Abstract
Organellar and cytosolic pH homeostasis is central to most cellular processes, including vesicular trafficking, post-translational modification/processing of proteins, and receptor-ligand interactions. SLC9A7 (NHE7) was identified as a unique (Na+, K+)/H+ exchanger that dynamically cycles between the trans-Golgi network (TGN), endosomes and the plasma membrane. Here we have used mass spectrometry to explore the affinity-captured interactome of NHE7, leading to the identification of cytoskeletal proteins, cell adhesion molecules, membrane transporters, and signaling molecules. Among these binding proteins, calcium-calmodulin, but not apo-calmodulin, binds to NHE7 and regulates the organellar transporter activity. Vimentin was co-immunoprecipitated with endogenous NHE7 protein in human breast cancer MDA-MB-231 cells. A sizable population of NHE7 relocalized to focal complexes in migrating cells and showed colocalization with vimentin and actin in focal complexes. Among the NHE7-binding proteins identified, CD44, a cell surface glycoprotein receptor for hyaluronate and other ligands, showed regulated interaction with NHE7. Pretreatment of the cells with phorbol ester facilitated the NHE7-CD44 interaction and the lipid raft association of CD44. When lipid rafts were chemically disrupted, the NHE7-CD44 interaction was markedly reduced. These results suggest potential dual roles of NHE7 in intracellular compartments and subdomains of cell-surface membranes.
Introduction
Since the initial discovery of SLC9A family Na+/H+ exchanger (NHE) as a cellular pH and cell volume regulator on the plasma membrane Citation[1], nine isoforms sharing similar amino acid sequences have been identified in mammals Citation[2]. The whole family of proteins was commonly recognized as Na+-H+ antiporters on the plasma membrane driven by the concentration gradients of these cations. Therefore, it was an unexpected discovery that NHE7 predominantly associates with mildly acidic intracellular compartments such as the trans-Golgi network (TGN), and triggers both Na+ and K+ fluxes into intracellular compartments in a pH-gradient dependent manner Citation[3]. These observations led to a model where NHE7 is responsible for fine-tuning the acidic luminal pH and cation homeostasis in the TGN, a key organelle for vesicular trafficking. Indeed, NHE7 is evolutionarily highly conserved among different species, and yeast Saccharomyces cerevisiae mutants lacking the functional NHE7-orthologue exhibited defective vesicular trafficking Citation[4].
Very few if any proteins work in isolation, therefore identification and characterization of binding partners for a protein of interest often provides insights into novel functions of the protein in question. We previously found that Secretory Carrier Membrane Proteins (SCAMPs) bind directly to NHE7 to control the targeting between recycling endosomes and the trans-Golgi network (TGN) Citation[5]. More recently, we reported caveolins as novel NHE7-binding proteins and proposed a model in which caveolins provide a platform for NHE7-containing protein complex on the cell surface Citation[6]. In the present study, we identified novel NHE7-binding proteins by co-immunoprecipitation and mass spectrometry. Among the candidates, calcium-calmodulin binds to NHE7 and regulates organellar transporter activity. We demonstrated that NHE7 relocalizes to focal complexes in migrating human breast cancer MDA-MB-231 cells, and actin and a type III intermediate filament protein vimentin associate with endogenous NHE7 in focal complexes. CD44, a type 1 transmembrane protein, associated with NHE7 predominantly on lipid rafts, and the CD44-NHE7 interaction was facilitated by phorbol ester. The NHE7 interactome profile identified in this study suggests that NHE7 may function in both intracellular organelles and on subdomains of cell surface membranes.
Materials and methods
Materials
All salts were of analytical grade or better and were obtained from Sigma-Aldrich (St Louis, MO, USA) unless otherwise indicated. All solvents were of HPLC grade and were obtained from ThermoFisher Scientific (Hampton, NH, USA). The following materials were obtained as indicated: Endopeptidase LysC – Wako Chemicals (Osaka, Japan), porcine modified trypsin – Promega (Nepean, ON), dithiothreitol and iodoacetamide – Sigma-Aldrich, loose ReproSil-Pur 120 C18-AQ 3 µm – Dr Maisch (Ammerbuch-Entringen, Germany), 96-well full skirt PCR plates – Axygen (Union City, CA), fused silica capillary tubing – Polymicro (Phoenix, AZ, USA). The purified mouse monoclonal antibody rho 1D4 was obtained from the National Cell Culture Center (Minneapolis, MN). The peptide corresponding to the 1D4 epitope sequence (Thr-Glu-Thr-Ser-Gln-Val-Ala-Pro-Ala) was synthesized in the UBC Nucleic Acid and Protein Service Unit (The University of British Columbia, Canada). Anti-actin (A2066), vimentin (Vim13.2, V5255), and GAP43 antibodies (GAP-7B10, G9264), and Calmodulin-Agarose (P4385) were obtained from Sigma-Aldrich. Anti-flotillin and clathrin heavy chain antibodies were purchased from BD Transduction Laboratories. An antibody to human TGN46 raised in sheep was obtained from Serotec (AHP500G). Rabbit polyclonal anti-SCAMP and cavolin antibodies were purchased from Affinity BioReagents. Anti-CD44 antibody DF1485 was obtained from a commercial source (sc-7297, Santa Cruz). Anti-CD44 antibodies Hermis 3 and Alexa-488 conjugated IM7 were kind gifts from Dr Pauline Johnson (The University of British Columbia). HRP-conjugated goat anti-mouse and anti-rabbit antibodies were purchased from Jackson Immuno Research Laboratories (West Grove, PA, USA). Alexa-568-conjugated and Alexa-488-conjugated goat antibodies, and Alexa-488 and Alexa-586 conjugated phalloidin were obtained from Molecular Probes/Invitrogen. Calmodulin inhibitor drugs N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide (W7), N-(4-aminobutyl)-5-chloro-2-naphthalenesulfonamide (W13) and trifluoperazine (TFP) were purchased from Calbiochem. Tissue culture media, serum and other reagents were purchased from Gibco/Invitrogen.
Cell culture
Human breast cancer MDA-MB-231 cells were maintained in DME/F12 media containing 5% FBS at 37°C with 5% CO2. PC12 cells were cultured in RPMI media containing 5% FBS. NHE7 tagged with 1D4 at the extreme C-terminus (NHE71D4) was transfected into MDA-MB-231 cells by standard calcium phosphate method. Single clones expressing NHE71D4 were selected by addition of 800 µg/ml G418, isolated and screened by Western blot using 1D4 antibody as described previously Citation[5]. The isolated clones stably expressing NHE71D4 (MDA-MB-231/NHE71D4 cells) were maintained in DME/F12 media plus 5% FBS containing 400 µg/ml of G418.
Liquid chromatography/tandem mass spectrometry
MDA-MB-231/NHE71D4 or MDA-MB-231 were lysed with 10 mM CHAPS/PBS plus protease inhibitor cocktail and clarified by centrifugation at 16,000 relative centrifugal force (r.c.f.) for 15 min at 4°C twice. Lysates were then incubated with 1D4 antibody conjugated sepharose beads for 20 min while rotating at 4°C. After extensive washing, NHE7 immune complexes were eluted from the Sepharose beads by 0.2 mg/ml 1D4 peptide plus 10 mM CHAPS in PBS twice for 10 min and digested in solution as described Citation[7]. After acidification of peptides with 1% trifluoroacetic acid, 0.5% acetic acid and 3% acetonitrile (Sample Buffer), the entire digest was purified and concentrated on Stop and Go Extraction-tips Citation[8], eluted in 80% acetonitrile, 0.5% acetic acid, dried in a vacuum concentrator (Eppendorf) and resuspended in Sample Buffer. Digested immune complexes were analyzed using an LTQ-Orbitrap hybrid mass spectrometer (ThermoFisher Scientific, Bremen, Germany) on-line coupled to a 1100 Series nanoflow HPLCs (Agilent, Mississauga, ON, Canada) using a nanospray ionization source (Proxeon Biosystems, Odense, Denmark) as described Citation[9]. Centroided fragment peak lists were processed to Mascot generic format using Extract_MSN.exe (ThermoFisher Scientific). Monoisotopic peak and charge state assignments were checked and corrected using DTA Supercharge, part of the MSQuant suite of open source software (http://msquant.sourceforge.net). Using Mascot v2.1 (www.matrixscience.com), peak lists were searched against the human IPI database (v3.18, 60090 sequences with bovine serum albumin, mouse immunoglobulins, trypsin and LysC sequences included). The following criteria were used in the Mascot search: peptide tolerance at 4 parts-per-million, fragment ion tolerance at 0.6 Da, ESI-TRAP scoring scheme, trypsin cleavage specificity with up to one missed cleavage. MSQuant v1.4.0a17 (http://msquant.sourceforge.net) was used for parsing Mascot files and iterative mass recalibration.
Production of anti-NHE7 antibody
Glutathione-S-transferase (GST) fused to the C-terminal tail of NHE7 corresponding to the amino acids 525-725 (GST-NHE7 [525-725]) was expressed in Escherichia coli BL21 and purified by affinity purification with glutathione-Sepharose (GE HealthCare) as described previously Citation[6]. The GST fusion protein of NHE7 was conjugated with Ribi Adjuvant System (Corixa Corporation, Hamilton, MT) and injected into New Zealand female rabbits four times at six-week intervals. Two weeks after each immunization, a small-scale test bleed was conducted and the titer of the antiserum was monitored by western blot. Anti-NHE7 antibody was immuno-affinity purified from serum using the maltose binding protein (MBP) fusion protein of the C-terminal tail of NHE7 [525-725] as described previously Citation[10]. For some experiments, 1 µg of anti-NHE7 antibody was pre-absorbed with 40 µg GST-NHE7 [525-725] fusion protein or MBP-NHE7 [525-725] fusion proteins at 4°C for 4 h with rotation.
Co-immunoprecipitation
MDA-MB-231/NHE71D4 cells or MDA-MB-231 cells were lysed by incubation in the cell lysis buffer (0.5% CHAPS plus protease inhibitor cocktail in PBS) for 20 min on ice. Cellular debris was removed by centrifugation at 16,000 r.c.f. for 15 min and 400 µg of lysate was incubated with 2 µg/ml of rabbit polyclonal anti-NHE7 antibody or preimmune serum for 4 h at 4°C. The lysate was then incubated with pre-washed protein A-conjugated Sepharose beads (GE HealthCare) overnight at 4°C with rotation. In some experiments, cell lysates were incubated with 1D4 antibody conjugated Sepharose beads. After 5 rinses with the pre-chilled cell lysis buffer and two washes with 10 min rotation each, the immobilized immune complex was eluted in the SDS sample buffer, resolved in SDS-PAGE and co-eluted NHE7-binding proteins were detected in western blot.
Immunofluorescence microscopy
MDA-MB-231/NHE71D4 cells or MDA-MB-231 cells were grown on collagen pre-coated glass coverslips, fixed with 4% paraformaldehyde/PBS (pH 7.4) for 5 min, quickly rinsed with PBS and the plasma membrane was permeabilized with 0.1% Triton X-100/PBS for 5 min. After blocking with 2% normal goat serum/PBS for 20 min, samples were incubated with primary antibodies. After washing with 0.01% Triton X-100/PBS, the samples were incubated with the Alexa-488 conjugated goat anti-rabbit and Alexa-562 conjugated goat anti-rabbit antibodies for 45 min. To concomitantly visualize NHE7 and CD44, Alexa-562 conjugated goat anti-rabbit antibody was used to visualize NHE7 in combination with rat anti-CD44 IM7 directly conjugated with Alexa-488 (a kind gift from Dr Pauline Johnson, UBC). Fluorescently labeled phalloidin was used to visualize the actin cytoskeleton. After extensive washing, the coverslips were mounted on a slide glass and analyzed by confocal microscopy. To induce cell migration, MDA-MB-231 cells were grown to confluence on glass coverslips and wounding was introduced by scratching the cell monolayer with a pipette tip. Cells were fixed after a 12 h-incubation and migration, and analyzed by immunofluorescence microscopy.
Calmodulin pull-down assays
Cell lysates were prepared from MDA-MB-231/NHE71D4 cells as described above. Rat brain was weighed, minced and homogenized with a Dounce homogenizer in a 4 volumes (vol/wt) of sonication buffer containing 250 mM sucrose, 20 mM HEPES pH 7.4 and protease inhibitor cocktail. The homogenate was then clarified by centrifugation at 800 r.c.f. for 15 min at 4°C and the supernatant was incubated with the same volume of 1% CHAPS in PBS plus protease inhibitor cocktail on ice for 20 min with rotation. Cell lysates were then centrifuged at 16,000 r.c.f. for 15 min at 4°C to eliminate cell debris, and the supernatant was kept at −80°C in small aliquots. Five hundred µg of MDA-MB-231 lysates or rat brain lysates were diluted five-fold with PBS plus protease inhibitor cocktail and split into two equal aliquots. Lysates were diluted with pre-chilled PBS plus protease inhibitor cocktail to adjust the CHAPS concentration to 0.1%, and either CaCl2 or EGTA was added to the each sample to a final concentration of 2 mM, representing Ca2 + (+) and Ca2 + (−) samples respectively. Cell lysates were then incubated with 20 µl bed-volume of pre-washed calmodulin-agarose beads in the presence (Ca2 + (+) samples) or absence (Ca2 + (−) samples) of Ca2 + . After extensive washing in the presence (Ca2 + (+) samples) or absence of CaCl2 (Ca2 + (−) samples), bound proteins were eluted with 20 µl of SDS-sample buffer, 7 µl of each sample was resolved in SDS-PAGE, and co-eluted proteins were detected in western blot. To determine whether the putative calmodulin binding site 195AGYSLKKRHFFRNLGSILA213 predicted by the calmodulin target database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/home.html) binds to calmodulin, sense and antisense oligonucleotides 5′-GATCCGGGTATTCCCTGAAAAAACGCCATTTCTTTCGCAACCTGGGGTCCATTCTGGCATAAGCTTG-3′ and 5′-AATTCAAGCTTATGCCAGAATGGACCCCAGGTTGCGAAAGAAATGGCGTTTTTTCAGGGAATACCCG-3′ were annealed and ligated into the BamHI and EcoRI sites of pGEX2T vector to generate the GST fusion construct. 0.5 µg of GST or the GST fusion protein expressed in E. coli BL21 strain was incubated with calmodulin-agarose beads and bound recombinant protein was analyzed by western blot using anti-GST antibody (StressGen).
Organellar 86RbCl influx assays
Organellar 86RbCl influx assays were conducted as previously described Citation[3]. In brief, PC12 cells stably expressing NHE71D4 (PC12/NHE71D4) were permeabilized with 50 µg/ml of saponin in K+-rich buffer (140 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 2 mM EGTA, 2 mM Mg2 + -ATP, and 20 mM HEPES pH = 7.2) at 23°C for 5 min. Cells were extensively washed with choline chloride buffer (140 mM choline chloride, 2 mM CaCl2, 1 mM MgCl2, 2 mM EGTA, 2 mM Mg2 + -ATP, and 20 mM HEPES pH = 7.8) and treated with 20 mM NH4Cl in choline chloride buffer for 5 min followed by a rapid washout to acutely acidify the organelles. 86RbCl influx measurement was initiated by adding 5 µCi/ml 86RbCl to the permeabilized cells. After 5-min incubation, the reaction was stopped by extensive washing with pre-chilled K+-rich buffer. In some experiments, inhibitor drugs were included in the 86RbCl influx buffer. The cells were then solubilized and liquid scintillation counting was used to assess the 86RbCl in the organelle preparation. The NHE7 activity was expressed as quinine-inhibitable 86RbCl uptake.
Flotation assay
MDA-MB-231/NHE71D4 cells were seeded to 50–80% confluency and serum-starved overnight. Cells were then treated or left untreated with 50 nM PMA in serum-free DMEM/F12 media for 4 h. Flotation assay was conducted as described previously Citation[6]. In brief, cells were broken by shearing through a 26 gauge needle on ice and homogenates were incubated with 0.5% saponin for 30 min on ice to disrupt lipid rafts for some experiments. The homogenate was solubilized with 1% Brij 58, precleaned, placed at the bottom of a 13×51 mm centrifuge tube (Beckman), and overlayed with 2.4 ml 30% and 0.8 ml 5% sucrose/1×MBS. The gradient was centrifuged at 170,000 r.c.f. in a SW50.1 rotor (Beckman) for 17 h at 4°C. Twelve fractions were isolated from the top of the tube.
Results
Identification of candidates for NHE7-binding proteins
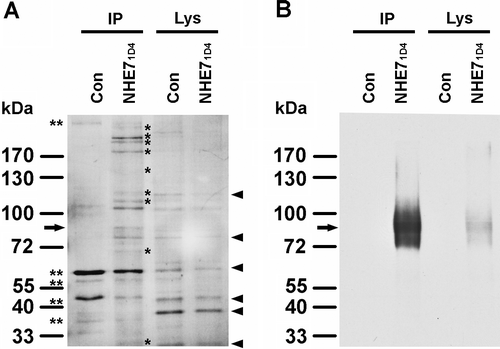
NHE7 is a unique (Na+, K+)/H+ exchanger that is expressed ubiquitously in most cell types, with the highest mRNA expression observed in secretory organs and brain Citation[3]. Our previous yeast two-hybrid screening of a human embryonic brain cDNA library identified SCAMPs Citation[5]. The goal of the current study was to identify physiologically-relevant direct and indirect NHE7-binding proteins in non-secretory cells by affinity capture. To this end, we first established a human breast cancer MDA-MB-231 cell line stably expressing 1D4-tagged NHE7 (MDA-MB-231/NHE71D4). Cell lysates from MDA-MB-231/NHE71D4 or control MDA-MB-231 cells were incubated with 1D4 antibody conjugated Sepharose beads to immunoprecipitate 1D4-tagged NHE7 protein. The eluted immune complex was resolved in SDS-PAGE and visualized with Coomassie staining (Supplementary 1A; please note: all Supplementary Figures and Supplementary , online version only). A parallel Western blot showed a major band with approximately 80 kDa in size, and fainter bands with slightly higher and lower molecular weights which may represent post-translational modifications, splicing variants and proteolytic cleavage Citation[3] (Supplementary B). Additional bands were detected in the immunoprecipitated sample from MDA-MB-231/NHE71D4 cell lysates (labeled with asterisks, Supplementary A IP), but not from the MDA-MB-231 cell lysate. Lysates taken from the two cell lines exhibited a similar Coomassie staining pattern (Supplementary A Lys). We reasoned that NHE7 forms stable complexes with multiple proteins and that putative NHE7-binding proteins might be detectable by characterizing the NHE7 immune complex under this experimental condition.
Supplementary Table I. Proteins specific to the NHE7 immune complex.
NHE7 immune complexes isolated from MDA-MB-231/NHE71D4 cell lysates were eluted from the Sepharose, digested in solution with trypsin, and analyzed by liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS, see Materials and methods). All proteins identified specifically in NHE7 immune complexes and not in parallel control preparations are listed in Supplementary . The high mass accuracy afforded by the LTQ-Orbitrap, the use of rigorous statistical cut-offs, the requirement for at least two peptide identifications per protein and manual inspection of each spectra for a contiguous y-ion series gave us extremely high confidence in the identity of all proteins in Supplementary (see Materials and methods). As expected from the staining pattern of Supplementary A, NHE7 itself was detected by LC-MS/MS. Several cytoskeletal proteins, cell adhesion molecules, a transporter/pump, and signaling molecules were also identified and are explored below.
Calcium-calmodulin binds to NHE7 and regulates the ion transporter activity
Calmodulin, a ubiquitous calcium-binding protein that regulates a number of cellular processes, was placed in the NHE7 interactome in our initial experiments (Supplementary ). Calmodulin (CaM) is a Ca2 + -binding protein that conveys signaling by interacting with a diverse range of proteins in two conformational forms, Ca2 + -bound CaM (Ca2 + -CaM) and Ca2 + -free CaM (ApoCaM) Citation[11]. It is well recognized that calmodulin directly binds to and up-regulates NHE1 Citation[12], but little is known about calmodulin's ability to regulate other NHE isoforms. To examine whether calmodulin binds to NHE7, we used a reciprocal pull-down approach with calmodulin as the bait. Lysates obtained from MDA-MB-231/NHE71D4 cells were incubated with calmodulin-agarose in the absence or presence of Ca2 + , and bound NHE71D4 was detected in western blot (PD). When MDA-MB-231/NHE71D4 cell lysates were incubated with calmodulin-agarose in the presence of Ca2 + , NHE7 was readily detectable after elution (A), whereas NHE7 was barely detectable in the absence of Ca2 + . In contrast, GAP43, also known as neuromodulin, was found to interact with calmodulin-agarose only when the lysate was incubated in the absence of Ca2 + as described previously Citation[13]. Although the CaM-binding motifs found in NHE1 Citation[14] are not present in NHE7, the calmodulin target database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/home.html) predicted a putative calmodulin binding site 195AGYSLKKRHFFRNLGSILA213 for NHE7. When incubated with calmodulin-agarose, this peptide fused with glutathione-S-transferase (GST), but not GST alone, exhibited binding in the presence of Ca2 + (B), suggesting that 195AGYSLKKRHFFRNLGSILA213 of NHE7 is at least partly responsible for CaM binding.
Figure 1. Ca2 + -CaM binds to NHE7 and regulates the transporter activity. (A) MDA-MB-231/NHE71D4 cell lysates were incubated with CaM-agarose in the presence (Ca2 + +) or absence (Ca2 + −) of CaCl2, bound NHE7 was resolved in SDS-PAGE and analyzed in Western blot by 1D4 antibody (PD). To define GAP43 binding to CaM, rat brain lysates were incubated with CaM-agarose and bound GAP43 was detected with anti-GAP43 antibody. 5% of each lysate was loaded as control (Lys). (B) GST alone or GST fused with NHE7 195AGYSLKKRHFFRNLGSILA213 was incubated with CaM-agarose beads in the presence (Ca2 + +) or absence (Ca2 + −) of CaCl2 and bound NHE7 was analyzed by Western blot using anti-GST antibody. (C–E) Permeabilized PC12/NHE71D4 cells were acutely acidified and then incubated with 86RbCl tracer in the absence or presence of inhibitor drugs as described in Materials and methods. Napthalenesulfonamide derivatives W7 and W13, and trifluoperazine (TFP) were used to inhibit Ca2 + -CaM. NHE7 activity was defined as quinine-inhibitable 86RbCl influx to intracellular compartments, and expressed as the mean percentage activity to the control +/− standard deviations of quadruplicate samples.
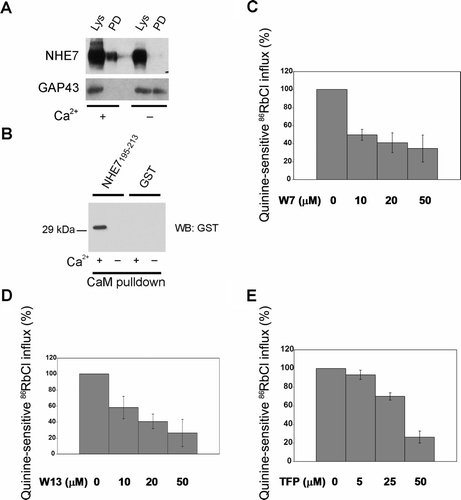
To explore the functional significance of calmodulin-NHE7 interaction, we next investigated the effect of different calmodulin inhibitors on NHE7 activity by using an organellar cation tracer influx assay, which was previously used to determine NHE7 activity Citation[3]. W7, W13 and trifluoperazine (TFP) were used as CaM inhibitors. While W7 and W13 are napthalenesulfonamide derivatives and share similar structures, TFP has chemically distinct structures from W7 and W13 Citation[15]. Although both napthalenesulfonamide derivatives (e.g., W7 and W13) and TFP bind to Ca2 + -CaM, binding-stoichiometry as well as their binding sites to CaM were reported to be different Citation[16–19]. PC12/NHE71D4 cells were permeabilized under mild conditions, and treated with NH4Cl followed by a rapid washout to acutely acidify the organellar compartments. Cells were then incubated with 86RbCl in the absence or presence of CaM inhibitors, or a NHE7 inhibitor quinine, and the quinine-inhibitable 86RbCl influxes into the acidified organelles driven by the pH-gradient were defined as NHE7 activity. 86RbCl instead of 22NaCl was used because NHE7 has a higher affinity for K+ and Rb+ than for Na+ Citation[3]. When permeabilized cells were incubated with 86RbCl in the presence of increasing concentrations of W7 (C), W13 (D), or TFP (E), NHE7 activity was suppressed in a dose-dependent manner. These results suggest that Ca2 + -CaM binds to NHE7 and regulates its transporter activity.
Characterization of the NHE7 antibody
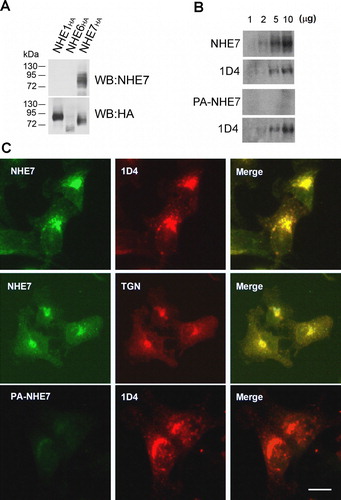
In order to validate the NHE7 interactome we required a reagent to detect native NHE7 protein. Thus, we immunized rabbits with a recombinant cytosolic C-terminal extension of human NHE7, and the antiserum was affinity-purified. To test the specificity of the affinity-purified antibody, Chinese hamster ovary (CHO) cells were transiently transfected with HA-tagged NHE1, NHE6 or NHE7 (NHE1HA, NHE6HA and NHE7HA) and the cell lysates were analyzed by Western blotting using the affinity-purified anti-NHE7 antibody. Endogenous NHE7 was undetectable in CHO cells probably because of the limited endogenous NHE7 expression and/or weak cross-species reactivity of this antibody with hamster NHE7. As shown in Supplementary A, only transfected NHE7HA yielded western blot signal, suggesting that this antibody specifically recognizes NHE7, but not NHE1 or NHE6. To further show the specificity of the antibody, the anti-NHE7 antibody was pre-incubated with or without recombinant NHE7, and their immunological reactivity was determined. When the blot was probed with the anti-NHE7 antibody, a predominant ∼80 kDa band was detected. An additional faint band slightly smaller than 80 kDa was also observed, which may represent endogenously expressed untagged NHE7 or a proteolytically cleaved product (NHE7, Supplementary B). When anti-NHE7 antibody was preabsorbed with the recombinant C-terminal extension of NHE7, no visible signal was observed (PA-NHE7), while re-probing this same blot with 1D4 antibody produced the typical NHE7 signal. Pre-absorbing anti-NHE7 with the immunogen prior to immunoprecipitation significantly reduced the amount of NHE7 in the immune complex (data not shown), further supporting the specificity of the antibody. Anti-NHE7 antibody was also used to label fixed MDA-MB-231/NHE71D4 cells grown on glass coverslips to test the efficiency of the antibody in immunocytochemistry. A predominant juxtanuclear accumulation was observed by immunofluorescence microscopy consistent with the previous findings (Citation[3], Citation[5] and Supplementary C, top panels). As previously reported Citation[3], Citation[5], Citation[6], NHE7 (green) co-localized with a TGN marker TGN46 (red). By double labeling with 1D4 antibody, NHE7 visualized with anti-NHE7 antibody (green) exhibited overlap with the 1D4 staining (red). In contrast, preabsorbed NHE7 antibody did not give any significant immunofluorescence signal (Supplementary C, bottom panels).
NHE7 associates with actin and vimentin in focal adhesion complexes
Actin either directly or indirectly binds to epithelial ion channels Citation[20] and transporters such as NHE3 Citation[21], Citation[22] and NHE5 Citation[23], and regulates their activity and intracellular targeting. In addition, ubiquitous NHE1 isoform may regulate the actin cytoskeleton Citation[24–26]. To verify the actin-NHE7 interaction identified by immuno-affinity trap and mass spectrometry, co-immunoprecipitations were conducted in MDA-MB-231/NHE71D4 cells. Actin was readily detectable by western blot in the NHE7 immune complex, confirming the mass spectrometric results (Supplementary 3). In reciprocal experiments, NHE7 was detectable in the actin immune-complex. We next attempted to confirm a native interaction between NHE7 and actin in untransfected MDA-MB-231 cells. However, due to high levels of background, most probably caused by IgG heavy chain, we were unable to assess their endogenous interaction.
Actin and intermediate filaments are structurally and functionally highly interdependent Citation[27], and the direct interaction between actin and a type III intermediate filament protein vimentin was recently shown Citation[28]. Vimentin is expressed in MDA-MB-231/NHE71D4 and MDA-MB-231 cells, and was readily detectable in the NHE7 immune complex (Supplementary B and 3C). When vimentin was immunoprecipitated from the lysate in reciprocal experiments, NHE7 was detected in the immune complex (Supplementary B). With the newly characterized anti-NHE7 antibody, we then asked whether the protein interactions reported above are detectable in native MDA-MB-231 cells. Vimentin was detected in the NHE7-immune complex by Western blot, but not in a control immune complex (Supplementary C). These results suggest that vimentin and actin compose a physiological complex with NHE7 in the cell.
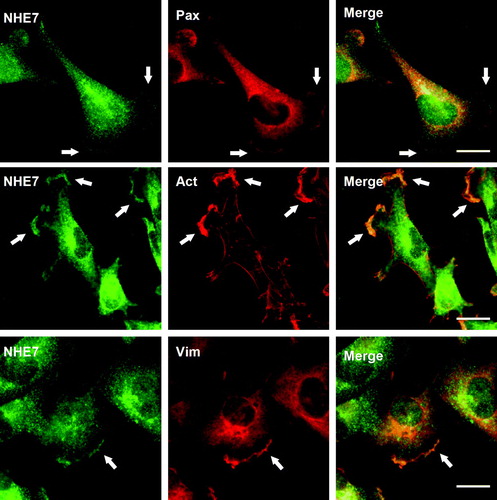
NHE1 has been suggested to associate with focal complexes and may regulate cell adhesion and migration Citation[29–31]. More recently, NHE5 that largely associates with recycling endosomes at steady state was shown to form a protein complex with RACK1 and integrins at the focal complex during cell spreading Citation[32]. Thus, multiple NHE-isoforms, despite their different intracellular localizations at steady state, appear to associate with focal complexes under certain circumstances. We next asked whether endogenous NHE7 also associates with focal complexes in migrating cells. Wounding was introduced in an MDA-MB-231 cell monolayer to induce cell migration. Migrating MDA-MB-231 cells were fixed and double-labeled with anti-NHE7 and anti-paxillin antibody as a focal complex marker and the intracellular localization was observed by immunofluorescence microscopy. In addition to the juxtanuclear signal, a significant population of NHE7 was now readily detectable in plasma membrane protrusions and coincided with the focal adhesion marker paxillin immunostaining as well as the actin cytoskeleton visualized by phalloidin staining (). Vimentin directly interacts with the integrin α2/β1 dimer in focal complexes of endothelial cells Citation[33], and regulates the focal complex structure, cell-matrix adhesion Citation[34] and possibly cell migration Citation[35–37]. Therefore, to test the hypothesis that vimentin colocalizes with NHE7 in focal complexes, their intracellular localization in migrating MDA-MB-231 cells was analyzed by double-labeled immunofluorescence microscopy. Immunofluorescence signals corresponding to NHE7 (green) and vimentin (red) showed most extensive co-localization in the plasma membrane protrusion in cell peripheries, whereas no appreciable co-localization was observed in intracellular compartments ().
Figure 2. NHE7 colocalizes with actin and vimentin in focal complexes of migrating MDA-MB-231 cells. Migrating MDA-MB-231 cells grown on glass coverslips were fixed and intracellular localization of NHE7, paxillin (Pax), and vimentin (Vim) were analyzed by double labeled immunofluorescence microscopy. NHE7 was visualized with Alexa 488-conjugated goat anti-rabbit antibody, and paxillin and vimentin were visualized with Alexa 568-conjugated goat anti-mouse antibody. Alexa 568 conjugated phalloidin was used to detect actin (Act). Arrows indicate co-localization signal of the plasma membrane. All the experiments were repeated three times and one set of representative results was shown. Bars, 10 µm.
The CD44-NHE7 interaction is facilitated by PMA-treatment
CD44 was one of the eleven proteins identified in the NHE7 interactome (Supplementary ). CD44 is a transmembrane receptor for extracellular ligands such as hyaluronan Citation[38], Citation[39], as well as a co-receptor for the c-Met receptor tyrosine kinase Citation[40]. To define the CD44-NHE7 interaction, we immunoprecipitated CD44 protein from MDA-MB-231 cells and bound NHE7 was detected in western blot. Our initial experiments yielded only a weak NHE7 signal in the immunoprecipitated CD44 immune complex. We therefore suspected that the NHE7-CD44 interaction might be weak at steady state, and that this interaction might be regulated. It was previously reported that CD44 is phosphorylated by protein kinase C (PKC) Citation[41], Citation[42], a key regulator of different NHE-isoforms Citation[2], and NHE7 contains several PKC consensus sequences in its cytosolic domain. To test the possibility that the CD44-NHE7 binding is regulated by PKC, MDA-MB-231 cells were treated with the phorbol ester PMA for 0, 30, 120 and 360 min, which did not alter NHE7 expression over the times tested (A, Lys). Immunoprecipitation of CD44 from lysates of PMA-treated cells revealed that the CD44-NHE7 interaction is enhanced by PMA challenge in a time dependent manner. After normalizing to CD44 intensity, which varied by up to 30%, PMA-treatment for 30, 120 and 360 min increased co-immunoprecipitated NHE7. In support of this, PMA-treatment triggered a partial redistribution of NHE7 to the plasma membrane, leading to increased co-localization with CD44 (B).
Figure 3. The NHE7-CD44 interaction is facilitated by PMA-treatment. (A) MDA-MB-231 cells were serum starved and treated with PMA for 0–360 min. Cell lysates were isolated from each time course, immunoprecipitated with pre-immune serum (IP [Con]) or anti-CD44 antibody (IP [CD44]), and co-immunoprecipitated NHE7 was detected in Western blot by anti-NHE7 antibody. The same blot was re-probed with anti-CD44 antibody. Five percent of lysate was loaded from each sample (Lys). The signal intensity for co-immunoprecipitated NHE7 measured by densitometry was normalized to the corresponding CD44 intensity, and the mean values and standard deviations of three independent experiments were shown. A representative result of three independent experiments was shown. (B) Serum-starved MDA-MB-231 cells grown on glass coverslips were treated with or without 50 nM PMA for 120 min. Intracellular localization of CD44 (green) was visualized with Alexa 488-conjugated anti-CD44, and NHE7 (red) was visualized with Alexa 568-conjugated anti-rabbit secondary antibody. Arrows indicate the co-localization of NHE7 and CD44 on the plasma membrane. Bar, 10 µm.
![Figure 3. The NHE7-CD44 interaction is facilitated by PMA-treatment. (A) MDA-MB-231 cells were serum starved and treated with PMA for 0–360 min. Cell lysates were isolated from each time course, immunoprecipitated with pre-immune serum (IP [Con]) or anti-CD44 antibody (IP [CD44]), and co-immunoprecipitated NHE7 was detected in Western blot by anti-NHE7 antibody. The same blot was re-probed with anti-CD44 antibody. Five percent of lysate was loaded from each sample (Lys). The signal intensity for co-immunoprecipitated NHE7 measured by densitometry was normalized to the corresponding CD44 intensity, and the mean values and standard deviations of three independent experiments were shown. A representative result of three independent experiments was shown. (B) Serum-starved MDA-MB-231 cells grown on glass coverslips were treated with or without 50 nM PMA for 120 min. Intracellular localization of CD44 (green) was visualized with Alexa 488-conjugated anti-CD44, and NHE7 (red) was visualized with Alexa 568-conjugated anti-rabbit secondary antibody. Arrows indicate the co-localization of NHE7 and CD44 on the plasma membrane. Bar, 10 µm.](/cms/asset/7f30f646-8216-462d-a646-0239b3e322a7/imbc_a_326471_f0003_b.jpg)
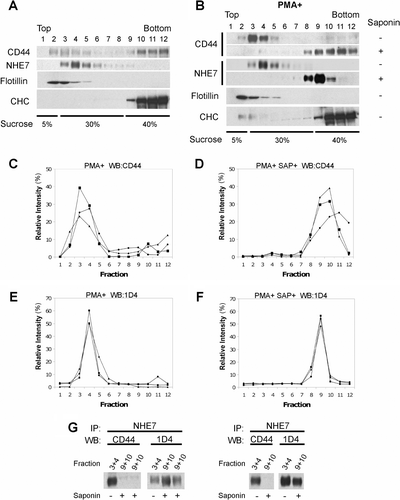
CD44 interacts with NHE7 predominantly in lipid rafts
Lipid rafts are membrane microdomains enriched with cholesterol and sphingolipids, and play a role in compartmentalized signaling. We previously showed that NHE7 is partially associated with lipid rafts in CHO cells and MCF-7 cells Citation[6]. To assess the lipid raft association of CD44 and NHE7 in MDA-MB-231/NHE71D4 cells, lysates were placed at the bottom of a discontinuous sucrose density gradient and subjected to ultracentrifugation. Twelve fractions were collected from the top of the gradient, resolved in SDS-PAGE and the presence of CD44 and NHE7 in each fraction was assessed by western blot. Because of the cholesterol/sphingolipid enriched nature, lipid rafts associate with lower-density (upper) fractions in this assay. As expected, a lipid raft marker flotillin was detected in lower-density (upper) fractions 2–5, and a non-raft marker clathrin heavy chain (CHC) in higher-density (bottom) fractions 9–12 (Supplementary 4A). While NHE7 predominantly associated with lower density (upper) fractions 3–6, CD44 was detected in both upper and lower fractions. When cells were pretreated with PMA, CD44 showed extensive association with lower density (upper) fractions 3–5, while cholesterol solubilization and the resulting lipid raft disruption by saponin shifted the signal to higher density (bottom) fractions 8–12 (Supplementary B–4D). PMA challenge did not affect the association of NHE7 with lower density (upper) fractions 3–6, which was redistributed to fractions 8–10 upon saponin-treatment (Supplementary B, E and F). These results suggest that both NHE7 and CD44 are associated at least partly with lipid rafts in MDA-MB-231/NHE71D4 cells and that PMA facilitates lipid raft targeting of CD44. We next addressed whether the NHE7-CD44 interaction occurs preferentially in lipid rafts. Lysates isolated from PMA-challenged MDA-MB-231/NHE71D4 cells were left untreated or treated with saponin, and lipid raft and non-raft fractions were separated through a sucrose density gradient. Fractions 9 and 10 (non-raft pool) from saponin-treated samples, and fractions 3 and 4 (lipid raft pool) from saponin-untreated samples were collected and subjected to co-immunoprecipitation to test the NHE7-CD44 interaction. CD44 was almost undetectable in the NHE7 complex in the non-raft pool after saponin-treatment, while CD44 was efficiently co-immunoprecipitated with NHE7 in the lipid raft pool without saponin-treatment (Supplementary G). When the same blots were stripped and reprobed with anti-1D4 antibody, similar levels of NHE7 proteins were detectable in all samples. These results suggest that PMA facilitates CD44 targeting to lipid rafts and that CD44 binds NHE7 in this membrane compartment.
SCAMPs and caveolin 1 bind to NHE7 in MDA-MB-231/NHE71D4 cells
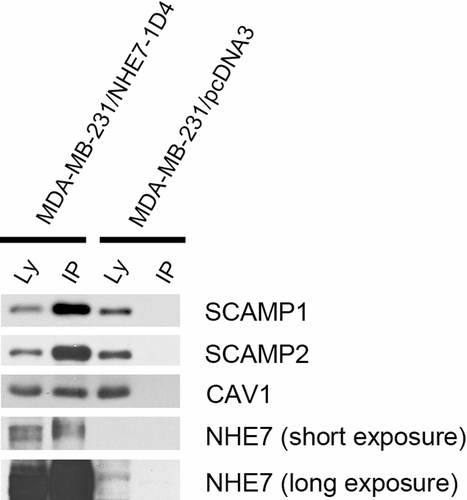
Although we previously identified Secretory Carrier Membrane Proteins (SCAMPs) and caveolin 1 as NHE7 binding proteins, these proteins were not identified by mass spectrometry. To clarify the discrepancies between the present and prior studies, we carried out co-immunoprecipitation experiments. Lysates isolated from MDA-MB-231/NHE71D4 or MDA-MB-231 cells were pre-cleaned, and the supernatant was incubated with 1D4 antibody conjugated Sepharose beads. After extensive washing, the immune complex was eluted out and analyzed in SDS-PAGE and Western blot. As shown in Supplementary 5, SCAMP1, SCAMP2 and caveolin 1 were readily detectable in the NHE7 immune complex.
Discussion and conclusion
SLC9A7 (NHE7) is a unique member of the NHE gene family, exhibiting (Na+, K+)/H+ exchange activity with a distinct pharmacological inhibitor profile. NHE7 predominantly associates with intracellular compartments such as the trans-Golgi network (TGN) and recycling endosomes in resting cells Citation[3], Citation[5]. In the present study, we used immuno-enrichment coupled with LC-MS/MS to define the interactome of NHE7. Among the 11 NHE7-binding proteins, vimentin was detected in NHE7 immune complexes in untransfected MDA-MB-231 cells, whereas myosin heavy chain 9, Na+-K+/ATPase α1 subunit, and plakoglobin (γ-catenin) were more weakly detectable (Supplementary and data not shown). The apparently weak binding suggests that these interactions may be transient or possibly indirect. Pull-down experiments showed that Ca2 + -calmoduin (Ca2 + -CaM) binds to full-length NHE7 expressed in cultured cells as well as to the recombinant GST fusion protein containing the putative CaM-binding domain 195AGYSLKKRHFFRNLGSILA213 predicted by a calmodulin target database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/home.html). NHE7 was readily detectable in CD44 immune complexes by western blot, and this interaction was enhanced by phorbol ester PMA. Four additional proteins were detected by LC-MS/MS in 1D4-immune complexes from NHE71D4-transfected cells but not in those from control MDA-MB-231 cells. These additional four were not confirmed by reciprocal immunoprecipitations or pull-down because of the unavailability of appropriate reagents. However, given their specificity and the positive confirmation of the other seven proteins it is likely that these four are also true components of the NHE7 interactome. Our current LC-MS/MS analysis did not reveal Secretory Carrier Membrane Proteins (SCAMPs) and caveolin, which were previously identified by yeast two-hybrid Citation[5] and co-immunoprecipitation in a heterologous expression system Citation[6], respectively. This is not surprising, because these methods are complementary and would not be expected to overlap completely. Indeed, we were able to detect SCAMPs and caveolin 1 by co-immunoprecipitation in MDA-MB-231/NHE71D4 cells (Supplementary ), suggesting that NHE7 binds to SCAMPs and caveolin 1, but the binding was under the detection limit of mass spectrometry. Since we were unable to detect protein-protein interaction motifs such as a typical type I, type II or type III PDZ binding motif at the C-terminus Citation[43], we used C-terminally tagged NHE7 to facilitate immunological experiments. However, there is still a possibility that the insertion of small artificial sequence interferes with certain protein-protein interaction. Thus, although affinity-trap LC-MS/MS is a powerful approach to define physiological interactome in the cell, it does have limitations.
NHE7 was originally identified as an organelle-membrane type (Na+, K+)/H+ exchanger, and the present study showed that Ca2 + -CaM associates with NHE7 and regulates the organellar ion transporter activity. Interestingly, however, we found that NHE7 changes its intracellular location to the plasma membrane upon cellular stimuli. In migrating cells, for example, a significant fraction of NHE7 redistributed to membrane protrusions and the NHE7 immunostaining overlapped with a focal complex marker paxillin staining. LC-MS/MS identified several plasma membrane proteins (Na+/K+-ATPase α1 subunit, glucose transporter GLUT1 and CD44) in the NHE7 immune complex, further suggesting a possible NHE7-function on the plasma membrane. We found that the majority of NHE7 and a partial CD44 population associate with lipid rafts in serum-starved MDA-MB-231/NHE71D4 cells. PMA-treatment caused a significant redistribution of CD44 to lipid rafts/caveolae fractions, and binding to NHE7 in this membrane fraction. CD44 binds to NHE1 in lipid rafts/caveolae and activates the transporter upon a cognitive ligand hyaluronan-binding Citation[44]. Thus, when cells are stimulated with PMA or hyaluoronan, CD44 may complex with NHE7, caveolins and NHE1 in lipid raft/caveolae. However, our initial attempt to complement the Chinese hamster ovary cell mutants that lack endogenous NHE-activity on the cell surface by heterologous NHE7 expression was unsuccessful. It is possible that physiological action of NHE7 differs from that of NHE1's and elicits a unique ion translocation or that the NHE7 activity might be spatially and temporally restricted. Specific association of NHE7, CD44 and NHE1 on lipid rafts/caveolae might represent novel signal scaffolding complex. Since vimentin Citation[45], Citation[46] and Na+/K+-ATPase Citation[47] have been suggested to associate with rafts/caveolae, these two proteins might also participate in these processes. Future studies will be needed to test these possibilities.
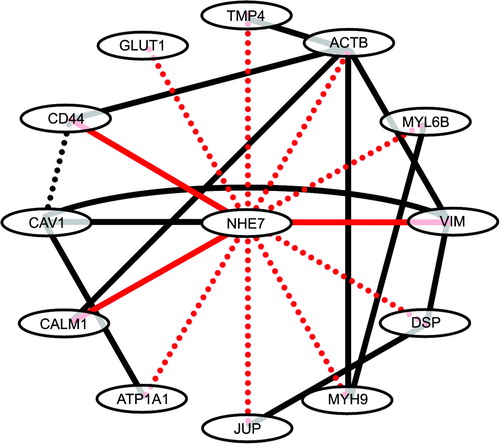
In conclusion, we report here that NHE7 localizes to the leading edge of migrating cells, while juxtanuclear localization predominates in resting cells. Concurrently, some of the newly identified NHE7 binding proteins associate with NHE7 on the cell surface membrane when cell migration is induced. As summarized in , components of NHE7-interactome identified in this study interact with each other and that NHE7 is placed in the center of this web. Our data suggest potential dual functions of NHE7 on intracellular compartments and the cell surface.
Figure 4. The NHE7 interactome. NHE7 binding proteins identified in the present study by affinity capture and MS was highlighted in red dotted lines, where solid red lines indicate binding was further confirmed by co-immunoprecipitation of endogenous proteins or pull-down assays. Black lines indicate previously identified protein-protein interactions. TPM4, tropomyosin 4; ACTB, β-actin; MYL6B, myosin light polypeptide 6; VIM, vimentin; DSP, desmoplakin; MYH9, myosin heavy chain 9; JUP, junction plakoglobin; ATP1A1, Na+/K+-ATPase α1 subunit; CALM1, calmodulin 1; CAV1, caveolin 1; GLUT1, glucose transporter 1.
Supplementary Figure 1. Visualization of co-immunoprecipitated proteins with 1D4-tagged NHE7. (A) One mg of cell lysate protein from MDA-MB-231 (Con) or MDA-MB-231/NHE71D4 (NHE71D4) was incubated with 1D4-antibody conjugated Sepharose beads and half of the eluted samples were resolved by SDS-PAGE and visualized with Coomassie staining. Asterisks indicate the co-eluted proteins that specifically appeared in the immunoprecipitated samples from MDA-MB-231/NHE71D4 cells, but not from the control MDA-MB-231 cells. Some bands appeared in the IP samples of both MDA-MB-231 (Con) and MDA-MB-231/NHE71D4 (NHE71D4), representing non-specific binding proteins (double asterisks). The arrow indicates the ∼80 kD band most probably corresponding to NHE7. Major bands in the lysate represent abundantly expressed proteins (arrowheads). IP, immunoprecipitated samples; Lys, lysates (20 µg). (B) The immunoprecipitated samples (IP) and lysates (Lys) from MDA-MB-231 (Con) and MDA-MB-231/NHE71D4 (NHE71D4) were resolved in SDS-PAGE and analyzed in western blot probed with 1D4 antibody.
Supplementary Figure 2. Characterization of anti-NHE7 antibody. A. HA-tagged NHE1, NHE6 or NHE7 was transiently transfected to CHO cells, and cell lysates from each transfectant was analyzed by Western blot. Anti-NHE7 antibody detected only NHE7HA, but not NHE1HA or NHE6HA, showing the specificity of this antibody (top panel). The same blot was stripped and reprobed with anti-HA antibody (bottom panel). (B) Increasing amounts (1–10 µg) of MDA-MB-231/NHE71D4 cell lysates were resolved in SDS-PAGE, transferred to a PVDF membrane and probed with anti-NHE7 antibody (NHE7, left panel) or pre-absorbed anti-NHE7 antibody (PA-NHE7, right panel) for Western blot. The same blot was stripped and re-probed with 1D4 antibody (bottom panel). (C) MDA-MB-231/NHE71D4 cells grown on glass coverslips were fixed and double stained with either 2 µg/ml anti-NHE7 antibody (NHE7, top panels) or pre-absorbed anti-NHE7 antibody (PA-NHE7, bottom panels) and 1D4 antibody. Anti-TGN46 antibody (20 µg/ml) was used as a TGN marker. NHE7 and 1D4 were visualized with Alexa 488- and Alexa 568-conjugated secondary antibodies respectively. Bars, 10 µm. A set of representative results of three independent experiments was shown.
Supplementary Figure 3. NHE7 associates with actin and vimentin in focal complexes of migrating MDA-MB-231 cells. (A) Top Panel: MDA-MB-231 lysates (IP Con) or MDA-MB-231/NHE71D4 lysates (IP 1D4) were incubated with 1D4 antibody-conjugated Sepharose beads and bound actin was detected in Western blot. Five percent of MDA-MB-231/NHE71D4 lysate (Lys) was used as a loading control. Bottom Panel: MDA-MB-231/NHE71D4 lysates were immunoprecipitated with preimmune serum (Con) or anti-actin antibody (Act) and bound NHE7 was probed with 1D4 antibody. Five percent of lysate (Lys) was used as a loading control. (B) Top Panel: MDA-MB-231/NHE71D4 lysates were immunoprecipitatd with preimmune serum (IP Con) or anti-NHE7 antibody (IP NHE7) and bound vimentin was analyzed by western blot. Five percent of lysate (Lys) was used as a loading control. Bottom Panel: MDA-MB-231/NHE71D4 lysates were immunoprecipitated with preimmune serum (Con) or anti-vimentin antibody (Vim), and bound NHE7 was probed with anti-NHE7 antibody. Five percent of lysate was used as a loading control (Lys). (C) Untransfected MDA-MB-231 cell lysates were immunoprecipitated with preimmune serum (IP Con) or anti-NHE7 antibody (IP NHE7) and bound vimetin was detected. Five percent of lysates (Lys) was used as a loading control.

Supplementary Figure 4. CD44 binds to NHE7 preferentially in lipid rafts. (A) MDA-MB-231/NHE71D4 cell lysates were placed at the bottom of a centrifugation tube and subjected to flotation assays. Twelve fractions were collected from the top of the tube and the presence of CD44, NHE71D4, flotillin and clathrin heavy chain (CHC) in each fraction was detected by western blot. Flotillin and CHC were used as lipid raft and non-raft markers, respectively. A set of representative results of three independent experiments was shown. (B) Serum-starved MDA-MB-231/NHE71D4 cells were stimulated with 50 nM PMA for 4 h and lipid raft association of CD44 and NHE71D4 was detected by Western blot. In some experiments, cell homogenates were incubated with 0.5% saponin on ice to disrupt lipid rafts. A set of representative results of three independent experiments was shown. (C–F) The intensities of CD44 (C and D) and NHE7 (E and F) signal in each fraction was measured by densitometry and the relative intensity was calculated. D and F represent saponin-treated samples. The data for three independent experiments were shown. (G) The fractions 9 and 10 of PMA-treated saponin-treated samples and fractions 3 and 4 of PMA-treated saponin-untreated samples were immunoprecipitated with anti-NHE7 rabbit polyclonal antibody, and bound CD44 was detected by mouse monoclonal anti-CD44 antibody (left panel). The same blot was reprobed with 1D4 mouse monoclonal antibody (right panel). PMA-pretreated saponin-treated non lipid raft pool (fractions 9 + 10) and PMA-pretreated saponin-untreated lipid raft pool (fractions 3 + 4) were analyzed.
Supplementary Figure 5. The same amount (150 µg) of lysates from MDA-MB-231/NHE71D4 cells or MDA-MB-231 cells were incubated with sepharose beads conjugated with 1D4-antibody, and bound SCAMP1, SCAMP2 and caveolin 1 (Cav1) were detected by Western blot. The blot was stripped and reprobed with anti-NHE7 antibody.
Acknowledgements
We thank Dr Pauline Johnson (University of British Columbia) for providing Alexa 488-conjugated IM7.8.1 rat anti-human monoclonal antibody and Dr Roger Brownsey (University of British Columbia) for rat brain. This work was supported by operating grants from Canadian Institutes of Health Research (CIHR) to MN and LJF and establishment grants from the Michael Smith Foundation for Health Research (MSFHR) to MN and LJF. MN and LJF are MSFHR Scholars and LJF is the Canada Research Chair in Organelle Proteomics and a Peter Wall Institute Early Career Scholar Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Pouyssegur J, Sardet C, Franchi A, L'Allemain G, Paris S. A specific mutation abolishing Na + /H+ antiport activity in hamster fibroblasts precludes growth at neutral and acidic pH. Proc Natl Acad Sci USA 1984; 81: 4833–4837
- Orlowski J, Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Arch 2004; 447: 549–565
- Numata M, Orlowski J. Molecular cloning and characterization of a novel (Na+, K+)/H+ exchanger localized to the trans-Golgi network. J Biol Chem 2001; 276: 17387–17394
- Bowers K, Levi BP, Patel FI, Stevens TH. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol Biol Cell 2000; 11: 4277–4294
- Lin PJ, Williams WP, Luu Y, Molday RS, Orlowski J, Numata M. Secretory carrier membrane proteins interact and regulate trafficking of the organellar (Na+, K+)/H+ exchanger NHE7. J Cell Sci 2005; 118: 1885–1897
- Lin PJ, Williams WP, Kobiljski J, Numata M. Caveolins bind to (Na+, K+)/H+ exchanger NHE7 by a novel binding module. Cell Signal 2007; 19: 978–988
- de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 2004; 117: 649–662
- Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 2003; 75: 663–670
- Chan QW, Howes CG, Foster LJ. Quantitative comparison of caste differences in honeybee hemolymph. Mol Cell Proteomics 2006; 5: 2252–2262
- Harlow E, Lane D. 1988. Antibodies: A laboratory manual Cold Spring Harbor Laboratory.
- Jurado LA, Chockalingam PS, Jarrett HW. Apocalmodulin. Physiol Rev 1999; 79: 661–682
- Wakabayashi S, Shigekawa M, Pouyssegur J. Molecular physiology of vertebrate Na+/H+ exchangers. Physiol Rev 1997; 77: 51–74
- Cimler BM, Andreasen TJ, Andreasen KI, Storm DR. P-57 is a neural specific calmodulin-binding protein. J Biol Chem 1985; 260: 10784–10788
- Bertrand B, Wakabayashi S, Ikeda T, Pouyssegur J, Shigekawa M. The Na+/H+ exchanger isoform 1 (NHE1) is a novel member of the calmodulin-binding proteins. Identification and characterization of calmodulin-binding sites. J Biol Chem 1994; 269: 13703–13709
- Matsushima N, Hayashi N, Jinbo Y, Izumi Y. Ca2 + -bound calmodulin forms a compact globular structure on binding four trifluoperazine molecules in solution. Biochem J 2000; 347: 211–215
- Osawa M, Swindells MB, Tanikawa J, Tanaka T, Mase T, Furuya T, Ikura M. Solution structure of calmodulin-W-7 complex: The basis of diversity in molecular recognition. J Mol Biol 1998; 276: 165–176
- Dalgarno DC, Klevit RE, Levine BA, Scott GM, Williams RJ, Gergely J, Grabarek Z, Leavis PC, Grand RJ, Drabikowski W. The nature of the trifluoperazine binding sites on calmodulin and troponin-C. Biochim Biophys Acta 1984; 791: 164–172
- Hidaka H, Yamaki T, Naka M, Tanaka T, Hayashi H, Kobayashi R. Calcium-regulated modulator protein interacting agents inhibit smooth muscle calcium-stimulated protein kinase and ATPase. Mol Pharmacol 1980; 17: 66–72
- Massom L, Lee H, Jarrett HW. Trifluoperazine binding to porcine brain calmodulin and skeletal muscle troponin C. Biochemistry 1990; 29: 671–681
- Mazzochi C, Benos DJ, Smith PR. Interaction of epithelial ion channels with the actin-based cytoskeleton. Am J Physiol Renal Physiol 2006; 291: F1113–1122
- Alexander RT, Furuya W, Szaszi K, O J., Grinstein S. Rho GTPases dictate the mobility of the Na+/H+ exchanger NHE3 in epithelia: Role in apical retention and targeting. Proc Natl Acad Sci USA 2005; 102: 12253–12258
- Cha B, Tse M, Yun C, Kovbasnjuk O, Mohan S, Hubbard A, Arpin M, Donowitz M. The NHE3 juxtamembrane cytoplasmic domain directly binds ezrin: Dual role in NHE3 trafficking and mobility in the brush border. Mol Biol Cell 2006; 17: 2661–2673
- Szaszi K, Paulsen A, Szabo EZ, Numata M, Grinstein S, Orlowski J. Clathrin-mediated endocytosis and recycling of the neuron-specific Na+/H+ exchanger NHE5 isoform. Regulation by phosphatidylinositol 3′-kinase and the actin cytoskeleton. J Biol Chem 2002; 277: 42623–42632
- Lagana A, Vadnais J, Le PU, Nguyen TN, Laprade R, Nabi IR, Noel J. Regulation of the formation of tumor cell pseudopodia by the Na+/H+ exchanger NHE1. J Cell Sci 2000; 113: 3649–3662
- Cardone RA, Casavola V, Reshkin SJ. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat Rev Cancer 2005; 5: 786–795
- Watts III BA, George T, Good DW. The basolateral NHE1 Na+/H+ exchanger regulates transepithelial HCO3- absorption through actin cytoskleton remodeling in renal thick ascending limb. J Biol Chem 2005; 280: 11439–11447
- Helfand BT, Chang L, Goldman RD. Intermediate filaments are dynamic and motile elements of cellular architecture. J Cell Sci 2004; 117: 133–141
- Esue O, Carson AA, Tseng Y, Wirtz D. A direct interaction between actin and vimentin filaments mediated by the tail domain of vimentin. J Biol Chem 2006; 281: 30393–30399
- Schwartz MA, Lechene C, Ingber DE. Insoluble fibronectin activates the Na + /H+ antiporter by clustering and immobilizing integrin alpha 5 beta 1, independent of cell shape. Proc Natl Acad Sci USA 1991; 88: 7849–7853
- Grinstein S, Woodside M, Waddell TK, Downey GP, Orlowski J, Pouyssegur J, Wong DC, Foskett JK. Focal localization of the NHE-1 isoform of the Na + /H+ antiport: Assessment of effects on intracellular pH. Embo J 1993; 12: 5209–5218
- Denker SP, Huang DC, Orlowski J, Furthmayr H, Barber DL. Direct binding of the Na+-H+ exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol Cell 2000; 6: 1425–1436
- Onishi I, Lin PJ, Diering GH, Williams WP, Numata M. RACK1 associates with NHE5 in focal adhesions and positively regulates the transporter activity. Cell Signal 2007; 19: 194–203
- Kreis S, Schonfeld HJ, Melchior C, Steiner B, Kieffer N. The intermediate filament protein vimentin binds specifically to a recombinant integrin alpha2/beta1 cytoplasmic tail complex and co-localizes with native alpha2/beta1 in endothelial cell focal adhesions. Exp Cell Res 2005; 305: 110–121
- Tsuruta D, Jones JC. The vimentin cytoskeleton regulates focal contact size and adhesion of endothelial cells subjected to shear stress. J Cell Sci 2003; 116: 4977–4984
- Eckes B, Dogic D, Colucci-Guyon E, Wang N, Maniotis A, Ingber D, Merckling A, Langa F, Aumailley M, Delouvee A, Koteliansky V, Babinet C, Krieg T. Impaired mechanical stability, migration and contractile capacity in vimentin-deficient fibroblasts. J Cell Sci 1998; 111: 1897–1907
- Nieminen M, Henttinen T, Merinen M, Marttila-Ichihara F, Eriksson JE, Jalkanen S. Vimentin function in lymphocyte adhesion and transcellular migration. Nat Cell Biol 2006; 8: 156–162
- Ivaska J, Vuoriluoto K, Huovinen T, Izawa I, Inagaki M, Parker PJ. PKCepsilon-mediated phosphorylation of vimentin controls integrin recycling and motility. Embo J 2005; 24: 3834–3845
- Liao HX, Lee DM, Levesque MC, Haynes BF. N-terminal and central regions of the human CD44 extracellular domain participate in cell surface hyaluronan binding. J Immunol 1995; 155: 3938–3945
- Yang B, Yang BL, Savani RC, Turley EA. Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. Embo J 1994; 13: 286–296
- Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev 2002; 16: 3074–3086
- Stapleton G, Malliri A, Ozanne BW. Downregulated AP-1 activity is associated with inhibition of Protein-Kinase-C-dependent CD44 and ezrin localisation and upregulation of PKC theta in A431 cells. J Cell Sci 2002; 115: 2713–2724
- Legg JW, Lewis CA, Parsons M, Ng T, Isacke CM. A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat Cell Biol 2002; 4: 399–407
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci 2001; 24: 1–29
- Bourguignon LY, Singleton PA, Diedrich F, Stern R, Gilad E. CD44 interaction with Na+ − H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J Biol Chem 2004; 279: 26991–27007
- Runembert I, Queffeulou G, Federici P, Vrtovsnik F, Colucci-Guyon E, Babinet C, Briand P, Trugnan G, Friedlander G, Terzi F. Vimentin affects localization and activity of sodium-glucose cotransporter SGLT1 in membrane rafts. J Cell Sci 2002; 115: 713–724
- Sprenger RR, Fontijn RD, Marle J, Pannekoek H, Horrevoets AJG. Spatial segregation of transport and signalling functions between human endothelial caveolae and lipid raft proteomes. Biochem J 2006; 400: 401–410
- Wang H, Haas M, Liang M, Cai T, Tian J, Li S, Xie Z. Ouabain assembles signaling cascades through the caveolar Na+/K+-ATPase. J Biol Chem 2004; 279: 17250–17259