Abstract
The mitochondrial carnitine/acylcarnitine carrier (CAC) is characterized by the presence of a distinct motif, RXXPANAAXF, within its sixth transmembrane α-helix. In this study, we analysed the role of the amino acids of this motif in the structure-function relationships of the human CAC by using two complementary approaches. First, we performed functional analysis in the model fungus Aspergillus nidulans of selected mutations with structural and functional relevance. Second, similar mutant human CACs were biochemically characterized after their reconstitution into liposomes. Both analyses have provided relevant information on the importance and role of the CAC motif residues in the activity and metabolic function of CAC. Only the two adjacent alanines, Ala281 and Ala282 in the human CAC, have been found not to be crucial for transport activity and in vivo function. Results obtained from amino acid substitutions of residues Arg275, Asn280 and Phe284 of human CAC together with structural analysis using molecular modelling of the carrier suggest that R275, N280 and F284 are involved in substrate binding during acylcarnitine/carnitine translocation. Furthermore, functional analysis of mutations of residues Pro278 and Ala279 in A. nidulans, together with kinetic data in reconstituted liposomes, suggest a predominant structural role for these amino acids.
Introduction
The carnitine/acylcarnitine carrier (CAC) is a member of the mitochondrial carrier family (Palmieri [Citation2004]). It catalyses the electroneutral exchange of cytosolic acylcarnitines for mitochondrial carnitine. In addition, CAC is able to perform unidirectional transport of carnitine into carnitine-depleted mitochondria to balance the matrix level of carnitine with the level present in the cytosol (Indiveri et al. [Citation1990], Indiveri et al. [Citation1991]).
Biochemical analyses carried out with CACs from humans (present study), rat (Indiveri et al. [Citation1998]), yeast Saccharomyces cerevisiae (Palmieri et al. [Citation1999]) and the filamentous fungus Aspergillus nidulans (unpublished results) after their reconstitution into liposomes have shown the ability of this carrier to transport a variety of acylcarnitines, ranging from acetylcarnitine to long-chain acylcarnitines. Physiologically, the type of acylcarnitines that CAC transports into the mitochondrial matrix varies from lower to higher eukaryotes (Jacobs & Wanders [Citation1995], De Lucas et al. 1999, Palmieri et al. [Citation1999]).
Eukaryotic microorganisms, such as A. nidulans, are able to grow on fatty acids (e.g., oleate) or C2 compounds (e.g., acetate or ethanol) as the sole source of carbon and energy (De Lucas et al. [Citation1997]). This implies that these cells must use the acetyl-CoA formed by the catabolism of these substrates for biosynthetic purposes and energy production. Consequently, when growing on fatty acids or C2 compounds, the A. nidulans CAC, encoded by the acuH gene, is an essential carrier that transports acetyl-CoA as acetylcarnitine into the mitochondria (De Lucas et al. [Citation1999]). In contrast, the acuH gene product is not essential for the utilization of alternative carbon sources, such as sugars or amino acids (De Lucas et al. [Citation1999], Perez et al. [Citation2003]). In humans, CAC, encoded by the cac gene (Iacobazzi et al. [Citation1998]), is an essential protein whose deficiency provokes a severe disorder of fatty acid β-oxidation that is usually lethal within a few months after birth (Huizing et al. [Citation1997] and references therein). In contrast to eukaryotic microorganisms, the physiological substrates of the human CAC (hCAC) are medium- and long-chain acylcarnitines. After transport into the mitochondria, these acylcarnitines are converted by carnitine palmitoyl transferase II into acyl-CoA esters that are subjected to final oxidation to CO2 and H2O. The resultant energy is crucial for humans during prolonged fasting as well as for cardiac and skeletal muscle during exercise.
Besides the typical features present in all members of the mitochondrial carrier family (Palmieri [Citation2004]), the CAC subfamily is characterized by the presence of a distinct motif, R-X-X-P-A-N-A-A-X-F, within its sixth transmembrane α-helix (De Lucas et al. [Citation1999]). This motif, which spans from amino acids 275 to 284 in the hCAC (301 amino acids) (Huizing et al. [Citation1997]), has been proven to be suitable for functional annotation of novel protein sequences appearing in the protein databases.
The conservation of this motif in all known CACs, from lower to higher eukaryotes, suggests an important role of this region in the structure-function relationships of this transport protein. In this work, we analysed the metabolic effect of selected missense mutations in this motif of the hCAC by using two complementary approaches. Firstly, we performed functional analysis of these hCAC variants in A. nidulans, a model organism in which quantification of mutant hCAC functionality can be accomplished (Perez et al. [Citation2003]). Among other reasons for utilizing A. nidulans to assay hCAC function is that the primary substrate of the A. nidulans CAC, acetylcarnitine, is also transported by hCAC and that CAC deficiency in A. nidulans is not lethal as in the metazoan lineage (De Lucas et al. [Citation1999]; Perez et al. [Citation2003]). Secondly, we determined the transport properties of the same mutants after their reconstitution into liposomes (Indiveri et al. [Citation1998], Indiveri et al. [Citation2002], Tonazzi et al. [Citation2005]). Both analyses have provided relevant information on the importance and role of the conserved amino acids of this motif.
Materials and methods
Materials
Sephadexes G-50, G-75 and G-200 were purchased from Pharmacia, l-[methyl-3H]carnitine from Amersham, egg-yolk phospholipids (l-α-phosphatidylcholine from fresh turkey egg yolk), Pipes, Triton X-100, cardiolipin, l-carnitine and N-dodecanoylsarcosine (sarcosyl) from Sigma. All other reagents were of analytical grade.
Aspergillus nidulans strains
A. nidulans PPA14 (biA1; argB2; ΔacuH::pyr4+) and A. nidulans ΔacuHT14 (pabaA1 pyrG89; ΔacuH::pyr4+; uaY9; fwA1) are two ΔacuH isolates in which the acuH ORF is replaced with the Neurospora crasa pyr-4 gene (Perez et al. [Citation2003]). Strain PPA14, that contains the argB2 marker, was utilised in transformation experiments with vectors derived from plasmid pARGHCC (Perez et al. [Citation2003]) (see below). Additionally, both ΔacuH strains were employed as negative controls in growth tests. The A. nidulans hCAC strain (biA1; argB2::argB+-cact+; ΔacuH::pyr4+), also used in this work, is an ArgB+ transformant that carries at the argB locus a single copy of the wild-type human cac cDNA under control of the acuH regulatory regions (Perez et al. [Citation2003]). This strain was used as positive control in growth tests.
Aspergillus media and culture conditions
A. nidulans was grown on Aspergillus minimal medium (Armitt et al. [Citation1976]) with the following carbon sources: 1% glucose, 0.1 M acetate or 6 mM oleate (Valenciano et al. [Citation1996]). The following nutritional requirements were added as necessary: 1 µg p-aminobenzoic acid ml−1, 0.02 µg biotine ml−1 and 200 µg arginine ml−1. For microcalorimetry studies, glass ampoules of 3 ml containing 0.4 ml of 0.1 M acetate minimal medium were inoculated with 30 µl of freshly harvested conidia (108 conidia/ml adjusted in a Neubauer chamber) of A. nidulans. Samples were incubated in a LKB Bioactivity Monitor at 37°C. Thermograms represent the heat output versus time (p-t curve) of each strain analysed (Perez et al. [Citation2003]).
Mutagenesis of the integrative plasmid pARGHCC
The integrative plasmid pARGHCC (Perez et al. [Citation2003]) used in this work drives transcription of the complete human cac cDNA under control of the acuH promoter and terminator. Also, this vector contains the A. nidulans argB gene as the fungal selection marker. Selected missense mutations were introduced in the human cac cDNA of plasmid pARGHCC by using appropriate primers and the QuikchangeSite-Directed Mutagenesis kit (Stratagene) according to the manufacturer′s instructions. Confirmation that the correct mutation had been introduced was performed by direct sequencing of the mutated human cac cDNA. The resultant pARGHCC-derived vectors were propagated in Escherichia coli DH5α and subsequently used to transform A. nidulans using the method described by Balance et al. ([Citation1983]).
Site-directed mutagenesis, overexpression and purification of wild-type and mutant hCACs
The coding region for the hCAC (Huizing et al. [Citation1997]) was amplified from total human liver cDNA by PCR. Mutations were introduced with complementary mutagenic primers using the overlap extension method (Ho et al. [Citation1989]) and the High Fidelity PCR System (Roche). The PCR products were purified by the Gene Clean Kit (La Jolla), digested with NdeI and HindIII (restriction sites added at the 5′ end of forward and reverse primers, respectively) and ligated into the pMW7 expression vector. All mutations were verified by DNA sequencing, and, except for the desired base changes, all of the sequences were identical to that of human cac cDNA. The resulting plasmids were transformed into E. coli C0214 (Indiveri et al. [Citation1998]). Bacterial overexpression, isolation of the inclusion body fraction, solubilization and purification of the wild-type hCAC and mutant hCAC proteins were performed as described previously (Indiveri et al. [Citation1998]).
Reconstitution of wild-type and mutant hCACs into liposomes and transport measurements
The purified recombinant proteins were reconstituted into liposomes in the presence of 13 mM carnitine, as described previously (Indiveri et al. [Citation1998]). The external substrate was removed from proteoliposomes on Sephadex G-75 columns. Transport at 25°C was started by adding 0.1 mM [3H]carnitine to proteoliposomes and terminated by the addition of 1.5 mM N-ethylmaleimide. In controls, the inhibitor was added together with the labeled substrate, according to the inhibitor stop method (Palmieri et al. [Citation1995]). Finally, the external substrate was removed by chromatography on Sephadex G-50 columns, and the radioactivity in the liposomes was measured. The experimental values were corrected by subtracting control values. All of the transport activities were determined by taking into account the efficiency of reconstitution (i.e., the share of successfully incorporated protein). The CAC reconstituted into liposomes was stable for at least 2 hours, i.e., it exhibited virtually the same transport rate after 2 h incubation at 25°C as at the beginning of the incubation period (immediately after the reaction mixture had reached 25°C).
Other methods
Plasmid DNA employed in sequencing and A. nidulans transformation experiments was isolated by using the Qiagen Plasmid Kit (Qiagen) following manufacturer′s instructions. Genomic DNA from A. nidulans was isolated as described by Raeder & Broder ([Citation1985]). The remaining molecular biology techniques were performed according to methods described by Sambrook et al. ([Citation1989]). The method used to obtain a 17000 g subcellular pellet (mitochondrial-enriched fraction) from A. nidulans cells was described elsewhere (Valenciano et al. [Citation1996]). In Western blot experiments specific polyclonal antibodies against the rat CAC (Indiveri et al. [Citation2002]) were utilised. The anti-rabbit peroxidase-conjugated secondary antibody and the enhanced chemiluminescence (ECL) immunodetection system were purchased from Amersham (GE Healthcare). The amount of recombinant protein was estimated on Coomassie blue-stained SDS-PAGE gels by the Bio-Rad GS-700 Imaging Densitometer equipped with the software Bio-Rad Multi-Analist, as previously described (Indiveri et al. [Citation2002]). The extent of incorporation of the recombinant proteins into liposomes was determined as described in Phelps et al. ([Citation1996]), with the modifications reported in Indiveri et al. ([Citation2002]). The homology model of the hCAC was built by the Swiss-Model protein modelling server (Schwede et al. [Citation2003]) using the X-ray structure of the carboxyatractyloside-ADP/ATP carrier complex as a template (Pebay-Peyroula et al. [Citation2003]).
Results
Functional analysis of mutant hCACs in A. nidulans
In A. nidulans, activity of CAC, encoded by the acuH gene, is essential for metabolism of substrates leading to acetyl-CoA, such as acetate or long-chain fatty acids. In contrast, activity of this transporter is dispensable when alternative compounds such as carbohydrates or amino acids act as the carbon and energy source (De Lucas et al. [Citation1997], De Lucas et al. [Citation1999]). Taking advantage of this important feature, we developed a functional metabolic assay based on the fully complementation of the deficiency of an A. nidulans ΔacuH strain by the wild-type human cac cDNA (Perez et al. [Citation2003]).
Using the integrative plasmid pARGHCC, that enables physiological expression of the human cac gene in A. nidulans, we replaced the fully conserved residues of the RX2PANAAXF motif of the hCAC by glycine or another amino acid with structural and/or functional relevance. The following amino acid substitutions were made: R275G, R275A and R275K, P278G and P278V, A279G, N280G and N280Q, A281G, A282G, and F284G. Mutated versions of the pARGHCC vector were transformed into the A. nidulans PPA14 (biA1; argB2; ΔacuH) strain, and argB+ transformants were selected and further purified. Genomic DNA from these arginine prototrophs was digested with EcoRI and analysed by Southern blotting to identify transformants carrying a single-copy integration of the site-directed mutagenized pARGHCC vector at the argB locus. Two different probes were employed in these Southern blotting experiments: a 1.7-kb HindIII internal fragment of the A. nidulans argB gene and the entire human cac cDNA (). Two positive transformants for each mutation were selected and further analysed.
Figure 1. Identification of A. nidulans transformants expressing a mutant version of the human cac cDNA at physiological level. (A) Schematic representation of the integration at the chromosomal argB locus of a single-copy of plasmid pARGHCC that enables expression of hCAC with a defined mutation in its RX2PANAAXF motif. Chromosomal and plasmid EcoRI sites are indicated. The EcoRI fragment of A. nidulans PPA14 (recipient strain) and the two EcoRI fragments of argB+ single-copy transformants detected by the argB probe used are shown by arrows. Sizes of these fragments are also given. Mutation at the argB2 loci is indicated by an open box. (B) Southern blotting of genomic DNA from PPA14 (ΔacuH; argB2) (lane1) and argB+ transformants (lanes 2–9) digested with EcoRI and hybridized with an 1.7-kb HindIII internal fragment of the A. nidulans argB gene. In this blot, transformants in lanes 2, 3 and 9 contain a single-copy of the cac transgene that encodes the R275K mutant hCAC. (C) Confirmation that previously selected argB+ single-copy transformants contain one copy of the human cac transgene was performed by Southern blotting using the cac cDNA as a probe. In this analysis, the PPA14 strain (lane 1) was employed as negative control, whereas correct transformants (lanes 2–9) give a 7.5-kb hybridization band. (D) Wild-type and mutant hCACs are similarly expressed and localized in A. nidulans mitochondria, as confirmed by Western blotting. A. nidulans cells expressing wild-type hCAC (lane 1) or the R275K (lane 2), P278G (lane 3), A279G (lane 4), N280Q (lane 5), A281G (lane 6), A282G (lane 7) and F284G (lane 8) mutated versions were grown for 20 h on 0.3% sucrose + 0.1 M acetate minimal medium and used to obtain a 17000 g mitochondrial-enriched fraction. Proteins (20 µg) from this fraction were separated on SDS-PAGE, blotted onto nitrocellulose membranes and immunodetected with specific anti-CAC antibodies.
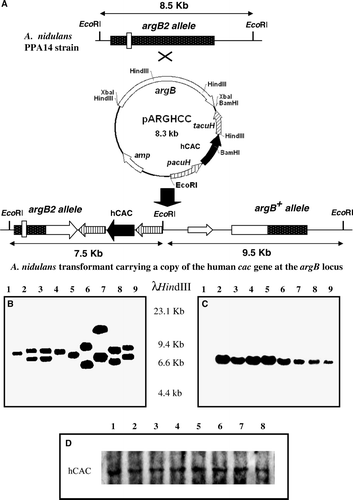
The growth of these transformants was scored after inoculation on plates of selective medium (0.1 M acetate or 6 mM oleate minimal medium) and non-selective medium (1% glucose minimal medium). Additionally, the A. nidulans ΔacuHT14 and PPA14 (ΔacuH) strains were used in these growth tests as negative controls, whereas the A. nidulans hCAC strain, expressing the human cac cDNA under control of the acuH promoter and terminator, was employed as positive control. The growth level of the latter strain is strictly similar to that of the wild-type strain of A. nidulans in selective and non-selective media (Perez et al. [Citation2003]).
A. nidulans strains carrying the substitutions R275K, P278G, A279G, N280Q and F284G in hCAC were unable to grow in selective medium but grew similarly to the hCAC strain in non-selective medium (). Similar results were obtained for mutant hCACs bearing the substitutions R275G, R275A, P278V and N280G (not shown). These results demonstrate that all these mutant hCACs result in a null phenotype similar to that of the A. nidulans ΔacuH strain. In contrast, A. nidulans strains carrying the A281G and A282G mutations in the hCAC were able to grow normally on acetate (or oleate) plates as well as on glucose minimal medium (). It is worth mentioning that wild-type and mutant hCACs were similarly expressed and localized in A. nidulans cells (D).
Figure 2. Growth analysis on glucose and acetate minimal medium of Aspergillus nidulans strains expressing different human CACs. A. nidulans single-copy transformants expressing the following versions of the hCAC: wild-type (hCAC) and R275K, P278G, A279G, N280Q, A281G, A282G, F284G mutants were streaked out on plates containing 1% glucose minimal medium (A) or 0.1 M acetate minimal medium (B) and incubated at 37°C for 3 days. The A. nidulans ΔacuHT14 (ΔACUH) strain was cultivated as a negative control. This Figure is reproduced in colour in Molecular Membrane Biology online.
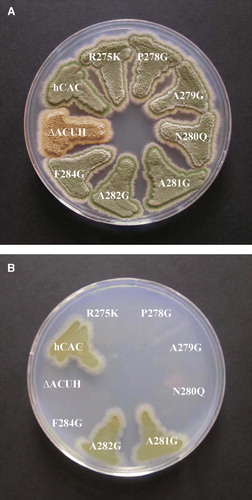
Quantification of the growth rate on acetate minimal medium dependent on the A281G and A282G non-lethal missense mutations in hCAC was performed by microcalorimetric analysis and compared to that of the wild-type hCAC.
The suitability of this system for functional characterization of hCAC mutations in A. nidulans was demonstrated in a previous work (Perez et al. [Citation2003]). In such a study, the importance of the Pro30 residue for hCAC structure and function was clearly proved. In contrast, the Asp32 residue was shown not to be crucial, since the Asp32Glu conservative substitution was physiologically tolerated permitting a significant growth rate (a third of that promoted by the wild-type hCAC) on acetate medium (Perez et al. [Citation2003]). As can be seen in , the power-time (p-t) curves of A. nidulans strains carrying the A281G or A282G substitutions were almost indistinguishable from each other. In addition, they resemble that of the A. nidulans hCAC strain (A). When comparing the semilogarithmic p-t representations during exponential growth on acetate medium of A. nidulans strains carrying the wild-type hCAC, the A281G or A282G mutated versions revealed that both amino acid substitutions lead only to a minor reduction (a 10% decrease) of the growth rate promoted by the A. nidulans strain expressing the wild-type hCAC (B). Results from this functional analysis unequivocally demonstrate that the A 281 and A 282 residues of the RX2PANAAXF motif of hCAC are not essential for transport and function, as they can be replaced by glycine. In contrast, microcalorimetric profiles obtained for the remaining hCAC mutants analysed were indistinguishable from that of the PPA14 (ΔacuH) strain (data not shown), confirming their null phenotype observed in selective medium plates.
Figure 3. Quantification of functionality of mutant hCACs in A. nidulans by microcalorimetric analysis. (A) Power-time (p-t) curve on 0.1 M acetate minimal medium of A. nidulans PPA14 (ΔacuH; argB2) strain and single-copy transformants expressing the following versions of the hCAC at the physiological level: wild-type (hCAC), R275K, A281G, A282G mutants. Symbols representing each (p-t) curve are indicated at the bottom of the plot. (B) Semilogarithmic p-t representation during exponential growth of A. nidulans single-copy transformants expressing the wild-type (hCAC) and the A281G or A282G mutant versions. Plot symbols are indicated at the bottom.
Functional analysis of wild-type and mutant hCACs in reconstituted liposomes
The mutations analysed with the A. nidulans system were introduced in the vector pMW7-hCAC and the corresponding mutant proteins as well as the wild-type hCAC were overexpressed in E.coli, purified and reconstituted into liposomes. The expression levels of the mutant hCACs in E. coli varied between 73% and 116% as compared to those of the wild-type hCAC. The efficiency of reconstitution of the wild-type and mutant hCACs ranged between 22% and 36% of the added protein, which is similar to the values found for recombinant rat CAC and other mitochondrial carrier proteins (Phelps et al. [Citation1996], Fiermonte et al. [Citation1998], Briggs et al. [Citation1999], Dolce et al. [Citation2001], Palmieri et al. [Citation2001], Stipani et al. [Citation2001], Indiveri et al. [Citation2002], Marobbio et al. [Citation2003], Fiermonte et al. [Citation2004]).
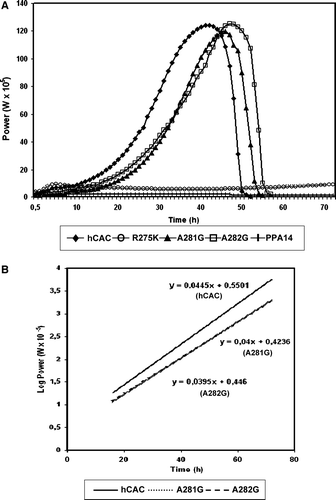
Reconstituted hCAC catalysed a first order kinetic exchange of labeled external carnitine for intraliposomal carnitine (). In the absence of the substrate inside the proteoliposomes, recombinant hCAC catalysed a unidirectional transport of carnitine at a rate eight times lower than that of the exchange (not shown). Approximately the same large difference between the initial rates of the exchange and the uniport modes of transport was previously observed for the rat CAC either expressed in E. coli (Indiveri et al. [Citation1998]) or purified from liver mitochondria (Indiveri et al. [Citation1991]). Furthermore, recombinant wild-type hCAC exhibited the same substrate specificity and inhibitor sensitivity (results not shown) as those determined previously for rat CAC (Indiveri et al. [Citation1990], Indiveri et al. [Citation1998]).
Figure 4. Time course of [3H]carnitine uptake by liposomes reconstituted with recombinant hCAC proteins. Transport was started by adding 0.1 mM [3H]carnitine to proteoliposomes containing 13 mM carnitine and stopped at the indicated times. In each panel the data relative to the wild-type hCAC (○) are shown with those relative to the mutants R275K (•) and R275A (□) in (A); P278G (•) and P278V (□) in (B); N278G (•) and N280Q (□) in (C); A279G (•), A281G (□), A282G (▪) and F284G (r) in (D). The data represent means±S.D. of three independent experiments carried out in duplicate.
![Figure 4. Time course of [3H]carnitine uptake by liposomes reconstituted with recombinant hCAC proteins. Transport was started by adding 0.1 mM [3H]carnitine to proteoliposomes containing 13 mM carnitine and stopped at the indicated times. In each panel the data relative to the wild-type hCAC (○) are shown with those relative to the mutants R275K (•) and R275A (□) in (A); P278G (•) and P278V (□) in (B); N278G (•) and N280Q (□) in (C); A279G (•), A281G (□), A282G (▪) and F284G (r) in (D). The data represent means±S.D. of three independent experiments carried out in duplicate.](/cms/asset/c229954c-78f2-4a8c-a02a-fe8eaebec993/imbc_a_269628_f0004_b.gif)
In , the time-courses of the [3H]carnitine/carnitine exchange in liposomes reconstituted with wild-type hCAC or with each of the mutant hCACs were compared. The substitutions R275K and R275A caused a strong inhibition of hCAC transport activity. The uptakes of [3H]carnitine mediated by R275K and R275A were about 35% and 10%, respectively, of the uptake mediated by the wild-type carrier (A). The P278G and P278V substitutions reduced the activity of hCAC to about 50% and 25%, respectively (B). When the Asn residue in position 280 was replaced by Gly, a nearly complete inactivation was observed, whereas the N280Q conservative replacement caused a transport inhibition of about 70% (C). The substitution of the Ala residues in positions 279, 281 and 282 with glycine is well tolerated since the activities of the respective mutants were about 85% (A281G), 70% (A282G) and 60% (A279G) of the value of the wild-type carrier (D). By contrast, the F284G substitution decreased carrier activity to about 10% of that of the wild-type protein (D). Taken together, these results indicated that the R275K, P278G, P278V, A279G and N280Q mutants display a significant transport activity in reconstituted liposomes, although they are unable to complement the phenotype of the A. nidulans ΔacuH strain.
To gain insight into this discrepancy, the kinetic characteristics of the [3H]carnitine/carnitine exchange catalysed by the wild-type and mutant hCACs were further investigated. The basic kinetic data of the hCACs proteins were determined by measuring the initial transport rate at various external carnitine concentrations in the presence of a constant internal concentration of 13 mM carnitine. The Km and Vmax values for carnitine exchange at 25°C are reported in .
Table I. Kinetic constants of reconstituted wild-type and mutant hCACs. The values were calculated from double-reciprocal plots of the rates of carnitine/carnitine exchange under variation of the external substrate concentration. The exchange was started by the addition of 0.12–2.0 mM [3H]carnitine to proteoliposomes containing 13 mM carnitine and reconstituted with the wild-type hCAC or one of the indicated mutants. The reaction time was 4 min. The Km values of mutant carriers that are severely affected in their transport activity by the indicated mutation were not assayed (n.d., not determined). The data represent the means±SD of four different experiments.
The Km of the wild-type hCAC (0.9±0.2 mM) was 1.95-fold that of the recombinant rat CAC (Indiveri et al. [Citation1998]) indicating a lower affinity of the human ortholog for carnitine. The Vmax of the hCAC (1.0±0.3 mmol/min×g protein) was 1.28-fold that of the rat ortholog (Indiveri et al. [Citation1998]). The Km for carnitine of the R275K, P278G and P278V mutants was 2.6-, 3.8- and 3.4-fold the value of the wild-type hCAC protein, respectively. A lower but still significant (1.7- and 1.9-fold) increase in the Km for carnitine was observed with the A279G and N280Q mutants, respectively, as compared to that of the wild-type protein. The Vmax values of the P278G, P278V, N280Q, A281G and A282G mutants ranged between 90% and 120% with respect to that of the wild-type carrier, whereas the Vmax values of the R275K and A279G mutants were 50% that of the wild-type hCAC.
Since transport of acetylcarnitine via CAC is the only catabolic pathway operating in A. nidulans cells growing on acetate or long-chain fatty acids (De Lucas et al. [Citation1999]), the ability of the hCAC mutants to transport acetylcarnitine was tested and compared to that of transporting carnitine. The data reported in show that wild-type hCAC and each of the mutant hCAC proteins transport acetylcarnitine as effectively as carnitine.
Table II. Transport of acetylcarnitine or carnitine in exchange for [3H]carnitine by liposomes reconstituted with recombinant hCAC proteins. Proteoliposomes were preloaded internally with 13 mM acetylcarnitine or 13 mM carnitine. Transport was started by the external addition of 0.1 mM [3H]carnitine and terminated after 10 min. The data represent means±SD of three independent experiments.
We also determined the inhibition constants (Ki) of acetylcarnitine for the recombinant and reconstituted hCAC proteins. Acetylcarnitine inhibited the hCAC-mediated carnitine/carnitine exchange competitively. The Ki values of acetylcarnitine, calculated from Dixon plots of the reciprocal [3H]carnitine/carnitine exchange versus acetylcarnitine concentration, were 0.8±0.1, 1.7±0.2, 2.8±0.3, 2.9±0.3, 1.2±0.1, and 1.5±0.1 mM for WT, R275K, P278G, P278V, A279G and N280Q, respectively (means of four experiments).
Discussion
The metabolic features of the model microorganism A. nidulans, together with its amenability for molecular genetic studies, permitted the development of a metabolic assay for functional characterization of mutations in the hCAC. This assay allows discrimination between CAC mutations that impair the in vivo function of this carrier and those having a neutral effect (Perez et al. [Citation2003]). On the other hand, the reconstitution into liposomes of over-expressed wild-type and mutant hCACs allows recognition of the biochemical basis of the growth impairment provoked by each deleterious mutation. In fact, while the in vivo system provides information on the effect of the mutation on the living cell, the reconstituted system allows the kinetic parameters of the mutated protein to be measured (Indiveri et al. [Citation1998], Indiveri et al. [Citation2002]). Consequently, the combined in vivo and in vitro approaches employed in this work provide exhaustive information on the effect of a defined mutation that can be used both for understanding the mechanism of metabolism impairment and for gaining new insight into the structure-function relationships of the protein. In fact, the results described in this study identified the amino acids of the characteristic motif (RX2PANAAXF) of the CAC subfamily that are crucial for biological function.
Functional analysis in A. nidulans of the R275G, R275A and R275K mutations in hCAC suggests that any amino acid substitution occurring at the R275 residue of hCAC, including the conserved replacement R275K, abolishes its biological function in this fungus. The same loss-of-function phenotype in selective medium plates was observed for the P278G, P278V, A279G, N280G, N280Q and F284G substitutions in hCAC. These results indicate that residues R275, P278, A279, N280 and F284 of hCAC play a crucial role in the in vivo transport of acetylcarnitine into the mitochondrial matrix of A. nidulans (De Lucas et al. [Citation1999]) and, thus, should be important for the maintenance of CAC structure and/or function. In addition, a severe clinical phenotype is to be expected for humans bearing these mutations in homozygosis or compound heterozygosis.
The results obtained in reconstituted liposomes are consistent with the observation that the R275A, N280G and F284G mutations lead to a non-viable A. nidulans phenotype, since these mutations inactivate the carrier protein. In the in vitro assay the conserved substitutions of R275 (R275K) and N280 (N280Q), and the A279G mutation result in proteins that still retain a consistent transport function. This finding seems to contrast with the non-viable A. nidulans phenotype obtained with these mutants on selective medium (0.1 M acetate or 6 mM oleate medium). However, these mutants showed a significant increase in the Km for carnitine compared to the wild-type protein and, in the case of R275K and A279G, a considerable decrease in Vmax. These changes likely produce an accumulation of acetyl-CoA that can reach toxic levels for the cells. The toxicity of acetyl-CoA is a well-known feature of A. nidulans mutants incapable of utilizing acetate, including the acuH mutants deficient in CAC (Armitt et al. [Citation1976], De Lucas et al. [Citation1997], De Lucas et al. [Citation1999], Martinez et al. [Citation2007]). Thus, we propose that the growth impairment resulting from the R275K, A279G and N280Q mutations is due to a toxic effect provoked by the accumulation of acetyl-CoA in the cytosol of these mutant cells. A similar interpretation may apply to the P278V and P278G substitutions because the corresponding hCAC mutant proteins have a very high Km for carnitine.
Some comments are in order regarding the functionally important residues identified within the characteristic motif of the mitochondrial carnitine carrier subfamily. In this section, these residues will be discussed in light of the comparative model of the hCAC based on the available crystallographic structure of the ADP/ATP carrier (Pebay-Peyroula et al. [Citation2003]). The mitochondrial ADP/ATP carrier and the other members of the family are highly homologous and must have the same overall structure. Therefore, homology models of mitochondrial carriers have been built for several members of the family (Walters & Kaplan [Citation2004], Morozzo della Rocca et al. [Citation2005], Robinson & Kunji [Citation2006], Cappello et al. [Citation2006], Cappello et al. [Citation2007], Perchiniak et al. [Citation2007]) including the CAC (Tonazzi et al. [Citation2005]). As shown in the homology-modeled structure of the hCAC (), R275, N280 and F284, which are located in the transmembrane α-helix VI (H6), protrude into the water-accessible cavity that, in the crystallographic structure of the ADP/ATP carrier-carboxyatractyloside complex, is exposed toward the cytosolic side of the mitochondrial membrane and is occupied by the inhibitor and possibly by the cytosolic ADP (Pebay-Peyroula et al. [Citation2003]).
Figure 5. Structural model of the hCAC protein. Ribbon diagrams viewing the carrier from the lateral side (A) and the cytosolic side (B). The transmembrane α-helices are coloured as follows: H1 red, H2 orange-green, H3 light-green, H4 dark-green, H5 light-blue and H6 blue. Stick representation highlights some residues of the motif R-X-X-P-A-N-A-A-X-F indicated by their number in the hCAC sequence.
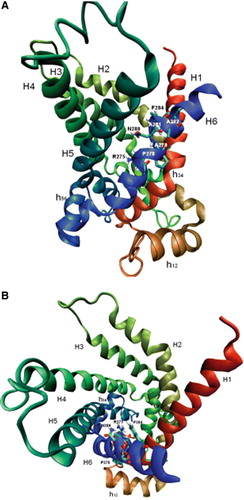
R275 is localized in the cavity at the midpoint of the membrane and is a highly conserved residue in the members of the mitochondrial carrier family. It corresponds to R288 in the bovine oxoglutarate carrier and R280 in the bovine ADP/ATP carrier (Runswick et al. [Citation1990], Aquila et al. [Citation1982]). As previously proposed for R288 of the oxoglutarate carrier (Cappello et al. [Citation2006]), it is likely that R275 of the hCAC is involved in the binding of carnitine to the carrier molecule. Like R288 in the oxoglutarate carrier (Cappello et al. [Citation2006]), R275 in hCAC cannot be replaced by other amino acids without the loss of the carrier activity, and even the conserved substitution R275K is not well tolerated. In addition, R275 in the hCAC is equivalent to R303 in the S. cerevisiae CAC (Palmieri et al. [Citation1999]). R303 is part of the mitochondrial carrier common substrate-binding site recently proposed by Robinson & Kunji using comparative models of carriers with known substrate specificity and correlating the chemical properties of a substrate to a site with the correct chemistry and geometry to bind the substrate (Robinson & Kunji, [Citation2006]). N280 and F284 could also be involved in substrate binding during acylcarnitine/carnitine translocation through the carrier molecule via H bonds and/or weaker interactions. It can be hypothesized that F284 plays a role in the early recognition of the substrate in view of its location in the carrier cavity near the membrane cytosolic face. Furthermore, secondary structure prediction using the Chou-Fasman algorithm indicates that the F284G substitution dramatically disturbs the structure of the carrier by creating a coil in transmembrane α-helix VI. P278 and A282 point towards the lipids of the membrane. Generally, amino acids interacting with the membrane bi-layer tolerate substitution quite well, as shown by cysteine-scanning mutagenesis experiments of the oxoglutarate carrier (Cappello et al. [Citation2006], Cappello et al. [Citation2007]) and by the replacement of the hCAC A282 with glycine (this study). However, mutagenesis of P278 in hCAC affects transport activity. Helix VI of the hCAC displays a small bend at the level of P278 with the concave side facing the lipids. It is likely that replacement of P278 with G or V eliminates the bend moving R275 and N280 away from the path of the substrate through the carrier protein. This explanation is supported by the substantial increase in the Km value of P278G and P278V mutations. A279 and A281 are localized between helix VI and helix I and between helix VI and helix V, respectively. These residues could therefore be important for inter-helical interactions and conformational changes occurring during substrate translocation. Accordingly, a small residue such as glycine is somewhat well tolerated, whereas the bulkier side chain of valine is not. In this context it is noteworthy that the A281V substitution, which displays a null phenotype in A. nidulans (not shown), is responsible for a severe clinical phenotype in humans (Iacobazzi et al. [Citation2004]).
Acknowledgements
This work was supported by grants from The University of Alcalá (Spain) to JRDL, the University of Bari (Italy), MUR, Apulia Region, CEGBA and by the EC contract LSHM-CT-2004-503116 to FP.
References
- Aquila H, Misra D, Eulitz M, Klingenberg M. Complete amino acid sequence of the ADP/ATP carrier from beef heart mitochondria. Hoppe Seylers Z Physiol Chem 1982; 363: 345–349
- Armitt S, McCullogh W, Roberts CF. Analysis of acetate non-utilizing (acu) mutants in Aspergillus nidulans. J Gen Microbiol 1976; 92: 263–282
- Balance DJ, Buxton FP, Turner G. Transformation of Aspergillus nidulans by the orotidine-5′-phsophate decarboxylase gene of Neurospora crassa. Biochem Biophys Res Commun 1983; 112: 284–289
- Briggs C, Mincone L, Wohlrab H. Replacements of basic and hydroxyl amino acids identify structurally and functionally sensitive regions of the mitochondrial phosphate transport protein. Biochemistry 1999; 38: 5096–5102
- Cappello AR, Curcio R, Miniero DV, Stipani I, Robinson AJ, Kunji ERS, Palmieri F. Functional and structural role of amino acid residues in the even-numbered transmembrane α-helices of the bovine mitochondrial oxoglutarate carrier. J Mol Biol 2006; 363: 51–62
- Cappello AR, Miniero DV, Curcio R, Ludovico A, Daddabbo L, Stipani I, Robinson AJ, Kunji ERS, Palmieri F. Functional and structural role of amino acid residues in the odd-numbered transmembrane α-helices of the bovine mitochondrial oxoglutarate carrier. J Mol Biol 2007; 369: 400–412
- De Lucas JR, Valenciano S, Domínguez AI, Turner G, Laborda F. Characterizaton of oleate-nonutilizing mutants of Aspergillus nidulans isolated by the 3-amino-1,2,4-triazole positive selection method. Arch Microbiol 1997; 168: 504–512
- De Lucas JR, Domínguez AI, Valenciano S, Turner G, Laborda F. The acuH gene of Aspergillus nidulans, required for growth on acetate and long-chain fatty acids, encodes a putative homologue of the mammalian carnitine/acyl-carnitine carrier. Arch Microbiol 1999; 171: 386–396
- Dolce V, Fiermonte G, Runswick MJ, Palmieri F, Walker JE. The human mitochondrial deoxynucleotide carrier and its role in toxicity of nucleoside antivirals. Proc Natl Acad Sci USA 2001; 98: 2284–2288
- Fiermonte G, Palmieri L, Dolce V, Lasorsa FM, Palmieri F, Runswick MJ, Walker JE. The sequence, bacterial expression and functional reconstitution of the rat mitochondrial dicarboxylate transporter cloned via distant homologs in yeast and Caenorhabditis elegans. J Biol Chem 1998; 273: 24754–24759
- Fiermonte G, De Leonardis F, Todisco S, Palmieri L, Lasorsa FM, Palmieri F. Identification of the mitochondrial ATP-Mg/Pi transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. J Biol Chem 2004; 279: 30722–30730
- Ho SN, Hunt HD, Hortnon RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989; 77: 51–59
- Huizing M, Iacobazzi V, Ijlst L, Savelkoul P, Rutienbeek W, van den Heuvel L, Indiveri C, Smeitink J, Trijbels F, Wanders RJA, Palmieri F. Cloning of the human carnitine-acylcarnitine carrier cDNA and identification of the molecular defect in a patient. Am J Hum Genet 1997; 61: 1239–1245
- Iacobazzi V, Naglieri MA, Stanley CA, Wanders RJA, Palmieri F. The structure and organization of the human carnitine/acylcarnitine translocase (CACT) gene. Biochem Biophys Res Commun 1998; 252: 770–774
- Iacobazzi V, Invernizzi F, Baratta S, Pons R, Chung W, Garavaglia B, Dionisi-Vici C, Ribes A, Parini R, Huertas MD, Roldan S, Lauria G, Palmieri F, Taroni F. Molecular and functional analysis of SLC25A20 mutations causing carnitine-acylcarnitine translocase deficiency. Hum Mutat 2004; 24: 312–320
- Indiveri C, Tonazzi A, Palmieri F. Identification and purification of the carnitine carrier from rat liver mitochondria. Biochim Biophys Acta 1990; 1020: 81–86
- Indiveri C, Tonazzi A, Palmieri F. Characterization of the unidirectional transport of carnitine catalyzed by the reconstituted carnitine carrier from rat liver mitochondria. Biochim Biophys Acta 1991; 1069: 110–116
- Indiveri C, Iacobazzi V, Giangregorio N, Palmieri F. Bacterial overexpression, purification, and reconstitution of the carnitine/acylcarnitine carrier from rat liver mitochondria. Biochim Biophys Res Commun 1998; 249: 589–594
- Indiveri C, Giangregorio N, Iacobazzi V, Palmieri F. Site-directed mutagenesis and chemical modification of the six native cysteine residues of the rat mitochondrial carnitine carrier: implications for the role of cysteine-136. Biochemistry 2002; 41: 8649–8656
- Jacobs BS, Wanders RJA. Fatty acid β-oxidation in perosixomes and mitochondria: the first, unquivocal evidence for the involvement of carnitine in shuttling propionyl-CoA from peroxisomes to mitochondria. Biochem Biophys Res Commun 1995; 213: 1035–1041
- Marobbio CMT, Agrimi G, Lasorsa FM, Palmieri F. Identification and functional reconstitution of yeast mitochondrial carrier for Sadenosylmethionine. EMBO J 2003; 22: 5975–5982
- Martinez O, Marco E, Gago F, Laborda F, De Lucas JR. Supression of the acuH13 and acuH31 nonsense mutations in the carnitine/acylcarnitine translocase (acuH) gene of Aspergillus nidulans by the G265S substitution in the domain 2 of the release factor eRF1. Fung Genet Biol 2007; 44: 139–151
- Morozzo della Rocca B, Miniero DV, Tasco G, Dolce V, Falconi M, Ludovico A, Cappello AR, Sanchez P, Stipani I, Casadio R, Desideri A, Palmieri F. Substrate-induced conformational changes of the mitochondrial oxoglutarate carrier: a spectroscopic and molecular modelling study. Mol Membr Biol 2005; 22: 443–452
- Palmieri F, Indiveri C, Bisaccia F, Iacobazzi V. Mitochondrial metabolite carrier proteins: purification, reconstitution, and transport studies. Methods Enzymol 1995; 260: 349–369
- Palmieri F. The mitochondrial transporter family (SLC25): physiologyical and pathological implications. Eur J Physiol 2004; 447: 689–709
- Palmieri L, Lasorsa FM, Iacobazzi V, Runswick MJ, Palmieri F, Walker JE. Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett 1999; 462: 472–476
- Palmieri L, Pardo B, Lasorsa FM, del Arco A, Kobayashi K, Iijima M. Citrin and aralar1 are Ca2 + -stimulated aspartate/glutamate transporters in mitochondria. EMBO J 2001; 20: 5060–5069
- Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJ, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 2003; 426: 39–44
- Perchiniak E, Lawrence SA, Kasten S, Woodard BA, Taylor SM, Moran RG. Probing the mechanism of the hamster mitochondrial folate transporter by mutagenesis and homology modeling. Biochemistry 2007; 46: 1557–1567
- Perez P, Martinez O, Romero B, Olivas I, Pedregosa AM, Palmieri F, Laborda F, De Lucas JR. Functional analysis of mutations in the human carnitine/acylcarnitine translocase in Aspergillus nidulans. Fung Genet Biol 2003; 39: 211–220
- Phelps A, Briggs C, Mincone L, Wohlrab H. Mitochondrial phosphate transport protein replacements of glutamic, aspartic, and histidine residues affect transport and protein conformation and point to a coupled proton transport path. Biochemistry 1996; 35: 10757–10762
- Raeder U, Broder P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol 1985; 1: 17–20
- Robinson AJ, Kunji ER. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Natl Acad Sci USA 2006; 103: 2617–2622
- Runswick MJ, Walker JE, Bisaccia F, Iacobazzi V, Palmieri F. Sequence of the bovine 2-oxoglutarate/malate carrier protein: structural relationship to other mitochondrial transport proteins. Biochemistry 1990; 29: 11033–11040
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 1989
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 2003; 31: 3381–3385
- Stipani V, Cappello AR, Daddabbo L, Natuzzi D, Miniero DV, Stipani I, Palmieri F. The mitochondrial oxoglutarate carrier: cysteine-scanning mutagenesis of transmembrane domain IV and sensitivity of cys mutants to sulphydryl reagents. Biochemistry 2001; 40: 15805–15810
- Tonazzi A, Giangregorio N, Indiveri C, Palmieri F. Identification by site-directed mutagenesis and chemical modification of three vicinal cysteine residues in rat mitochondrial carnitine/acylcarnitine transporter. J Biol Chem 2005; 280: 19607–19612
- Valenciano S, De Lucas JR, Pedregosa A, Monistrol I, Laborda F. Induction of β-oxidation enzymes and microbody proliferation in Aspergillus nidulans. Arch Microbiol 1996; 166: 336–341
- Walters DE, Kaplan RS. Homology-modeled structure of the yeast mitochondrial citrate transport protein. Biophys J 2004; 87: 907–911