance from southern periglacial regions and/or the mainland. It is not immediately apparent why northern European populations (Orkney, Norway, Iceland) exhibited consistent heterozygote deficiencies, but this pattern does not seem random. Its regional scope could indicate some shift in mating-system or habitat characteristics (e.g. lower tidal amplitudes) at higher latitudes leading to increased inbreeding or a Wahlund effect (e.g. individuals reproducing in distinct reproductive windows), but in the absence of data on reproductive ecology, the underlying cause remains speculative.
The clear genetic discontinuity and the high number of non-shared (private) regional alleles suggests that fine-scale geographic structuring across the older Brittany/Ireland range dates back to the LGM, even if more contemporary oceanographic regimes help fine-tune the actual boundaries of each cluster (Nicastro et al. Citation2020). This evolutionary pattern was previously found in Palmaria palmata, Fucus serratus and Ascophyllum nodosum (Provan et al., Citation2005; Hoarau et al., Citation2007; Olsen et al., Citation2010), and pinpoints how genetic structure in seaweeds can arise and be maintained at relatively small spatial scales within continuous refugial areas (see also Neiva et al., Citation2012). This segregation might also suggest that Brittany has contributed little to the recolonization of northern Europe, with most shared alleles probably representing unsorted polymorphisms.
Both Brittany and N Europe clusters were further subdivided into smaller genetic sectors, corresponding to single populations or pairs of contiguous populations. Distance may help explain the pattern in N Europe, and finer-scale studies may show additional structuration. This pattern was striking in Brittany, where minimum distances between contiguous sampled populations from different clusters (e.g. HOU/LIN, RAN/BIG) were of the order of 200 km, a pattern previously observed in this region even between populations sampled at smaller scales (Billot et al., Citation2003; Robuchon et al., Citation2014). Obvious differentiation was also detected between Cornish and Scottish populations in Britain (King et al., Citation2019), but not between populations separated by less than 20 km in Northern Ireland (Brennan et al., Citation2014), where instead of clear subdivisions only a signal of isolation-by-distance was apparent.
The clear differentiation of the NW Atlantic and European clusters seems to favour the hypothesis, amply supported by niche modelling (i.e. presence of suitable habitat (Assis et al., Citation2018b)), of local LGM survival over post-glacial trans-Atlantic colonization from northern Europe. LGM survival however would be reflected in higher regional genetic diversity and unique private alleles, which was not the case. Due to field misidentifications, the northern parts of the Gulf of Saint Laurence and Newfoundland were not sampled, thus the set of populations analysed here may not represent the entire regional diversity pool. Nonetheless, such low diversity is unexpected; the Canadian Maritimes, like Brittany and Ireland, are in the vicinity of periglacial regions just south of the Laurentide ice sheet, and of formerly emerged and unglaciated areas of the Grand Banks shelf to the north-east. Several scenarios can account for such a severe genetic bottleneck in this region.
The first, and perhaps the most plausible, scenario entails the area actually undergoing post-glacial colonization. In a stepping-stone, trans-Atlantic migration scenario, presumably involving a sequential colonization of Iceland, southern Greenland and – after reaching the American continent – Labrador and Newfoundland, regional genetic diversity would have been extremely depleted by the time L. digitata reached the Canadian Maritimes and New England. Glacial eradication is interesting, since suitable habitat was seemingly available and so many other amphi-Atlantic seaweeds survived glacial advances there (Li et al., Citation2016; Bringloe & Saunders, Citation2018; Neiva et al., Citation2018). Perhaps its absence was not related to lack of suitable habitat, but to not being able to get there in the first place. L. digitata is not buoyant and lacks long-lived dispersive stages (Valero et al., Citation2011). Long-distance colonization across the open sea is probably very rare and contingent upon entanglement in drifting algal rafts and/or extreme events. These seem necessary to explain the colonization of remote sites such as Iceland and Greenland, but may have never occurred (or rare early colonizers never survived the initial establishment stages) in the NW Atlantic.
It may even be possible, as for F. serratus and the snail Littorina littorea, that trans-Atlantic crossing has occurred only in the past few centuries associated with maritime traffic (Brawley et al., Citation2009). The relatively wide recorded distribution of L. digitata in the western Atlantic (Sharp et al., Citation2008; Adey & Hayek, Citation2011; Merzouk & Johnson, Citation2011; Assis et al., Citation2018b, their fig. S1e) appears to suggest an older establishment, but modern (and probably historical) ranges also seem to have been overestimated due to confusion with Hedophyllum nigripes (Longtin & Saunders, Citation2015; Filbee-Dexter et al., Citation2019). Genetically confirmed records of L. digitata on the NW Atlantic are restricted to temperate New England and the Canadian Maritimes (Bringloe & Saunders, Citation2018, fig. S16), whereas its presence along colder subarctic regions has only been confirmed around Churchill (Hudson Bay), but not in Labrador or the Canadian Arctic (Gary Saunders, pers. comm.) where digitate forms of H. nigripes also occur. Likewise, the species has never been genetically confirmed from Greenland, a presumably obligatory trans-Atlantic stepping stone. Recent literature records and consultations with local seaweed experts suggest its absence there (Dorte Krause-Jensen, Susse Wegeberg, pers. comm.; see also Krause-Jensen et al., Citation2012; Filbee-Dexter et al., Citation2019), although additional efforts would be required to confirm this. This apparent absence is striking because in the NE Atlantic L. digitata reaches the high Arctic archipelagos of Svalbard (genetically confirmed, Inka Bartsch, pers. comm.) and Novaya Zemlya (Filbee-Dexter et al., Citation2019).
Genetic bottlenecks may alternatively have occurred despite the continued (but not static) presence of L. digitata in the NW Atlantic. Its former glacial range may have been extremely narrow, i.e. a true refugium. Erosion of pre-glacial variation may have been intensified by periodic displacements associated with the shoreline’s transgressions and regressions, which in this shallow basin are of the scale of hundreds of kilometres. Speculatively, sea-level effects may have been further aggravated by shifts in substratum availability and/or productivity (Graham et al., Citation2003; Fauvelot et al., Citation2008). Finally, the mid-Holocene warming pulse (around 6 ka BP) may have also played a role – niche models, for instance, support temporary extirpation from its modern rear edge in New England (Assis et al., Citation2018b). The hypothesized regional genetic bottleneck was not supported by tests (BOTTLENECK), because the NW Atlantic displayed significant heterozygote deficiency and not, as expected, heterozygote excess. However, heterozygosity and allele size-class departures are quite transient, and thus much more appropriate for detecting recent demographic shifts rather than historical demography. The fixed (or nearly so) microsatellite alleles that ‘defined’ the NW Atlantic cluster (Supplementary fig. S3b) were not identified as private because they were also detected (generally at much lower frequencies) in European populations. These loci however show many consecutive alleles across their respective size ranges in European populations, so there is a possibility that allele sharing across the Atlantic results, at least for some of them, from size homoplasy rather than from recent shared ancestry.
Disentangling these competing scenarios of bottlenecked LGM survival versus recent trans-Atlantic colonization may require additional NW Atlantic samples, both from nearby periglacial areas (e.g. Newfoundland) and from potential migration routes along Labrador (and potentially southern Greenland). Independent molecular data may also help clarify this issue. Laminaria digitata, unlike so many other amphi-Atlantic seaweeds (Li et al., Citation2016; Bringloe & Saunders, Citation2018; Neiva et al., Citation2018), does not show any trans-Atlantic divergence at organelle markers, specifically at the barcoding locus mt cox1 (McDevit & Saunders, Citation2010; Bringloe & Saunders, Citation2018, fig. S16). Unfortunately, monomorphism (or very low variability) across the entire range, and also within the NE Atlantic, shows that their resolution at the infraspecific level is not adequate for phylogeographic inference.
It is prudent to consider the potential negative effects of ongoing climatic change at both southern range edges. The distribution of L. digitata is predicted to contract in the 21st century at lower latitudes, particularly in high CO2 emission scenarios. In the NE Atlantic, these contractions appear particularly problematic, because they forecast the complete extirpation of L. digitata from Brittany and southern England (Raybaud et al., Citation2013; Assis et al., Citation2018b), and therefore the loss of the unique (and generally diverse) genetic cluster present there. Fine-scale analyses of sea-surface temperature shifts in Brittany over the past two decades (0.7°C on average) reveal that Brittany constitutes a mosaic of thermal conditions, with colder western and north-western regions being less affected by climatic change (Gallon et al., Citation2014). Common-garden experiments also suggest the existence of locally adapted thermal ecotypes in L. digitata. Populations from south-western UK (Cornwall), for instance, show higher resilience to marine heat waves – as assessed in short-term heat-shock experiments – than more northern ones (King et al., Citation2019). Environmental heterogeneity, local refugia and local adaptation might delay the decline of L. digitata at its trailing edge, particularly in more benign greenhouse-gas emission scenarios, but may not be sufficient to ensure survival in the long term.
In the NW Atlantic, L. digitata is also predicted to undergo a northward shift of uncertain magnitude. Depending on climate model and study, the species is projected to contract from Long Island to the northern Gulf of Maine/Bay of Fundy (Wilson et al., Citation2019) or even to the Gulf of St. Lawrence/Newfoundland (Assis et al., Citation2018b). The global redistribution (and thus vulnerability) of the unique NW Atlantic cluster is uncertain, as is, in the absence of samples beyond the Maritimes, the existence of additional genetic groups. The consequences are thus harder to forecast, but assuming a more extended distribution into the Gulf of St. Lawrence/Newfoundland, this genetic cluster, similar to the European cluster, seems safeguarded from regional extinction.
A few concluding remarks: L. digitata and H. nigripes bear great morphological resemblance and co-occur at the same intertidal/subtidal depths throughout the entire confirmed range of L. digitata in the NW Atlantic and in the European Arctic. Researchers should be aware of this potential problem, and double-check their material for established barcoding (cox1) markers. Barcoding also seems the most cost-effective approach to establish the actual range limits of L. digitata in the NW Atlantic, where low genetic diversity is apparently accompanied by some degree of niche unfilling, as the species seems absent (unlike in the NE Atlantic) from more Arctic environments. Accurate distributional data and niche comparisons across the Atlantic are necessary to explore this hypothesis. Other markers (or populations) may be required to settle the issue of glacial persistence versus post-glacial colonization of the NW Atlantic, because the extreme regional genetic bottleneck contradicts ENM-based predictions. How European pools segregate and/or admix throughout Cornwall and southern Ireland is another interesting, open question. Monitoring this southern range is advised given its ecological and economic value and vulnerability, and because range shifts are already being documented for related kelp species (Schoenrock et al., Citation2019). Partial incongruence between modelling and genetic-based biogeographic inferences highlight the benefits of comparing both approaches to understand how shifting climatic conditions affect marine species distributions and explain large-scale patterns of spatial genetic structure.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at http://xxx.
Supplementary table S1. Primer sequences and amplification details of amplified microsatellite markers.
Supplementary table S2. Calibration of the Roscoff and CCMAR genotyping data.
Supplementary Table S3. Bottleneck results.
Supplementary fig. S1. Calibration of allele sizes between the Centro de Ciências do Mar (CCMAR) and Station Biologique de Roscoff (SBR) data-sets.
Supplementary fig. S2. ‘Best’ number of genotypic clusters (k) in sampled Laminaria digitata according to multiple criteria.
Supplementary fig. S3. Genetic discrimination of the NW Atlantic cluster.
Supplementary fig. S4. Heatmaps of pairwise differentiation between Laminaria digitata populations.
Author contributions
J. Neiva: analysis of molecular data, led manuscript drafting and editing; E.A. Serrão: original concept, funding and supervision, sampling, editing manuscript; C. Paulino, L. Gouveia, M. Ballenghien, S. Mauger: laboratory work, data analysis assistance; A. Want and É. Tamigneaux: sampling, editing manuscript; L. Fouqueau: data analysis assistance, editing manuscript; C. Engel-Gautier and C. Destombe: original concept, sampling, data analysis assistance, manuscript revision and editing; M. Valero: original concept, funding and supervision, sampling, analysis of molecular data, editing manuscript.
Availability of data and materials
Genotypic data was deposited and is available in the Figshare repository at http://dx.doi.org/10.6084/m9.figshare.10255475.
TEJP-2019-0135-File006.docx
Download MS Word (869.1 KB)Acknowledgments
We are grateful to Marta Valente (CCMAR DNA sequencing platform) and to the Biogenouest genomics core facility (Genomer Plateforme génomique at the Station Biologique de Roscoff) for their technical support. We warmly thank several phycologists and the Marine Observation Department and diving team at the Station Biologique de Roscoff for sampling, and Inka Bartsch, Gary Saunders, Dorte Krause-Jensen and Susse Wegeberg for their valuable insights regarding distribution and depth range.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental Material
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at https://doi.org/10.1080/09670262.2020.1750058
References
- Adey, W.H. & Hayek, L.C. (2011). Elucidating marine biogeography with macrophytes: quantitative analysis of the North Atlantic supports the thermogeographic model and demonstrates a distinct subarctic region in the Northwestern Atlantic. Northeastern Naturalist, 18: 1–128.
- Assis, J., Araújo, M.B. & Serrão, E.A. (2018b). Projected climate changes threaten ancient refugia of kelp forests in the North Atlantic. Global Change Biology, 24: e55–e66.
- Assis, J., Coelho, N.C., Lamy, T., Valero, M., Alberto, F. & Serrão, E.A. (2016a). Deep reefs are climatic refugia for genetic diversity of marine forests. Journal of Biogeography, 43: 833–844.
- Assis, J., Lucas, A.V., Bárbara, I. & Serrão, E.A. (2016b). Future climate change is predicted to shift long-term persistence zones in the cold-temperate kelp Laminaria hyperborea. Marine Environmental Research, 113: 174–182.
- Assis, J., Serrão, E.A., Claro, B., Perrin, C. & Pearson, G.A. (2014). Climate-driven range shifts explain the distribution of extant gene pools and predict future loss of unique lineages in a marine brown alga. Molecular Ecology, 23: 2797–2810.
- Assis, J., Serrão, E.A., Coelho, N.C., Tempera, F., Valero, M. & Alberto, F. (2018a). Past climate changes and strong oceanographic barriers structured low‐latitude genetic relics for the golden kelp Laminaria ochroleuca. Journal of Biogeography, 45: 2326–2336.
- Bermejo, R., Chefaoui, R.M., Engelen, A.H., Buonomo, R., Neiva, J., Ferreira-Costa, J., Pearson, G.A., Marbà, N., Duarte, C.M., Airoldi, L., Hernández, I., Guiry, M.D. & Serrão, E.A. (2018). Marine forests of the Mediterranean-Atlantic Cystoseira tamariscifolia complex show a southern Iberian genetic hotspot and no reproductive isolation in parapatry. Scientific Reports, 8: 10427.
- Billot, C., Engel, C.R., Rousvoal, S. Kloareg, B. & Valero, M. (2003). Current patterns, habitat discontinuities and population genetic structure: the case of the kelp Laminaria digitata in the English Channel. Marine Ecology Progress Series, 253: 111–121.
- Billot, C., Rousvoal, S., Estoup, A., Epplen, J.T., Saumitou-Laprade, P., Valero, M. & Kloareg, B. (1998). Isolation and characterization of microsatellite markers in the nuclear genome of the brown alga Laminaria digitata (Phaeophyceae). Molecular Ecology, 7: 1778–1780.
- Brawley, S.H., Coyer, J.A., Blakeslee, A.M.H., Hoarau, G., Johnson, L.E., Byers, J.E., Stam, W.T. & Olsen, J.L. (2009). Historical invasions of the intertidal zone of Atlantic North America associated with distinctive patterns of trade and emigration. Proceedings of the National Academy of Sciences USA, 106: 8239–8244.
- Brennan, G., Kregting, L., Beatty, G.E., Cole, C., Elsäßer, B., Savidge, G. & Provan, J. (2014). Understanding macroalgal dispersal in a complex hydrodynamic environment: a combined population genetic and physical modelling approach. Journal of the Royal Society, Interface, 11: 20140197.
- Bringloe, T.T. & Saunders, G.W. (2018). Mitochondrial DNA sequence data reveal the origins of postglacial marine macroalgal flora in the Northwest Atlantic. Marine Ecology Progress Series, 589: 45–58.
- Brochmann, C., Gabrielsen, T.M., Nordal, I., Landvik, J.Y. & Elven, R. (2003). Glacial survival or tabula rasa? The history of North Atlantic biota revisited. Taxon, 52: 417–450.
- Carlson, A.E. & Winsor, K. (2012). Northern hemisphere ice-sheet responses to past climate warming. Nature Geoscience, 5: 607–613.
- Coelho, N.C., Serrão, E.A. & Alberto, F. (2014). Characterization of fifteen microsatellite markers for the kelp Laminaria ochroleuca and cross species amplification within the genus. Conservation Genetics Resources, 6: 949–950.
- Coyer, J.A., Hoarau, G., Van Schaik, J., Luijckx, P. & Olsen, J.L. (2011). Trans-Pacific and trans-Arctic pathways of the intertidal macroalga Fucus distichus L. reveal multiple glacial refugia and colonizations from the North Pacific to the North Atlantic. Journal of Biogeography, 38: 756–771.
- Engelen, A.H., Lévèque, L., Destombe, C. & Valero, M. (2011). Spatial and temporal patterns of recovery of low intertidal Laminaria digitata after experimental spring. Cahiers de Biologie Marine, 52: 441–453.
- Evanno, G., Regnaut, S. & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14: 2611–2620.
- Fauvelot, A.C., Bernardi, G. & Planes, S. (2008). Reductions in the mitochondrial DNA diversity of coral reef fish provide evidence of population bottlenecks resulting from holocene sea-level change. Evolution, 57: 1571–1583.
- Filbee-Dexter, K., Wernberg, T., Fredriksen, S., Norderhaug, K.M. & Pederson, M.F. (2019). Arctic kelp forests: diversity, resilience and future. Global and Planetary Change, 172: 1–14.
- Fraser, C.I., Thiel, M., Spencer, H.G. & Waters, J.M. (2010). Contemporary habitat discontinuity and historic glacial ice drive genetic divergence in Chilean kelp. BMC Evolutionary Biology, 10: 203.
- Gallon, R.K., Robuchon, M., Leroy, B. Le Gall, L., Valero, M., & Feunteun, E. (2014). Twenty years of observed and predicted changes in subtidal red seaweed assemblages along a biogeographical transition zone: inferring potential causes from environmental data. Journal of Biogeography, 41: 2293–2306.
- Gómez, A., Hughes, R.N., Wright, P.J., Carvalho, G.R. & Lunt, D.H. (2007). Mitochondrial DNA phylogeography and mating compatibility reveal marked genetic structuring and speciation in the NE Atlantic bryozoan Celleporella hyalina. Molecular Ecology, 16: 2173–2188.
- Graham, M.H., Dayton, P.K. & Erlandson, J.M. (2003). Ice ages and ecological transitions on temperate coasts. Trends in Ecology & Evolution, 18: 33–40.
- Hoarau, G., Coyer, J.A., Veldsink, J.H., Stam, W.T. & Olsen, J.L. (2007). Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Molecular Ecology, 16: 3606–16.
- Hop, H., Wiencke, C., Barbara, V. & Kovaltchouk, N.A. (2012). Species composition, zonation, and biomass of marine benthic macroalgae in Kongsfjorden, Svalbard. Botanica Marina, 55: 399–414.
- Hubbard, A., Sugden, D., Dugmore, A., Norddahl, H. & Pétursson, H.G. (2006). A modelling insight into the Icelandic Last Glacial Maximum ice sheet. Quaternary Science Reviews, 25: 2283–2296.
- Ingólfsson, A. (2009). A marine refugium in Iceland during the last glacial maximum: fact or fiction? Zoologica Scripta, 38: 663–665.
- Jombart, T., Lyon, D. & Biome, L. De. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics, 24: 1403–1405.
- Kain, J.M. & Jones, N.S. (1975). Algal recolonization of some cleared subtidal areas. Journal of Ecology, 63: 739–765.
- Keenan, K., Mcginnity, P., Cross, T.F., Crozier, W.W., & Prodöhl, P.A. (2013). Diversity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods in Ecology and Evolution, 4: 782–788.
- King, N.G., Mckeown, N.J., Smale, D.A., Wilcockson, D.C., Hoelters, L., Groves, E.A., Stamp, T. & Moore, P.J. (2019). Evidence for different thermal ecotypes in range centre and trailing edge kelp populations. Journal of Experimental Marine Biology and Ecology, 514–515: 10–17.
- Krause-Jensen, D., Marbà, N., Olesen, B., Sejr, M.K., Christensen, P.B., Rodrigues, J., Renaud, P.E., Balsby, T.J.S. & Rysgaard, S. (2012). Seasonal sea ice cover as principal driver of spatial and temporal variation in depth extension and annual production of kelp in Greenland. Global Change Biology, 18: 2981–2994.
- Li, J.-J., Hu, Z.-M. & Duan, D.-L. (2015). Genetic data from the red alga Palmaria palmata reveal a mid-Pleistocene deep genetic split in the North Atlantic. Journal of Biogeography, 42: 902–913.
- Li, J.-J., Hu, Z.-M. & Duan, D.-L. (2016). Survival in glacial refugia versus postglacial dispersal in the North Atlantic: The cases of red seaweeds. Pp. 309–330 in: Seaweed Phylogeography - Adaptation and Evolution of Seaweeds under Environmental Change.
- Longtin, C.M. & Saunders, G.W. (2015). On the utility of mucilage ducts as a taxonomic character in Laminaria and Saccharina (Phaeophyceae) – the conundrum of S. groenlandica. Phycologia, 54: 440–450.
- Lourenço, C.R., Zardi, G.I., McQuaid, C.D., Serrão, E.A., Pearson, G.A. & Nicastro, K.R. (2016). Upwelling areas as climate change refugia for the distribution and genetic diversity of a marine macroalga. Journal of Biogeography, 43: 1595–1607.
- Lüning, K. (1979). Growth strategies of three Laminaria species (Phaeophyceae) inhabiting different depth zones in the sublittoral region of Helgoland (North Sea). Marine Ecology Progress Series, 1: 195–207.
- Lüning, K. (1990). Seaweeds: Their Environment, Biogeography, and Ecophysiology. Revised Ed. Wiley, New York. 527 pp.
- Luttikhuizen, P.C., Heuvel, F.H.M. Van Den, Rebours, C., Witte, H.J., Bleijswijk, J.D.L. Van & Timmermans, K. (2018). Strong population structure but no equilibrium yet: genetic connectivity and phylogeography in the kelp Saccharina latissima (Laminariales, Phaeophyta). Ecology and Evolution, 8: 4265–4277.
- Maggs, C.A., Castilho, R., Foltz, D., Henzler, C., Jolly, M.T., Kelly, J., Olsen, J., Perez, K.E., Stam, W., Väinölä, R., Viard, F. & Wares, J. (2008). Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology, 89: S108–S122.
- Mauger, S., Couceiro, L. & Valero, M. (2012). A simple and cost-effective method to synthesize an internal size standard amenable to use with a 5-dye system. Prime Research on Biotechnology, 2: 40–46.
- McDevit, D.C. & Saunders, G.W. (2010). A DNA barcode examination of the Laminariaceae (Phaeophyceae) in Canada reveals novel biogeographical and evolutionary insights. Phycologia, 49: 235–248.
- Merzouk, A. & Johnson, L.E. (2011). Kelp distribution in the northwest Atlantic Ocean under a changing climate. Journal of Experimental Marine Biology and Ecology, 400: 90–98. Elsevier B.V.
- Neiva, J., Assis, J., Fernandes, F., Pearson, G.A. & Serrão, E.A. (2014). Species distribution models and mitochondrial DNA phylogeography suggest an extensive biogeographical shift in the high-intertidal seaweed Pelvetia canaliculata. Journal of Biogeography, 41: 1137–1148.
- Neiva, J., Paulino, C., Nielsen, M.M., Krause-Jensen, D., Saunders, G.W., Assis, J., Bárbara, I., Tamigneaux, É., Gouveia, L., Aires, T., Marbà, N., Bruhn, A., Pearson, G.A. & Serrão, E.A. (2018). Glacial vicariance drives phylogeographic diversification in the amphi-boreal kelp Saccharina latissima. Scientific Reports, 8: 1112.
- Neiva, J., Pearson, G.A., Valero, M. & Serrão, E.A. (2012). Fine-scale genetic breaks driven by historical range dynamics and ongoing density-barrier effects in the estuarine seaweed Fucus ceranoides L. BMC Evolutionary Biology, 12: 78.
- Neiva, J., Serrão, E.A., Assis, J., Pearson, G.A., Coyer, J.A., Olsen, J.L., Hoarau, G. & Valero, M. (2016). Climate oscillations, range shifts and phylogeographic patterns of North Atlantic Fucaceae. In Seaweed Phylogeography – Adaptation and Evolution of Seaweeds under Environmental Change (Hu, Z.-M. & Fraser, C., editors), 279–308. Springer, Dordrecht.
- Nicastro, K.R., Assis, J., Serrão, E.A., Pearson, G.A., Neiva, J., Valero, M., Jacinto, R. & Zardi, G.I. (2020). Congruence between fine-scale genetic breaks and dispersal potential in an estuarine seaweed across multiple transition zones. ICES Journal of Marine Science, 77: 371–378.
- Olsen, J.L., Zechman, F.W., Hoarau, G., Coyer, J., Stam, W., Valero, M. & Åberg, P. (2010). The phylogeographic architecture of the fucoid seaweed Ascophyllum nodosum: an intertidal “marine tree” and survivor of more than one glacial-interglacial cycle. Journal of Biogeography, 37: 842–856.
- Oppliger, L.V., von Dassow, P., Bouchemousse, S., Robuchon, M., Valero, M., Correa, J.A., Mauger, S. & Destombe, C. (2014). Alteration of sexual reproduction and genetic diversity in the kelp species Laminaria digitata at the southern limit of its range. PLoS ONE, 9: e102518.
- Petit, R.J., Aguinagalde, I., Beaulieu, J.-L., Bittkau, C., Brewer, S., Cheddadi, R., Ennos, R., Fineschi, S., Grivet, D., Lascoux, M., Mohanty, A., Müller-Starck, G., Demesure-Musch, B., Palmé, A., Martín, J.P., Rendell, S., & Vendramin, G.G. (2003). Glacial refugia: Hotspots but not melting pots of genetic diversity. Science, 300: 1563–1565.
- Piry, S., Luikart, G. & Cornuet, J.-M. (1999). BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity, 90: 502–503.
- Provan, J. & Maggs, C. A. (2012). Unique genetic variation at a species’ rear edge is under threat from global climate change. Proceedings of the Royal Society B, Biological Sciences, 279: 39–47.
- Provan, J., Wattier, R.A. & Maggs, C.A. (2005). Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Molecular Ecology, 14: 793–803.
- Raybaud, V., Beaugrand, G., Goberville, E., Delebecq, G., Destombe, C., Valero, M., Davoult, D., Morin, P. & Gevaert, F. (2013). Decline in kelp in West Europe and climate. PLoS ONE, 8: e66044.
- Robuchon, M., Le Gall, L., Mauger, S. & Valero, M. (2014). Contrasting genetic diversity patterns in two sister kelp species co-distributed along the coast of Brittany, France. Molecular Ecology, 23: 2669–2685.
- Schoenrock, K.M., Callaghan, T.O., Callaghan, R.O. & Krueger-Hadfield, S.A. (2019). First record of Laminaria ochroleuca Bachelot de la Pylaie in Ireland in Béal an Mhuirthead, county Mayo. Marine Biodiversity Records, 12: 9.
- Sharp, G., Allard, M. & Lewis, A. (2008). The potential for seaweed resource development in subarctic Canada; Nunavik, Ungava Bay. Journal of Applied Phycology, 20: 491–498.
- Spalding, M.D., Fox, H.E., Allen, G.R., Davidson, N., Ferdaña, Z.A., Finlayson, M., Halpern, B.S., Jorge, M.A., Lombana, A., Lourie, S.A., Martin, K.D., McManus, E., Molnar, J., Recchia, C.A. & Robertson, J. (2007). Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. BioScience, 57: 573–583.
- Toonen, R.J., & Hughes, S. (2001). Increased throughput for fragment analysis on an ABI Prism® 377 automated sequencer using a membrane comb and STRand software. BioTechniques, 3: 1320–1324.
- Valero, M., Destombe, C., Mauger, S., Ribout, C., Engel, C.R., Daguin-thiebaut, C. & Tellier, F. (2011). Using genetic tools for sustainable management of kelps: a literature review and the example of Laminaria digitata. Cahiers de Biologie Marine, 52: 467–483.
- Vermeij, G.J. (1991). Anatomy of an invasion: the trans arctic interchange. Paleobiology, 17: 281–307.
- Waltari, E. & Hickerson, M.J. (2013). Late Pleistocene species distribution modelling of North Atlantic intertidal invertebrates. Journal of Biogeography, 40: 249–260.
- Wares, J.P. & Cunningham, C.W. (2001). Phylogeography and historical ecology of the North Atlantic intertidal. Evolution, 55: 2455–2469.
- Wilson, K.L., Skinner, M.A. & Lotze, H.K. (2019). Projected 21st‐century distribution of canopy‐forming seaweeds in the Northwest Atlantic with climate change. Diversity and Distributions, 25: 582–602.
