ABSTRACT
The green seaweeds Ulva australis and U. pertusa were described from southern Australia and Japan, respectively. They are conspecific and U. australis, the currently accepted taxon, is native to temperate marine waters in north-eastern Asia, and known to be introduced overseas into Australasia, the Americas and Europe. Although the genetics of U. australis have been investigated elsewhere, along French coasts the origins and history of the introduction of this species need to be clarified. We used mitochondrial, plastid and nuclear markers to differentiate introduced populations of U. australis along the French Atlantic coasts. The plastid tufA gene used as a barcoding marker revealed a well-defined species with a higher haplotype diversity in native vs. introduced areas. The ITS2 region (nuclear) and rbcL (plastid) were used to compare French specimens with the lectotype of U. australis. Putative geographic origins of the genetically determined U. australis were examined using genetic markers with better resolution, the plastid atpI-H combined with the mitochondrial trnA-N. Origin(s) and introduction history of French specimens were inferred from the comparison between their haplotypes and those previously described in native and non-native temperate areas worldwide. Our results indicate that the presence of U. australis along the French Atlantic and Mediterranean coasts is the result of multiple introductions and independent pathways, and suggest that historical oyster transfers from Japan and British Columbia can only partially explain the observed patterns in genetic markers. Alternative hypotheses for the timing and pathways of introductions are proposed in the light of the historic background of maritime transport networks and trade between north-eastern Asia and Europe.
Introduction
The green macroalgal genus Ulva Linnaeus, which includes species previously named under the genus Enteromorpha Link (Hayden et al., Citation2003; Shimada et al., Citation2003), is distributed worldwide in different marine, brackish and freshwater ecosystems. According to AlgaeBase, amongst the 407 historical names presently reported as Ulva sp. in the database, only 86 have been flagged as taxonomically accepted species (Guiry & Guiry, Citation2020). This spectacular number of synonyms reflects, to some extent, the notorious problems in distinguishing Ulva species on the basis of their habitat, morphology, anatomy, cytology and physiology (Bliding, Citation1968; Hoeksema & Van Den Hoek, Citation1983; Phillips, Citation1988; Fort et al., Citation2019, Citation2020a). Difficulties in the delineation of Ulva species are linked to their high degree of phenotypic plasticity, but have also been enhanced by human-mediated biological invasions in coastal waters. Ulva species are common biofoulers on ship hulls (Mineur et al., Citation2007), mariculture structures and products (Verlaque et al., Citation2002; Boudouresque et al., Citation2011), and hitchhikers in ballast waters (Flagella et al., Citation2010). International trans-oceanic shipping and marine aquaculture are recognized as major vectors of introductions of non-indigenous species (NIS) in coastal waters (Carlton, Citation1987; Katsanevakis et al., Citation2013). Distinguishing between native species and NIS, which often remained overlooked (Haydar & Wolff, Citation2011), is still a major challenge in the context of marine Ulva species (Baamonde López et al., Citation2007; Hofmann et al., Citation2010; Kraft et al., Citation2010; Wolf et al., Citation2012; Steinhagen et al., Citation2019).
Molecular methods have been employed for a few decades to overcome these difficulties and new light has been shed on Ulva spp. taxonomy, to identify and delineate species (Coat et al., Citation1998; Baamonde López et al., Citation2007; Flagella et al., Citation2010), critically re-examine inventory records (Wolf et al., Citation2012; Steinhagen et al., Citation2019), infer phylogenetic relationships among taxa (Hayden et al., Citation2003; Shimada et al., Citation2003), elucidate introduction history (Couceiro et al., Citation2011) and characterize type materials to clarify species taxonomy and biogeography (Hanyuda & Kawai, Citation2018; Hughey et al., Citation2019). Both the nuclear ribosomal internal transcribed spacer DNA (ITS nDNA) and the chloroplast-encoded rbcL gene have been tested alone (Coat et al., Citation1998; Flagella et al., Citation2010) or in tandem (e.g. Shimada et al., Citation2003; Couceiro et al., Citation2011). However, compared with these two molecular markers, the plastid elongation factor tufA was recommended by Saunders & Kucera (Citation2010) as a more suitable barcode marker for most marine green macroalgae including the genus Ulva. This was further confirmed by Kirkendale et al. (Citation2013), Lee et al. (Citation2019) and Fort et al. (Citation2020b) for Australian, Korean and west European specimens, respectively, as they provided estimates of intra- and inter-specific divergence in Ulva species. Our current sequencing survey of hundreds of Ulva specimens along the French Atlantic coast found that many tufA sequences fully matched GenBank sequences of U. australis Areschoug and U. pertusa Kjellman, a junior synonym (see details in Couceiro et al., Citation2011), from Korea in the native area (Kang et al., Citation2019), and disjunct areas where the species has been introduced e.g. Australasia (Heesch et al., Citation2009; Kirkendale et al., Citation2013), the Italian North Adriatic (Wolf et al., Citation2012) and German North Sea (Steinhagen et al., Citation2019). U. australis, recognized to be of north-eastern Asian origin (Hanyuda et al., Citation2016; Hanyuda & Kawai, Citation2018) was thought to have been introduced into the Mediterranean Sea and along North European coasts through multiple introduction events and different vectors including maritime transport and mariculture (Baamonde López et al., Citation2007; Couceiro et al., Citation2011; Verlaque & Breton, Citation2019).
To compare our specimens unequivocally to the type material of U. australis, recently characterized with molecular markers (Hanyuda & Kawai, Citation2018), and reanalyse earlier records of the species in Brittany (Coat et al., Citation1998), we conducted molecular analyses based on ITS2 and rbcL sequences. We also used both plastid and mitochondrial non-coding sequences, atpI-H and trnA-N, as suggested by Hanyuda et al. (Citation2016). This latter combination of markers has allowed significant differences in genetic diversity to be resolved among native (Japan and Korea) and non-native populations of U. australis (Hanyuda et al., Citation2016, Citation2018) and contributed to clarifying the introduction history of U. australis in Australia in terms of pathways and timing (Hanyuda & Kawai, Citation2018). This approach is used here to provide biogeographic information about the origin of our U. australis specimens identified based on ITS2, rbcL and tufA and to re-evaluate commonly accepted hypotheses concerning the vectors and timing of introductions of U. australis along the French coasts in view of the history of maritime transport between Europe and North-east Asia.
Materials and methods
Sample collection
Our sampling strategy was based on collecting large sample sizes (> 100 specimens) from three sites separated by over 460 km. Green algae with large and flat rigid thalli, pale to dark green in colour and with a stiff base, were sampled from rocky shores at three sites during low spring tides from 22 January to 21 February 2019. The three sites are in Brittany at Roscoff (48.7327′N, 3.9868′W, Parmentier dock, Estacade, Ile Verte) and Concarneau (47.8601′N, 3.9148′W, Pointe du Cabellou) and in Vendée at La Tranche-sur-Mer (46.3461′N, 1.4189′W, La Grière). This last site is a sand-influenced rocky shore and sampling was restricted to mid-shore pools. At each site, c. 120 living specimens (blade up to 7 cm2) growing attached to rocks were collected by hand and placed individually in small plastic bags. Samples were separated by at least several metres. In the lab, each specimen was rinsed with filtered seawater to remove epiphytes and debris. Following individual photography (Olympus Stylus TG-830), specimens were preserved at −80°C in individual plastic bags numbered with letters (sampling sites) and serial numbers.
DNA extraction, amplification and sequencing
Frozen tissue from each thallus was ground to a powder in liquid nitrogen. Whole genomic DNA was extracted from 0.3 mg of powder using the NucleoSpin Tissue Kit (Macherey-Nagel). The manufacturer’s standard protocol for tissues was followed, except for the following steps: (1) an overnight tissue digestion in proteinase K and (2) DNA was eluted in two steps, each consisting of a 3 min incubation with 25 μl of dH2O pre-heated at 70°C, for a final volume of 50 μl. DNA quality and quantity were assessed using a Nanodrop ND-2000 spectrophotometer (Thermo Scientific), a Qubit 1.0 (Thermo Scientific) fluorometer (dsDNA HS Assay Kit), and 1× agarose gel electrophoresis. Primers for tufA were designed based on Saunders & Kucera (Citation2010) but modified to reduce the number of degenerate bases using the complete Ulva chloroplast genomes available on GenBank on 16 March 2019 (Supplementary table S1). The nuclear internal transcribed spacer 2 (ITS2) together with the plastid ribulose 1, 5-bisphosphate carboxylase large chain (rbcL) gene were used to compare our sequences with the type material of U. australis housed in the Swedish Museum of Natural History (http://herbarium.nrm.se/specimens/A2025). The lectotype was characterized by Hanyuda & Kawai (Citation2018) based on partial sequences of ITS2 (LC331301, 242 bp) and rbcL (LC331300, 197 bp). Following ITS2 and rbcL validations, further analyses of genetic variability used the intergenic regions of chloroplast atpI and atpH (atpI-H) and mitochondrial trnA and trnN (trnA-N) genes amplified with primers from Hanyuda et al. (Citation2016). Primer sequences and PCR amplification cycles are presented in Supplementary table S1 and fig. S1. PCRs were carried out with a Sensoquest labcycler using the TaKara ExTaq reaction kit (Takara Bio). PCR amplicons were checked on a 1× agarose gel electrophoresis prior to purification and Sanger sequencing in both forward and reverse directions by Eurofins Genomics (Ebersberg-Germany: www.eurofinsgenomics.eu/).
Data analysis
For each DNA sequence, chromatograms were cleaned manually with Geneious Prime 2019 (www.geneious.com); primer sequences were trimmed and sequences were checked for the presence of ambiguities and stop codons. Forward and reverse chromatograms were then assembled and consensus sequences were submitted to GenBank (accession numbers in Supplementary table S2). Newly generated consensus sequences were compared with sequences available on GenBank and aligned using Muscle 3.8.425 (Edgar, Citation2004). Our last search on GenBank (17 October 2020) returned 181 tufA sequences identified as U. australis (174 sequences) or U. pertusa (7 sequences) including the two recent releases of complete chloroplast genomes of U. pertusa LC507117 (Mitsuhashi et al., Citation2020) and MN853875 (Han, direct submission 9 December 2019). Fifty-five tufA sequences not yet available on GenBank were also utilized including our sequences (MT078952 from Roscoff and MT078953 from Concarneau), and 10 sequences manually retrieved from the doctoral dissertation of Melton (Citation2017). Our final data set contained 219 sequences since 2 and 15 sequences were excluded due to inconsistencies and mismatches in sequence length, respectively (see details in Supplementary table S3). DnaSP v6 (Rozas et al., Citation2017) was used to identify haplotypes from the 659 bp-long tufA alignment performed on our data set. Uncorrected p distances were calculated from the tufA alignment matrix using PAUP* v.4.0 (Swofford, Citation2002) and are presented as percentages. A median-joining network (Bandelt et al., Citation1999) was constructed to visualize relationships among tufA haplotypes using PopART with default settings (http://popart.otago.ac.nz). Two tufA sequences of U. fenestrata were used as an outgroup as there is consistent support for a close taxon relationship of tufA between U. australis and U. fenestrata (Kirkendale et al., Citation2013; Hughey et al., Citation2019; Kang et al., Citation2019): HQ610325 from Saunders & Kucera (Citation2010) and MK456404 as the holotype of U. fenestrata (Hughey et al., Citation2019). Sequence similarity between the type material and our specimens for ITS2 and rbcL was calculated using BLASTn (Zhang et al., Citation2000) with default parameters (http://blast.ncbi.nlm.nih.gov/Blast.cgi). BLASTn was also used for Coat et al.’s (1998) molecular data for testing the two ITS sequences U.rot.4 (MT078968) and U.rot.5 (MT078969) erroneously labelled as U. rotundata. These two sequences with corrected labels were submitted to GenBank on behalf of B. de Reviers (Supplementary table S2). Correspondence between our specimens and the type material of U. australis using ITS2 and rbcL allowed the atpI-H and trnA-N intergenic sequences of our specimens to be aligned with, respectively, the 15 (haplotypes cH1 to cH15) and 27 (haplotypes mH1 to mH27) sequences defined by Hanyuda et al. (Citation2016) as descriptive of the genetic diversity of native U. australis populations. All sequences were concatenated and analysed following Hudson et al. (Citation1992) using DnaSP v6 (Rozas et al., Citation2017) in order to detect haplotypes identical to any of the 50 combined haplotypes H1-H48 and H49-H50 described by Hanyuda et al. (Citation2016) and Hanyuda et al. (Citation2018), respectively. The metadata associated with Ulva sequences deposited in GenBank, including geographic location, were retrieved from flatfiles. Geographic information, when absent, was manually compiled from published references (Supplementary table S3). Maps were edited with ArcGIS ArcMap 10.6 (ESRI).
Results
Sequencing success across markers
A total of 339 Ulva samples were sequenced for tufA. Forty-one samples matched the U. australis (or U. pertusa) tufA sequences, and 298 represented other Ulva species (). Amplification and/or sequencing of ITS2 and rbcL were less successful. Of the 41 samples putatively assigned to U. australis, only 32 and 28 were successfully sequenced at ITS2 and rbcL, respectively. However, sequencing of the atpH-I and trnA-N intergenic regions was successful for all of the same 41 samples (). This last dataset was used to identify which combined haplotypes among those previously described from Japan were also present in France.
Table 1. Combined haplotypes (H) of chloroplast (cH) and mitochondrial (mH) haplotypes found in Ulva australis following numbering by Hanyuda et al. (Citation2016). *: new combination neither described by Hanyuda et al. (Citation2016) nor Hanyuda et al. (Citation2018), -: no record
tufA sequences
The 41 specimens identified as U. australis were unevenly distributed among localities: 31 samples were from Roscoff and 10 from Concarneau; no U. australis specimens were detected from La Tranche in Vendée (). All 41 tufA sequences (659 bp) are identical to 106 GenBank sequences, and differ by 1, 2 and 3 substitutions from 105, 6 and 1 sequences, respectively. Uncorrected p distances calculated between our sequences (from Roscoff and Concarneau) and U. australis sequences produced by others ranged from 0–0.46%.
The haplotype network revealed two well-separated groups, the U. fenestrata outgroup, and the other incorporating all U. australis haplotypes. Thirty-eight substitutions separated the U. fenestrata outgroup from its closest relative U. australis. Within the alignments for U. australis, there were 8 variable sites (Supplementary fig. S2) but only up to 3 substitutions per sequence (). The network of U. australis is star-like and included two very common haplogroups here named tuH0 and tuH1, which are associated with six rare haplotypes (). Our tufA sequences from Brittany matched tuH0, the commonest (163 samples), recorded from Northern Asia (27%), Australasia (18%), North and South America (9%) and Western Europe (46%) including the Mediterranean Sea (). TufA sequences differing from tuH0 by one transition constitute the second haplogroup tuH1 (103 samples), from Northern Asia (91%), British Columbia (BC), California (5%) and Western Europe (4%). All other tufA sequences (7 samples) with at least 2 mutations from tuH0 were from north-eastern Asia () within the Yellow Sea from Donggang, China (Du et al., Citation2014) to Jeju Island, Korea (Kang et al., Citation2019; Lee et al., Citation2019). For simplicity, these tufA sequences were labelled tuH2 and tuH3, as they had 2 and 3 substitutions, respectively. Substitutions in tuH2a and tuH2b haplotypes were at the 5’ end of the tufA gene and tuH2c, tuH2d, tuH2e and tuH3 haplotypes substitutions were at the 3’ end (see Supplementary fig. S2). The two commonest haplogroups and five of the six rare haplotypes were all recorded in Asia (), from Jeju Island (, Yellow Sea map).
Fig. 1. Median-joining network of tufA haplotype (659 bp) in Ulva australis. Grey: U. fenestrata as the outgroup with MK456404 as the holotype of U. fenestrata (Hughey et al., Citation2019) and HQ610325 from Saunders & Kucera (Citation2010); tick mark: one mutation; U. australis tufA haplotype labels and GenBank accession numbers: see Supplementary figure S2 and table S3
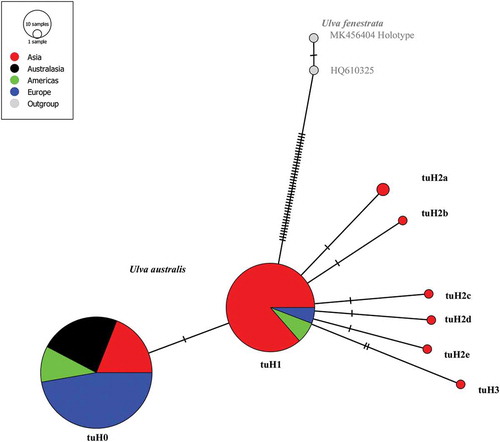
Fig. 2. Map of sampling sites showing the relative occurrence of the tufA haplotypes (659 bp) detected for Ulva australis. Labels tuH0–tuH3 are named based on the number (0–3) of substitutions in tufA sequences (see Supplementary figure S2). Other markers than tufA and studies based on morphological observations: red and blue cross, respectively
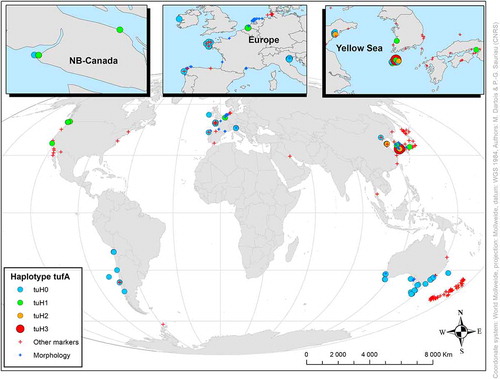
Visual inspection of 14 aligned sequences shorter than 659 bp (excluded from our data set) and two sequences from BC positioned in 349–1122 bp of the complete tufA sequence (see GenBank numbers in Supplementary table S3) concurred with the assumptions that (1) both tuH0 and tuH1 haplogroups are present within the Bohai Sea, (2) tuH0 is recorded in Chile and (3) tuH1 occurred along both sides of the North Atlantic (Germany and Virginia, USA). However, one sequence (MH538644) collected in Schleswig-Holstein, Germany, near Cuxhaven at the mouth of the Elbe River (Steinhagen et al., Citation2019) and two identical sequences (HQ610379 and HQ610380) from BC (Saunders & Kucera, Citation2010) differed from tuH0 by one and two transitions, respectively. They would represent two new additional haplotypes not shown here () as the nucleotide substitutions are out of the 659 bp range of our analysis.
ITS2 and rbcL regions
Thirty-one of the 35 ITS2 sequences, 25 from Roscoff (MT078941) and 6 from Concarneau (MT078940), were identical to the ITS2 sequence from the lectotype of U. australis (Supplementary table S4). Two other sequences from Roscoff and one from Concarneau were poorer quality (50–92% of untrimmed based with phred score > 40). Two were < 184 bp (134 and 164 bp, respectively) and one Roscoff sequence had three ambiguities (S instead of C or G). Another sequence, from Concarneau, had one substitution (T instead of G at position 84). These variants could be due to the co-amplification of multiple alleles/copies of ITS2. The ITS1-5.8S-ITS2 sequences MT078968 (U.rot.4) and MT078969 (U.rot.5) originated from Coat et al. (Citation1998) and incorporated ITS2 sequences identical to that of the lectotype U. australis.
Among all the ITS2 sequences related to Ulva australis and/or U. pertusa available in GenBank (Supplementary table S4), 172 sequences from the literature (80 U. australis and 92 U. pertusa) fully matched the ITS2 of lectotype material. Only six additional ITS2 sequences revealed intra- and inter-individual variation with one (5 specimens) to three (1 specimen) substitutions or indels. AJ234321, initially labelled U. pertusa but misidentified as U. fasciata (Hayden et al., Citation2003), differed from the lectotype of U. australis at 25 positions, and has the lowest percentage identity with the lectotype ITS2.
In contrast to ITS2, the 28 rbcL sequences from France (195 bp) displayed no variation, were identical between Roscoff (MT078958) and Concarneau (MT078959), and showed 100% similarity with the lectotype rbcL gene sequence of U. australis.
atpI-H and trnA-N marker regions
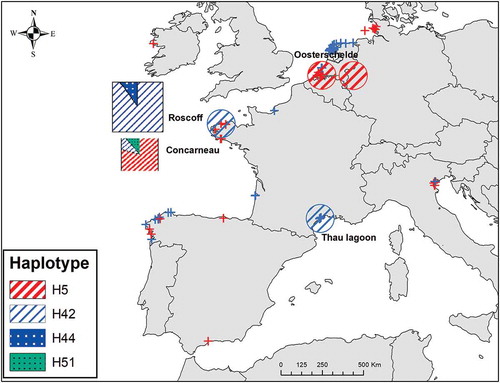
For each marker, three haplotypes were detected, resulting in four combined haplotypes, three identified as H5, H42, H44 by Hanyuda et al. (Citation2016), and a new combination H51 (). Two combined haplotypes, H42 and H44, were recorded at Roscoff, the former with 90% relative abundance. At Concarneau, H5, H42 and H51 were recorded. Both H42 and H51 are rare (10% relative abundance; , ). H42, shared by Roscoff and Concarneau, was previously reported from the Thau Lagoon (Mediterranean Sea) by Hanyuda et al. (Citation2016); haplotype H5 was previously observed in the Netherlands ().
Fig. 3. Map of combined atpI-H and trnA-N haplotypes in western Europe. Square: this study, circle: data from Hanyuda et al. (Citation2016), red and blue crosses: as in
Discussion
Conspecificity of Ulva australis and Ulva pertusa
Since its first morphological description by Kjellman from Japan in 1897, and until the recent work of Couceiro et al. (Citation2011), the specific status of Ulva pertusa has not been seriously challenged (see discussion in Tanner, Citation1979). Very common in Japan, U. pertusa is polymorphic (Okamura, Citation1921) even including ‘stalked-Ulva‘ forms (Hiraoka et al., Citation2003) and floating green tides (Yoshida et al., Citation2015). In contrast, the status of the South Australian U. australis, which was described before U. pertusa, has long been controversial. Several nomenclatural synonyms have been proposed, including U. rigida C. Agardh (Agardh, Citation1883) and U. lactuca Linnaeus by Womersley (Citation1956, p. 354) following the view that U. lactuca is a cosmopolitan species with multiple varieties. Contradictory opinions on U. australis were finally clarified using ITS1 and rbcL analyses on Spanish, Japanese and Australian specimens (Couceiro et al., Citation2011). Molecular evidence of the identity of U. australis and U. pertusa presented by Couceiro et al. (Citation2011) corroborates previous results of Kraft et al. (Citation2010), who distinguished a clade of U. australis and U. pertusa from a more speciose clade with U. laetevirens, U. rigida and U. scandinavica using ITS and rbcL markers. They reinstated U. australis as a valid species and suggested U. pertusa as its junior synonym (see details in the Appendix S1 of Kraft et al., Citation2010). Kirkendale et al. (Citation2013) confirmed this view using rbcL and tufA markers.
Despite known limitations associated with the use of ITS (Saunders & Kucera, Citation2010), more recent advances in secondary structural analysis of ITS2 have advocated the use of ITS2 as a potential DNA barcode for the class Chlorophyceae (Buchheim et al., Citation2011). Species identification of Ulva specimens based on ITS2 (Hanyuda et al., Citation2016) also supports the conspecificity of U. australis and U. pertusa. All available ITS2 sequences labelled as U. australis or U. pertusa in GenBank (Supplementary table S4) are identical or nearly identical to the historical lectotype material of U. australis characterized with ITS2 and rbcL by Hanyuda & Kawai (Citation2018). The inter-specimen variability i.e. one to six substitution(s) here reported from this large data set is consistent with previous analyses made on a more restricted data set (Melton, Citation2017, fig. 3.11, p. 98). Our synopsis of ITS2 data suggests misidentification of two specimens previously labelled U. fasciata (AF099728) and U. lactuca (AF099729), and their molecular identification as U. australis (100% match with the lectotype ITS2 sequence). Both were collected in Victoria, Australia (Woolcott & King, Citation1999). Until the recent study of Hughey et al. (Citation2021), the type material of U. pertusa was thought to be unavailable (Tanner, Citation1979, p. 219). However, both ITS and rbcL sequences are available from specimens collected at Yokohama (Shimada et al., Citation2003), one of three syntype Japanese localities, together with Hakodate and Yenoshima (Kjellman, Citation1897): ITS2 (AB097657) and rbcL (AB097627) sequences from the specimen voucher SAP:095069 were identical to ITS2 (LC331301) and rbcL (LC331300) sequences from the lectotype of U. australis, respectively. Most recently, Hughey et al. (Citation2021) retrieved the type material of U. pertusa from the Museum of Evolution (UPS) at Uppsala University, and confirmed that U. pertusa is conspecific with U. australison the basis of rbcL gene sequences from one newly designated lectotype (MT815855) from Yenoshima and two syntype specimens (MT815856, MT815857).
A large body of literature shows the high level of genetic similarity in rbcL gene sequences between populations of U. australis and U. pertusa from Australia (Kraft et al., Citation2010; Kirkendale et al., Citation2013), BC-Canada (Saunders & Kucera, Citation2010), California (Hayden et al., Citation2003; Hayden & Waaland, Citation2004), Chile (Oróstica et al., Citation2017), China (Du et al., Citation2014; Kang et al., Citation2014), Japan (Matsumoto & Shimada, Citation2015), Korea (Kang et al., Citation2014), Italy (Wolf et al., Citation2012) and Spain (Baamonde López et al., Citation2007; Couceiro et al., Citation2011).
Synopsis of French records
Ulva australis is now widespread within the Thau lagoon (Mediterranean Sea), particularly within oyster farming zones (Verlaque et al., Citation2002), and is one of the commonest macroalgal NIS species, as U. pertusa, in the lagoon (Boudouresque et al., Citation2011). The first record is thought to be the herbarium specimen labelled Ulva sp. from Thau in 1984 collected by Ben Maiz (see details and references in Verlaque et al., Citation2002). Molecular identification of U. australis at Thau was recently confirmed (Hanyuda et al., Citation2016). Along the French Atlantic coast, U. australis was first reported as U. pertusa from Arcachon Bay in 2005–2006 (Verlaque et al., Citation2008), and as U. australis from Port of Le Havre in 2012 (Verlaque & Breton, Citation2019). All these records were based on morphological diagnoses (Okamura, Citation1921; Verlaque et al., Citation2002), and as pointed out by Verlaque & Breton (Citation2019), still required molecular confirmation. To the best of our knowledge, the first molecular sequences came from Brittany (Coat et al., Citation1998), including two ITS1-5.8S-ITS2 sequences misidentified as U. rotundata Bliding. They are representative of at least six specimens collected from Roscoff (attached thalli from Île Verte, Embarcadère and Pointe du Bloscon) plus one specimen from Lannion Bay (floating thalli during a green tide event at Plestin-les-Grèves) in 1994–1995. Woolcott & King (Citation1999) clearly suspected the U. rotundata specimens to be misidentified as they wrote ‘There are again strong molecular similarities between Australian isolates here‘ i.e. U. australis ‘and European Ulva species in Coat et al. (Citation1998)’. This taxonomic inconsistency was also highlighted by Malta et al. (Citation1999) following a comparison of ITS between U. rotundata from Coat et al. (Citation1998) and specimens labelled U. rigida from Flinders, Australia (UrigAus: AF153494, collected by van Oppen) and Oosterschelde, the Netherlands (UrigOS: AF153490). Blastn comparisons among these sequences confirmed their similarity (Supplementary table S4). On the other hand, Shimada et al. (Citation2003) noted that U. rotundata from Brittany clustered with U. pertusa from Japan in phylogenetic ITS1-5.8S-ITS2 trees. Hayden & Waaland (Citation2004) highlighted a similar grouping in ITS and rbcL trees of U. rotundata from Britanny and U. pertusa specimens from Japan and California. Baamonde López et al. (Citation2007) first recognized the presence of U. pertusa in Europe based on the molecular data of Coat et al. (Citation1998) following Shimada et al. (Citation2003) and Hayden & Waaland (Citation2004). This conclusion was also accepted by Flagella et al. (Citation2010) before the final suggestion that U. pertusa is a synonym of U. australis (Couceiro et al., Citation2011). Molecular identification of U. australis at Roscoff was also confirmed by Hanyuda et al. (Citation2016).
Our observations of U. australis at Roscoff from the same sampling sites as those selected by Coat et al. (Citation1998) suggest that the species has been present at Roscoff over the past 25 years. Concarneau (Pointe du Cabellou) is a new locality for U. australis in southern Brittany (, ). Observations here supplement previous reports of U. australis in Brittany (Fort et al., Citation2020a, Citation2020b) during spring and summer 2018 green tides at BegMeil (near Concarneau), Bay of Brest (Moulin Blanc), Roscoff and Lannion Bay (Plestin les Grèves) (), demonstrating the relative dominance of U. australis in green tides in Brittany. These observations are scattered both temporally and spatially () suggesting that the distribution of U. australis along the French Channel, Bay of Biscay (from Brest to Biarritz) and Mediterranean coasts might be more continuous than presently reported.
Current distribution of Ulva australis in Europe
Ulva australis has been reported from Germany, the Netherlands, Ireland, Spain and Italy incorporating the oldest records and corrected misidentifications as U. pertusa (Baamonde López et al., Citation2007; Couceiro et al., Citation2011). The first records in the German Wadden Sea were in Helgoland and Schleswig-Holstein in 2014 (Steinhagen et al., Citation2019). Records from the Netherlands came from the Oosterschelde at least from 1995 (Stegenga et al., Citation2006), the Dutch Delta area (Fort et al., Citation2018, Citation2019, Citation2020a), where U. pertusa was recognized in herbarium specimens collected in the area as early as 1993 (Wolff, Citation2005 and references therein), the entrance of Rotterdam port (Gittenberger et al., Citation2014) and the Dutch Wadden Sea including major ports (Gittenberger et al., Citation2015). Stegenga et al. (Citation1997, p. 42 note no. 8) mentioned the occurrence of an aberrant Ulva species with a thicker thallus than U. rigida in Zeeland, referencing a 1984 collection, but it may not be represented in the material housed at the Leiden Herbarium (Stegenga, pers. comm. 19 Feb. 2020). Ulva australis has also been reported from Galway, Ireland since 2018 (Fort et al., Citation2020a). In Spain, the oldest record was morphologically recognized from a herbarium specimen collected from La Coruña (Galicia) in 1990 (Baamonde López et al., Citation2007). Spanish records of live specimens came from both the Atlantic (Fort et al., Citation2020b) and Mediterranean coasts (Couceiro et al., Citation2011). The first Italian records were from Venice in 2010 (Manghisi et al., Citation2011) and U. australis was widespread within the lagoon in 2011 (Wolf et al., Citation2012). Ulva pertusa at Naples was mentioned by Wolf et al. (Citation2012) and Cormaci et al. (Citation2014) according to Flagella et al. (Citation2010). A closer examination of the Flagella et al. (Citation2010) data suggests an erroneous interpretation of their ITS1 phylogenetic tree. Only U. ohnoi Hiraoka and Shimada and U. fasciata Delile, today known as U. lactuca Linnaeus (Hughey et al., Citation2019), were reported from ballast waters in the Neapolitan harbour.
Worldwide genetic variability in U. australis tufA
Among the eight tufA haplotypes (659 bp) found in this study (), seven matched those reported from Jeju Island (Kang et al., Citation2019; Lee et al., Citation2019). The eighth was found only at Donggang Rizhao, China, some 700 km north-east in the Yellow Sea (Du et al., Citation2014). At Jeju Island, Lee et al. (Citation2019) reported seven haplotypes for tufA including one haplotype with three substitutions from tuH0 not included in our analysis. All these results support a scenario of a drastic population bottleneck following the introduction of U. australis outside its native range: (1) a high level of genetic diversity for tufA within the native area (Kang et al., Citation2019; Lee et al., Citation2019), (2) the predominance of a unique haplogroup (tuH0) worldwide and (3) the occurrence of a second one (tuH1) in a few European and North American countries (). According to Rius et al. (Citation2015), a founder effect is not the most common scenario in marine invasions. Introduced populations exhibiting similar or even higher genetic diversity than native populations have been described, as multiple introductions from large native regions with highly genetically structured populations can lead to increased diversity at introduction sites (e.g. Simon-Bouhet et al., Citation2006). Nonetheless, all results for tufA from Australia (Kirkendale et al., Citation2013) and New Zealand (Lawton et al., Citation2013, Citation2020) showed a lack of genetic diversity (only tuH0, ). This supports previous findings that U. australis, despite its abundance and spread along these temperate coasts, is a NIS in Australia (Kirkendale et al., Citation2013) and New Zealand (Heesch et al., Citation2009). A similar conclusion was reported for combined genetic markers atpI-H and trnA-N in U. australis from Australasia compared with Japan and Korea (Hanyuda et al., Citation2016, Citation2018): 1 and 5 combinations in Australia and New Zealand, respectively vs. 48 in both Japan and Korea.
The first records of U. australis as U. pertusa in North America are recent and were validated by molecular analyses in BC in 2005–2006 (Saunders & Kucera, Citation2010) and California in 1999 (Hayden & Waaland, Citation2004). Aguilar-Rosas et al. (Citation2008) also recorded U. australis (determined by ITS2 sequences) from Baja California, Mexico in 2006–2007 and suggested the species to be a NIS in range expansion via transoceanic shipping. Along the Eastern Pacific coast, two tufA haplotypes (tuH0 and tuH1) were observed (), in Canada (Saunders & Kucera, Citation2010) and Chile (Melton, Citation2017; Oróstica et al., Citation2017), but only tuH1 in California (Saunders, Citation2014). However, visual checks of Saunders & Kucera’s (Citation2010) tufA sequences (774 bp) suggested the occurrence of a third haplotype exclusive to BC, Canada, and not recorded from Korea (Kang et al., Citation2019; Lee et al., Citation2019). This may support the view of repeated introductions into BC) waters from different Asian populations. Scarcity of full-length tufA data from Japan (Mitsuhashi et al., Citation2020) hinders the discussion of this hypothesis for now. The status of U. australis along the North-eastern Pacific coast as a NIS or a native (Amphi-Pacific with disjunct temperate areas) was already questioned by Miller et al. (Citation2011). Among vectors involving transoceanic transport of NIS from the western to the eastern North Pacific ocean, the oyster trade from Japan and international shipping are major invasion vectors (Carlton, Citation1987; Wonham & Carlton, Citation2005), and are well documented in terms of biogeography and timing. Ulva australis is a common fouling alga on the oyster Magallana gigas (Thunberg) cultivated on rafts in Japan (Miyazaki, Citation1938). Successful introductions of oyster seed from Japan to BC and Puget Sound started at the beginning of the 19th century and lasted to the 1970s (Ruesink et al., Citation2005). Oysters originated from the main Japanese farming centres i.e. Miyagi-Sendai (East Japan), Nagasaki-Kumamoto and Hiroshima (SW Japan) (Fujiya, Citation1970). Maritime transport between Japan and the USA is well-documented in terms of historical changes in port hierarchy and geographic specializations (Maurette, Citation1922; Ducruet, Citation2015). Yokohama and Osaka-Kobe have been the main Japanese ports involved in the trade with Western and Eastern overseas countries since the mid-19th century (Maurette, Citation1922). However, natural dispersal of U. australis may also have contributed to its wide amphi-Pacific distribution as tsunamis are known to occur on the scale of geological times (Dawson & Stewart, Citation2007), particularly in seismically active coasts like those of Japan (Garrett et al., Citation2016). The 2011 Great East Japan Earthquake and Tsunami was responsible for the arrival of huge amounts of marine debris on North-eastern Pacific shores, including derelict boats fouled with U. australis. Use of combined haplotypes atpI-H and trnA-N (Hanyuda et al., Citation2016) elucidated the biogeographic origin of these U. australis stranded on the Oregon shores since their combined haplotypes matched those found in Iwate Pref.-Northern Japan (Hanyuda et al., Citation2018). Finally, the amphi-Pacific map of these combined haplotypes in U. australis highlighted the wide distribution of the unique haplotype H5 (Hanyuda et al., Citation2016, ). This surprisingly fully matches the aquaculture history of oyster transfers from the East coast of Japan (Miyagi Pref.) to Australasia and the Americas (Ruesink et al., Citation2005 and references therein for Australia, New Zealand and the Americas). Known secondary transfers of oysters within American countries, e.g. USA to Chile (Ruesink et al., Citation2005) do not contradict the view of contemporary oyster transfers as a major vector for U. australis within the Pacific region.
Challenging the introduction history of Ulva australis in France
In Europe (France and the Netherlands), two combined haplotypes H5 and H42 () were previously found, but their distinct geographic distribution suggested repeated introduction events (Hanyuda et al., Citation2016). Haplotype H5 was found in the Netherlands ( in Hanyuda et al., Citation2016) in U. australis collected in 2005 within the Oosterschelde in the vicinity of Yerseke, which has been the main Magallana gigas culture centre since the mid 1960s in the Netherlands (Haydar & Wolff, Citation2011). Haplotype H5 was found along the Miyagi (Japan), BC and west USA coasts ( in Hanyuda et al., Citation2016), regions known to be the biogeographic primary sources of imported M. gigas into the Oosterschelde (Ruesink et al., Citation2005). At Concarneau, 80% of U. australis specimens harboured the same H5 (, ), in line with this interpretation. Indeed, the rebound of the French oyster industry was linked to the national RESUR operation based on massive and repeated imports of M. gigas adults from BC and spat from Miyagi in the 1970s (Grizel & Héral, Citation1991).
However, all other combined haplotypes we found at Concarneau (H42 and H51) and Roscoff (H42 and H44) () conflict with the above view of the oyster trade as a unique vector of introduction of U. australis into Brittany. Previous results obtained from Roscoff and Thau by Hanyuda et al. (Citation2016) (only H42 found in specimens collected in 2005 and 2002 at Roscoff and Thau, respectively, ) also strengthen this conflict and furthermore challenge the initial suggestion that U. australis could have been introduced into the Thau lagoon as early as the 1970s via oyster M. gigas transfers in 1971–1976 (Verlaque et al., Citation2002). Based on the distribution map of the combined haplotypes H42 and H44 in Japan and Korea ( in Hanyuda et al., Citation2016), it is suggested that U. australis found at Concarneau, Roscoff and Thau originated from Kobe (Japan) and/or South-eastern Korea. A closer examination of microsatellite genotypes ( in Hanyuda et al., Citation2016) identifies Kobe Port as the most likely origin of U. australis populations at Roscoff, although the genotype found at Thau was more geographically spread (from Kobe to Kumamoto in Japan and in South-eastern Korea). The finding of a new H51 haplotype at Concarneau does not contradict this view. According to Hanyuda et al. (Citation2016) this may reflect a mutation that arose since the first introductions or, more likely, insufficient sampling in the native range as further exemplified by the study of tsunami marine debris (Hanyuda et al., Citation2018). These results indicate maritime transport between Japan (at least) and France as another vector for the introduction of U. australis into Mediterranean and Atlantic waters. Aguilar-Rosas et al. (Citation2008) and Couceiro et al. (Citation2011) similarly suggested the arrival of U. australis was linked to maritime vessel traffic in Baja California and the Strait of Gibraltar, respectively.
We thus speculate that these repeated and independent events have been linked to the maritime trade between France and South-east Asia in the late 19th or early 20th centuries. The maritime history of Japan was profoundly reorganized during the Meiji era (1867–1912) and a new foreign trade policy was initiated for 1868 (Horie, Citation1952) that actively promoted cooperation and business with the USA and Colonial Countries. The opening of the Suez Canal on November 1869 also offered new shipping routes between Europe and Asia (Fletcher, Citation1958; Berneron-Couvenhes, Citation2007), and favoured maritime companies from leading European countries i.e. Great Britain, France, the Netherlands, Italy and Germany (Miller, Citation2012). For instance, the ‘Compagnie des Messageries Maritimes’ set up in 1851 at Marseille operated several ship lines among Yokohama, Kobe, Shanghai, Hong-Kong, Singapore, Calcutta, Mumbai and Marseille (Berneron-Couvenhes, Citation2007; Metzger, Citation2009). Historical analysis of global maritime transports since the late 19th century de facto highlighted the leading role of Yokohama and Kobe ports together with the emerging role of Chinese ports in the early 20th century (Ducruet, Citation2013), and the leading role of Marseille and Le Havre, as the main French port at that time (Ducruet & Marnot, Citation2016). Long-term port activities are under the control of regional conflicts and world wars (Ducruet & Marnot, Citation2016). Numerous wrecks from trading ships from Japan from WWI are known from the Mediterranean and the Atlantic (https://uboat.net/wwi/ships_hit/; see also Hiroaki, Citation2015). Changes in the north-eastern Asian port hierarchy between Japan, Korea and China have also been noticeable since the 1980s (Ducruet et al., Citation2011; Ducruet, Citation2013), so contemporary European records of U. australis may be linked to vessel traffic originating from ports within the Bohai and Yellow Seas. More phylogeographic studies along Korean and Chinese coasts within the native area of U. australis are required to test these hypotheses.
The patchy records of U. australis along the French coasts suggest that its presence is underestimated, mostly due to historical misidentifications as Ulva species show great morphological diversity and plasticity. The use of molecular markers for population genetics and biogeography is a key tool contributing to a better interpretation of the introduction history of U. australis. Using markers alongside systematic field sampling using large sample sizes and analysing herbarium specimens will lead to a much deeper understanding of the biogeographic origin of introduced populations, vector hierarchy and timing of introductions. Including an interdisciplinary exploration of human maritime activities is crucial since coastal macroalgal biodiversity has been heterogeneously reshaped over centuries by transport and aquaculture activities.
Supplementary table S1. Primers used for amplification and sequencing.
Supplementary table S2. List of specimens deposited in GenBank.
Supplementary table S3. List of GenBank sequences at tufA used in the haplotype network.
Supplementary table S4. List of GenBank sequences at ITS2.
Supplementary fig. S1. Conditions of PCR amplification cycle for each DNA region.
Supplementary fig. S2. Schematic diagram of tufA sequence variability in U. australis
Author contributions
P.-G. Sauriau: original concept, fieldwork, analysis of molecular data, drafting and editing manuscript; M. Dartois: original concept, fieldwork, molecular analysis, analysis of molecular data, drafting and editing manuscript; V. Becquet: molecular analysis, drafting and editing manuscript; F. Aubert: fieldwork, laboratory analysis; V. Huet: fieldwork, laboratory analysis; M. Bréret: macroalgae taxonomy, laboratory analysis; A. Viricel: analysis of molecular data, drafting and editing manuscript; E. Pante: original concept, molecular analysis, analysis of molecular data, drafting and editing manuscript.
Supplemental Material
Download MS Word (57.7 KB)Supplemental Material
Download MS Word (95.8 KB)Supplemental Material
Download MS Word (35.3 KB)Supplemental Material
Download MS Word (38.1 KB)Supplemental Material
Download MS Word (37 KB)Supplemental Material
Download MS Word (36.7 KB)Acknowledgements
We are grateful to Drs Bruno de Reviers (Museum National d’Histoire Naturelle, Paris), Takeaki Hanyuda (Kobe University), Antoine Fort and Ronan Sulpice (National University of Ireland, Galway), Hyung Woo Lee (Department of Biology, Jeju), Herre Stegenga (Naturalis Biodiversity Centre, Leiden), Sophie Steinhagen (University of Gothenburg, Strömstad), Stacy Krueger-Hadfield (University of Alabama at Birmingham), Wei Song (Key Laboratory of Science and Engineering for Marine Ecology and Environment, Qingdao), Marc Verlaque and Fred Mineur (MIO, Marseille) for their kind advice, personal data communication and comments on Ulva species. Nadia Améziane, director of the Marine Station at Concarneau, kindly provided access to the Marine Station facilities during our field trips in Brittany. We also thank Michel Prineau, Laureen Beaugeard, Aya Ghedmasi and Tony Robinet for help in the field and in the laboratory. The authors thank the molecular core facility of the LIENSs laboratory. We also gratefully acknowledge comments from Prof. Christine Maggs and anonymous reviewers that allowed us to greatly improve the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at https://doi.org/10.1080/09670262.2021.1876249
Additional information
Funding
References
- Agardh, J.G. (1883). Till algernes systematik. Nya bidrag. (Tredje afdelningen)Lunds Universitets.
- Aguilar-Rosas, R., Aguilar-Rosas, L.E. & Shimada, S. (2008). First record of Ulva pertusa Kjellman (Ulvales, Chlorophyta) in the Pacific coast of Mexico. Algae, 23: 201–207.
- Baamonde López, S., Baspino Fernández, I., Barreiro Lozano, R. & Cremades Ugarte, J. (2007). Is the cryptic alien seaweed Ulva pertusa (Ulvales, Chlorophyta) widely distributed along European Atlantic coasts? Botanica Marina, 50: 267–274.
- Bandelt, H.-J., Forster, P. & Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16: 37–48.
- Berneron-Couvenhes, M.-F. (2007). Les Messageries Maritimes. L’essor d’une grande compagnie de navigation française, 1851–1894. PUPS – Presse de l’Université Paris-Sorbonne, Paris.
- Bliding, C. (1968). A critical survey of European taxa in Ulvales, Part II. Ulva, Ulvaria, Monostroma, Kornmannia. Botaniska Notiser, 121: 535–629.
- Boudouresque, C.-F., Klein, J., Ruitton, S. & Verlaque, M. (2011). Biological invasion: the Thau lagoon, a Japanese biological island in the Mediterranean Sea. In Global Change: Mankind Marine Environmental Interactions. Proceedings of the 13th French-Japanese Oceanography Symposium (Ceccaldi, H.-J., editor), 151–156. Springer Science, Berlin.
- Buchheim, M.A., Keller, A., Koetschan, C., Förster, F., Merget, B. & Wolf, M. (2011). Internal transcribed spacer 2 (nu ITS2 rRNA) sequence-structure phylogenetics: towards an automated reconstruction of the green algal tree of life. PLoS ONE, 6: e16931–e16931.
- Carlton, J.T. (1987). Patterns of transoceanic marine biological invasions in the Pacific Ocean. Bulletin of Marine Science, 41: 452–465.
- Coat, G., Dion, P., Noailles, M.C., de Reviers, B., Fontaine, J.M., Berger-Perrot, Y. & Loiseaux-De Goér, S. (1998). Ulva armoricana (Ulvales, Chlorophyta) from the coasts of Brittany (France). II. Nuclear rDNA ITS sequence analysis. European Journal of Phycology, 33: 81–86.
- Cormaci, M., Furnari, G. & Alongi, G. (2014). Flora marina bentonica del Mediterraneo: Chlorophyta. Bollettino delle sedute della Accademia gioenia di scienze naturali in Catania, 47: 11–436.
- Couceiro, L., Cremades, J. & Barreiro, R. (2011). Evidence for multiple introductions of the Pacific green alga Ulva australis Areschoug (Ulvales, Chlorophyta) to the Iberian Peninsula. Botanica Marina, 54: 391–402.
- Dawson, A.G. & Stewart, I. (2007). Tsunami deposits in the geological record. Sedimentary Geology, 200: 166–183.
- Du, G., Wu, F., Mao, Y., Guo, S., Xue, H. & Bi, G. (2014). DNA barcoding assessment of green macroalgae in coastal zone around Qingdao, China. Journal of Ocean University of China, 13: 97–103.
- Ducruet, C. (2013). Asian cities in the global maritime network since the late nineteenth century. In Asian Cities: from Colonial to Global (Bracken, G., editor), 173–186. Amsterdam University Press, International Institute for Asian Studies & TU Delft Seminar, Leiden.
- Ducruet, C. (2015). Inside the pond: an analysis of Northeast Asia’s long‐term maritime dynamics. International Journal of Maritime Affairs and Fisheries, 7: 25–40.
- Ducruet, C. & Marnot, B. (2016). Analyser les trafics portuaires mondiaux en 1890 et 1925 à partir des registres du Lloyd’s. In La Maritimisation du Monde de la Préhistoire à nos Jours, 383–398. Presses Universitaires de Paris-Sorbonne, Paris.
- Ducruet, C., Lee, S.-W. & Ng, A.K.Y. (2011). Port competition and network polarization in the East Asian maritime corridor. Territoire en mouvement. Revue de géographie et aménagement, 10. https://doi.org/https://doi.org/10.4000/tem.1327.
- Edgar, R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32: 1792–1797.
- Flagella, M.M., Andreakis, N., Hiraoka, M., Verlaque, M. & Buia, M.C. (2010). Identification of cryptic Ulva species (Chlorophyta, Ulvales) transported by ballast water. Journal of Biological Research–Thessaloniki, 13: 47–57.
- Fletcher, M.E. (1958). The Suez Canal and world shipping, 1869–1914. Journal of Economic History, 18: 556–573.
- Fort, A., Guiry, M.D. & Sulpice, R. (2018). Magnetic beads, a particularly effective novel method for extraction of NGS-ready DNA from macroalgae. Algal Research, 32: 308–313.
- Fort, A., Lebrault, M., Allaire, M., Esteves-Ferreira, A.A., McHale, M., Lopez, F., Fariñas-Franco, J.M., Alseekh, S., Fernie, A.R. & Sulpice, R. (2019). Extensive variations in diurnal growth patterns and metabolism among Ulva spp. strains. Plant Physiology, 180: 109–123.
- Fort, A., Mannion, C., Fariñas-Franco, J.M. & Sulpice, R. (2020a). Green tides select for fast expanding Ulva strains. Science of The Total Environment, 698: 134337.
- Fort, A., McHale, M., Cascella, K., Potin, P., Usadel, B., Guiry, M.D. & Sulpice, R. (2020b). Foliose Ulva species show considerable inter-specific genetic diversity, low intra-specific genetic variation, and the rare occurrence of inter-specific hybrids in the wild. Journal of Phycology, in press.
- Fujiya, M. (1970). Oyster farming in Japan. Helgoländer Wissenschaftliche Meeresuntersuchungen, 20: 464–479.
- Garrett, E., Fujiwara, O., Garrett, P., Heyvaert, V.M.A., Shishikura, M., Yokoyama, Y., Hubert-Ferrari, A., Brückner, H., Nakamura, A. & De Batist, M. (2016). A systematic review of geological evidence for Holocene earthquakes and tsunamis along the Nankai-Suruga Trough, Japan. Earth-Science Reviews, 159: 337–357.
- Gittenberger, A., Rensing, M., Niemantsverdriet, P., Schrieken, N. & Stegenga, H. (2014). Port of Rotterdam survey and monitoring non-native species conform HELCOM/OSPAR protocol. (31), 111. Office for Risk Assessment and Research, The Netherlands Food and Consumer Product Safety Authority, Leiden.
- Gittenberger, A., Rensing, M., Dekker, R., Niemantsverdriet, P., Schrieken, N. & Stegenga, H. (2015). Native and non-native species of the Dutch Wadden Sea in 2014. (08), 94. Office for Risk Assessment and Research, The Netherlands Food and Consumer Product Safety Authority, Leiden.
- Grizel, H. & Héral, M. (1991). Introduction into France of the Japanese oyster (Crassostrea gigas). ICES Journal of Marine Science, 47: 399–403.
- Guiry, M.D. & Guiry, G.M. (2020). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http//:www.algaebase.org.
- Hanyuda, T. & Kawai, H. (2018). Genetic examination of the type specimen of Ulva australis suggests that it was introduced to Australia. Phycological Research, 66: 238–241.
- Hanyuda, T., Heesch, S., Nelson, W., Sutherland, J., Arai, S., Boo, S.M. & Kawai, H. (2016). Genetic diversity and biogeography of native and introduced populations of Ulva pertusa (Ulvales, Chlorophyta). Phycological Research, 64: 102–109.
- Hanyuda, T., Hansen, G.I. & Kawai, H. (2018). Genetic identification of macroalgal species on Japanese tsunami marine debris and genetic comparisons with their wild populations. Marine Pollution Bulletin, 132: 74–81.
- Haydar, D. & Wolff, W.J. (2011). Predicting invasion patterns in coastal ecosystems: relationship between vector strength and vector tempo. Marine Ecology Progress Series, 431: 1–10.
- Hayden, H.S. & Waaland, J.R. (2004). A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific. Phycologia, 43: 364–382.
- Hayden, H.S., Blomster, J., Maggs, C.A., Silva, P.C., Stanhope, M.J. & Waaland, J.R. (2003). Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. European Journal of Phycology, 38: 277–294.
- Heesch, S., Broom, J.E.S., Neill, K.F., Farr, T.J., Dalen, J.L. & Nelson, W.A. (2009). Ulva, Umbraulva and Gemina: genetic survey of New Zealand taxa reveals diversity and introduced species. European Journal of Phycology, 44: 143–154.
- Hiraoka, M., Shimada, S., Serisawa, Y., Ohno, M. & Ebata, H. (2003). Two different genetic strains of stalked-Ulva (Ulvales, Chlorophyta) grow on intertidal rocky shores in Ebisujima, central Japan. Phycological Research, 51: 161–167.
- Hiroaki, M. (2015). Les naufragés du Nagata Maru. In Zoom Japon55, 7–8. Editions Ilyfunet, Paris.
- Hoeksema, B.W. & van den Hoek, C. (1983). The taxonomy of Ulva (Chlorophyceae) from the coastal region of Roscoff (Brittany, France). Botanica Marina, 26: 65.
- Hofmann, L.C., Nettleton, J.C., Neefus, C.D. & Mathieson, A.C. (2010). Cryptic diversity of Ulva (Ulvales, Chlorophyta) in the Great Bay Estuarine System (Atlantic USA): introduced and indigenous distromatic species. European Journal of Phycology, 45: 230–239.
- Horie, Y. (1952). Foreign trade policy in the early Meiji era. Kyoto University Economic Review, 22: 1–21.
- Hudson, R.R., Boos, D.D. & Kaplan, N.L. (1992). A statistical test for detecting geographic subdivision. Molecular Biology and Evolution, 9: 138–151.
- Hughey, J.R., Maggs, C.A., Mineur, F., Jarvis, C., Miller, K.A., Shabaka, S.H. & Gabrielson, P.W. (2019). Genetic analysis of the Linnaean Ulva lactuca (Ulvales, Chlorophyta) holotype and related type specimens reveals name misapplications, unexpected origins, and new synonymies. Journal of Phycology, 55: 503–508.
- Hughey, J.R., Gabrielson, P.W., Maggs, C.A., Mineur, F. & Miller, K.A. (2021). Taxonomic revisions based on genetic analysis of type specimens of Ulva conglobata, U. laetevirens, U. pertusa and U. spathulata (Ulvales, Chlorophyta). Phycological Research, in press.
- Kang, E.J., Kim, J.-H., Kim, K., Choi, H.-G. & Kim, K.Y. (2014). Re-evaluation of green tide-forming species in the Yellow Sea. Algae, 29: 267–277.
- Kang, J.H., Jang, J.E., Kim, J.H., Byeon, S.Y., Kim, S., Choi, S.K., Kang, Y.H., Park, S.R. & Lee, H.J. (2019). Species composition, diversity, and distribution of the genus Ulva along the coast of Jeju Island, Korea based on molecular phylogenetic analysis. PLoS ONE, 14: e0219958.
- Katsanevakis, S., Zenetos, A., Belchior, C. & Cardoso, A.C. (2013). Invading European Seas: assessing pathways of introduction of marine aliens. Ocean & Coastal Management, 76: 64–74.
- Kirkendale, L., Saunders, G.W. & Winberg, P. (2013). A molecular survey of Ulva (Chlorophyta) in temperate Australia reveals enhanced levels of cosmopolitanism. Journal of Phycology, 49: 69–81.
- Kjellman, F.R. (1897). Marina Chlorophyceer från Japan [in Swedish]. Norstedt P.A. & Söner, Stockholm.
- Kraft, L.G.K., Kraft, G.T. & Waller, R.F. (2010). Investigations into southern Australian Ulva (Ulvophyceae, Chlorophyta) taxonomy and molecular phylogeny indicate both cosmopolitanism and endemic cryptic species. Journal of Phycology, 46: 1257–1277.
- Lawton, R.J., Mata, L., de Nys, R. & Paul, N.A. (2013). Algal bioremediation of waste waters from land-based aquaculture using Ulva: selecting target species and strains. PLoS ONE, 8: e77344.
- Lawton, R.J., Mata, L., de Nys, R. & Paul, N.A. (2020). Correction: Algal bioremediation of waste waters from land-based aquaculture using Ulva: selecting target species and strains. PLoS ONE, 15: e0231281.
- Lee, H.W., Kang, J.C. & Kim, M.S. (2019). Taxonomy of Ulva causing blooms from Jeju Island, Korea with new species, U. pseudo-ohnoi sp. nov. (Ulvales, Chlorophyta). Algae, 34: 253–266.
- Malta, E.-J., Draisma, S. & Kamermans, P. (1999). Free-floating Ulva in the southwest Netherlands: species or morphotypes? A morphological, molecular and ecological comparison. European Journal of Phycology, 34: 443–454.
- Manghisi, A., Minicante, S.A., Bertuccio, C., Morabito, M., Torricelli, P. & Genovese, G. (2011). A cryptic alien seaweed spreading in Mediterranean coastal lagoons. Transitional Waters Bulletin, 5: 1–7.
- Matsumoto, K. & Shimada, S. (2015). Systematics of green algae resembling Ulva conglobata, with a description of Ulva adhaerens sp. nov. (Ulvales, Ulvophyceae). European Journal of Phycology, 50: 100–111.
- Maurette, F. (1922). Progrès des ports de Yokohama et de Kobé. Annales de Géographie, 31: 91–93.
- Melton, J.T. (2017). Molecular systematics and organellar genomics of the green macroalgal genus Ulva (Ulvophyceae, Chlorophyta) with a phylogenomic analysis of the “core chlorophyta”. Ph.D. Thesis, 408. University of Alabama, Tuscaloosa.
- Metzger, L. (2009). Relations between Marseille and East and Southeast Asia. In Port Cities in Asia and Europe (Graf, A. & Huat, C.B., editors), 19–30. Routledge, Abingdon.
- Miller, K.A., Aguilar-Rosas, L.E. & Pedroche, F.F. (2011). A review of non-native seaweeds from California, USA and Baja California, Mexico. Hidrobiológica, 21: 365–379.
- Miller, M.B. (2012). Europe and the Maritime World: A Twentieth Century History. Cambridge University Press, Cambridge.
- Mineur, F., Johnson, M.P., Maggs, C.A. & Stegenga, H. (2007). Hull fouling on commercial ships as a vector of macroalgal introduction. Marine Biology, 151: 1299–1307.
- Mitsuhashi, C., Teramura, H. & Shimada, H. (2020). Construction of genomic marker sets based on the chloroplast genome of a green alga, Ulva pertusa (syn. Ulva australis), leads to simple detection of Ulva species. Genes and Genetic Systems, 95: 55–63.
- Miyazaki, I. (1938). On fouling organisms in the oyster farm. 日本水産学会誌, 6: 223–232.
- Okamura, K. (1921). Icones of Japanese Algae. Kazamashobo Publ., Tokyo.
- Oróstica, M., Calderon, M.S., Boo, S.M., Sandoval, C. & Edding, M. (2017). Nuevo registro de Ulva australis (Ulvaceae, Chlorophyta) en el norte de Chile. Revista de Biología Marina y Oceanografía, 52: 621–630.
- Phillips, J.A. (1988). Field, anatomical and development studies on southern Australian species of Ulva (Ulvaceae, Chlorophyta). Australian Systematic Botany, 1: 411–456.
- Rius, M., Turon, X., Bernardi, G., Volckaert, F.A. & Viard, F. (2015). Marine invasion genetics: from spatio-temporal patterns to evolutionary outcomes. Biological Invasions, 17: 869–885.
- Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J.C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S.E. & Sánchez-Gracia, A. (2017). DnaSP v6: DNA Sequence Polymorphism analysis of large datasets. Molecular Biology and Evolution, 34: 3299–3302.
- Ruesink, J.L., Lenihan, H.S., Trimble, A.C., Heiman, K.W., Micheli, F., Byers, J.E. & Kay, M.C. (2005). Introduction of non-native oysters: ecosystem effects and restoration implications. Annual Review of Ecology and Systematics, 36: 643–689.
- Saunders, G.W. (2014). Long distance kelp rafting impacts seaweed biogeography in the Northeast Pacific: the kelp conveyor hypothesis. Journal of Phycology, 50: 968–974.
- Saunders, G.W. & Kucera, H. (2010). An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie Algologie, 31: 487–528.
- Shimada, S., Hiraoka, M., Nabata, S., Iima, M. & Masuda, M. (2003). Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva. Phycological Research, 51: 99–108.
- Simon-Bouhet, B., Garcia-Meunier, P. & Viard, F. (2006). Multiple introductions promote range expansion of the mollusc Cyclope neritea (Nassariidae) in France: evidence from mitochondrial sequence data. Molecular Ecology, 15: 1699–1711.
- Stegenga, H., Mol, I., Prud’homme van Reine, W.F. & Lokhorst, G.M. (1997). Checklist of the marine algae of the Netherlands. Gorteria, Supplement 4: 3–60.
- Stegenga, H., Karremans, M. & Simons, J. (2006). Zeewieren van de voormalige oesterputten bij Yerseke. Gorteria, 32: 125–143.
- Steinhagen, S., Karez, R. & Weinberger, F. (2019). Cryptic, alien and lost species: molecular diversity of Ulva sensu lato along the German coasts of the North and Baltic Seas. European Journal of Phycology, 54: 466–483.
- Swofford, D.L. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4.0 b10. Sinauer Associates, Sunderland, MA.
- Tanner, C.E. (1979). The taxonomy and morphological variation of distromatic ulvaceous algae (Chlorophyta) from the Northeast Pacific. Ph.D. Thesis, 249. The University of British Columbia, Vancouver.
- Verlaque, C., Belsher, T. & Deslous-Paoli, J.-M. (2002). Morphology and reproduction of Asiatic Ulva pertusa (Ulvales, Chlorophyta) in Thau Lagoon (France, Mediterranean Sea). Cryptogamie Algologie, 23: 301–310.
- Verlaque, M. & Breton, G. (2019). Biological invasion: long term monitoring of the macroalgal flora of a major European harbor complex. Marine Pollution Bulletin, 143: 228–241.
- Verlaque, M., Auby, I., Plus, M. & Belsher, T. (2008). Etude de la flore introduite dans le Bassin d’Arcachon. In PNEC “Lagunes Méditerranéennes”, Atelier 2.3 Espèces introduites – Traçabilité des espèces algales introduites en milieu ostréicole. Rapport 35. CNRS, Université de la Méditerranée, Ifremer, Marseille.
- Wolf, M.A., Sciuto, K., Andreoli, C. & Moro, I. (2012). Ulva (Chlorophyta, Ulvales) biodiversity in the North Adriatic Sea (Mediterranean, Italy): cryptic species and new introductions. Journal of Phycology, 48: 1510–1521.
- Wolff, W.J. (2005). Non-indigenous marine and estuarine species in the Netherlands. Zoologische Mededelingen (Leiden), 79: 1–116.
- Womersley, H.B.S. (1956). A critical survey of the marine algae of Southern Australia. I. Chlorophyta. Marine and Freshwater Research, 7: 343–383.
- Wonham, M.J. & Carlton, J.T. (2005). Trends in marine biological invasions at local and regional scales: the Northeast Pacific Ocean as a model system. Biological Invasions, 7: 369–392.
- Woolcott, G.W. & King, R.J. (1999). Ulva and Enteromorpha (Ulvales, Ulvophyceae, Chlorophyta) in eastern Australia: comparison of morphological features and analyses of nuclear rDNA sequence data. Australian Systematic Botany, 12: 709–725.
- Yoshida, G., Uchimura, M. & Hiraoka, M. (2015). Persistent occurrence of floating Ulva green tide in Hiroshima Bay, Japan: seasonal succession and growth patterns of Ulva pertusa and Ulva spp. (Chlorophyta, Ulvales). Hydrobiologia, 758: 223–233.
- Zhang, Z., Schwartz, S., Wagner, L. & Miller, W. (2000). A greedy algorithm for aligning DNA sequences. Journal of Computational Biology, 7: 203–214.
