Abstract
Membrane Protein Structure Initiative (MPSI) exploits laboratory competencies to work collaboratively and distribute work among the different sites. This is possible as protein structure determination requires a series of steps, starting with target selection, through cloning, expression, purification, crystallization and finally structure determination. Distributed sites create a unique set of challenges for integrating and passing on information on the progress of targets. This role is played by the Protein Information Management System (PIMS), which is a laboratory information management system (LIMS), serving as a hub for MPSI, allowing collaborative structural proteomics to be carried out in a distributed fashion. It holds key information on the progress of cloning, expression, purification and crystallization of proteins. PIMS is employed to track the status of protein targets and to manage constructs, primers, experiments, protocols, sample locations and their detailed histories: thus playing a key role in MPSI data exchange. It also serves as the centre of a federation of interoperable information resources such as local laboratory information systems and international archival resources, like PDB or NCBI. During the challenging task of PIMS integration, within the MPSI, we discovered a number of prerequisites for successful PIMS integration. In this article we share our experiences and provide invaluable insights into the process of LIMS adaptation. This information should be of interest to partners who are thinking about using LIMS as a data centre for their collaborative efforts.
Introduction
A Laboratory information management system (LIMS) for structural biology has been developed for the 12 UK institutions funded by Biotechnology and Biological Sciences Research Council (BBSRC) Structural Proteomics of Rational Targets (SPoRT) initiative Citation[1]. Having such a large number of parties involved made the development and integration of a LIMS system very complex, but at the same time has ensured that the LIMS is developed to cater for a variety of laboratory practices and gives the Protein Information Management System (PIMS) a better chance of being universally accepted within structural proteomics laboratories.
We explore the process of LIMS development and integration into individual laboratory practices using the example of the Membrane Protein Structure Initiative (MPSI), BBSRC funded consortium of eight institutions. This is one of the SPoRT programmes and was established in 2005 to develop high throughput methods for membrane protein structure determination and to determine the structure of a number of integral membrane transporters and ion-channels.
At MPSI, each institution has specific expertise in membrane protein research, and these are shared within the consortium. The various laboratories therefore act as nodes of expertise. This does not imply that they are exclusively performing a task as would be expected of a pipeline process. Instead the nodes are experts in a particular step of the process and share their expertise within the consortium as well as providing access to specialized equipment and materials. For example, Leeds University group act as a node with expertise in high-throughput cloning and large volume fermentation. The University of Sheffield group undertake EM imaging of 2-D crystals and rapid screening of membrane protein solutions for mono-dispersity. Oxford University group is a node exclusively performing modelling simulations on membrane transporters in native membranes.
Although PIMS is generic software which can be used equally well by any structural genomics consortium it has developed its own flavour which reflects the interests of the consortium members. For example it is common that membrane proteins are expressed in cells at low levels (<1 mg/l) and this requires large scale fermentation. It is therefore important to track this process for each target so as to develop an efficient protocol for maximum protein production.
Methods
PIMS is a multilayered multi-component system written in the Java programming language. To run PIMS two server components are required. The first one is the database; the second is a web application server. A single user PIMS version is planned, and it may not require a separate database server, instead a simple database will be supplied with the PIMS distribution. PIMS uses a relational database which allows better manageability and a fuller set of features which are especially important in the long run.
The database access in PIMS is abstracted using Hibernate object relational mapping Citation[2]. Therefore PIMS is largely database independent. The PIMS officially supports two databases one of them is an open source, freely available production level data-base server PostgreSQL version 8 or above Citation[3], another one is a database from the major commercial vendor – Oracle version 10 or higher Citation[4]. MPSI PIMS is run on Postgresql.
Any web application server which comply with Sun servlet 2.4 and JSP 2.0 Citation[5] specifications will be able to run PIMS. MPSI PIMS is run on the freely available open source web application server Tomcat versions 5.5 or 6.0 Citation[6].
Both the web application server and the database server are available for many platforms including but not limited to MS Windows and Linux. The application part of PIMS and the database part can be configured to be run on separate machines. In addition both the database and the web application server support clustering therefore PIMS can be scaled up and used successfully within large organizations with thousands of users.
MPSI PIMS is run on a single machine. MPSI server has 16 Gb of memory, 2 dual core 2.4 Ghz AMD Opteron processors ordinary SATA hard drive and run under OpenSuSe Linux. According to the results of the performance tests this server should be able to serve well up to 500 users given a small size of the database [unpublished work, Troshin PV]. Under the small database we assume the database which contains one thousand targets, twice the amount of experiments, and up to 10000 samples. We do not have experience of running PIMS on large databases (e.g., the hundreds of thousands of experiments).
From the client side nothing apart from a java enabled web browser is required to access PIMS. PIMS can be accessed through any web browser, however some of it most advanced functionality has been verified to work correctly with MS Internet Explorer version 7.0 and Mozilla Firefox version 2 and above. The ‘client’, – a web browser is responsible only for displaying the user interface; none of the application runs on the client side and only the processing relevant for displaying the page is performed there.
At the moment of writing PIMS installation requires a qualified IT technician and is unlikely to be successfully installed by scientists. The work is ongoing to simplify the PIMS installation. The technical part of the configuration, which has to be done, involves things like defining users, groups and their access rights for, updating PIMS. Although not required is beneficial to have a PIMS administrator who is able to perform configuration tasks for the laboratory. This configuration fine turning is a little more scientifically loaded and involves adjusting experiments types (expression, purification etc), sample categories (Buffer, Antibiotic etc), plate types (96-deep well plate, 24-well etc), target statuses (selected, cloned, purified etc). PIMS distribution has all these things defined but due to the fact that the practices are so diverse, some fine turning of the scientific part of the PIMS may be required.
Finally PIMS is updated about four times a year. Once the new version of PIMS application is downloaded, the updating procedure is simplistic and consists of two steps. The first step is to run a database updater which updates the database without human intervention. Secondly the Web Application aRchive (.war) file needs to be replaced and redeployed on the server. All update procedures completed on the server, client computers do not need updating. All specific settings like users, groups, access rights, web addresses, database settings remain the same and will not be changed during the update. At this stage of PIMS development (PIMS version 2.2), we recommend to run the update procedure by a qualified IT technician as it involves working with unfamiliar server applications.
Results and discussion
Structural proteomics efforts are generating a vast amount of data, and more structures are solved each year for example in 2007 nearly 20% more structures were deposited than 2006 Citation[7]. This underpins the need for LIMS that can store information in a structured manner Citation[8]. PIMS Citation[9] is designed to coordinate the efforts of researchers and was funded at the same time as MPSI as a part of a SPoRT initiative to cater for needs of the SPoRT-funded laboratories. Simultaneous funding has allowed the PIMS development to be driven by scientific needs and to benefit from the scientists’ needs very early in the PIMS project. The functionality has expanded, through many revisions, from a simple target tracking facility to advanced functionalities that include the tracking of targets, constructs, primers, experiments, people and sample management facilities, thereby forming a complete framework for capturing information at all stages in high-throughput protein production and structure determination.
In general, a Laboratory Information Management System (LIMS) is software that is used for storing and retrieving the information on targets, experiments, chemicals, samples and other laboratory functions such as invoicing, work flow automation and also holders, boxes and plate's management. MPSI have been contributing to the development of a LIMS targeted specifically for protein production called PIMS. PIMS is loaded with the information on all steps of a structure determination pipeline up to crystal production. That means that PIMS is supplied with many commonly used protocols for cloning, expression, purification, concentration and crystallization preloaded. Due to the variety of techniques used the number of protocols supplied with PIMS is more than 50 and new protocols are constantly introduced. As the PIMS protocols are very flexible it is easy to design new ones for less frequently used techniques. To simplify the data entry PIMS also comes with about a thousand most frequently used chemicals preloaded which are categorized according to their type. Also the navigation system is centred on the targets which, in the case of MPSI, are the gene products under investigation. PIMS also supports research into complexes, and DNA sequences other than ORFs, e.g., promoters. Once the user is logged on the system she/he can immediately see what targets are ‘active’, what samples are ready for the next experiments and what stage of a pipeline each targets is at. Therefore the PIMS, by and large, is an out of the box solution for protein production laboratories, in which the expertise of many scientists has been distilled and implemented.
There are many benefits that LIMS can potentially bring to the laboratory. Among them are:
Helps to keep continuity in the work after a PostDoc leaves.
Improves communication when a technician is doing work for several scientists, especially when combining their targets in a single plate.
Ensures that important information about the provenance of samples is retained.
The lab process can be supervised thus the bottlenecks in workflow can be identified and reacted upon much quicker
Ensures that the regulatory demands are met by keeping the history of records, as to who made the first record when it was made and when it was modified later
External participants can place work requests and review results
It helps in managing high throughput data by (i) providing a centralized data repository accessible from anywhere, (ii) automatically capturing the data, (iii) mining the data, and (iv) avoiding data duplication and expensive human data entry errors
Helps with many housekeeping tasks like location tracking and labelling e.g., barcoding of samples, plates, boxes etc …
In the longer term, since a LIMS records negative results as well as positive, it can enable a review of methods, leading to improvements. Several groups have published improved crystallization screens, by using their records of crystallization experiments Citation[10]. It has not yet been possible to review expression screening in this way. A typical LIMS includes search facilities so that it is much easier to find past records than in laboratory note books.
Despite the number of benefits that LIMS can bring introducing the LIMS can be a daunting experience. Perhaps, the hardest part of it is matching the LIMS to the current work practice. At MPSI we were fortunate that we do not need to do it, as PIMS has been developed to match the MPSI scientist needs. Other example of organizations where LIMS has been successfully introduced seems to prove this point.
Phazer's customized version of LabVantage SAPPHIRE LIMS Citation[11] has been developed specifically to suit Phazers’ scientists established workflow. The LIMS had been developed for several years before the integration work with Phazer was started. The integration and roll out process has taken more than a year for a system which initially was used by about 30 scientists from several Phazer's sites [unpublished work, Brown D., A global crystallography tracking system, 2008, Protein Crystallography, The European Laboratory Robotics Interest Group meeting]. Some of the largest academic efforts have resorted to the LIMS help. For instance US funded Protein Structure Initiative (PSI) has had a LIMS component. However, different PSI sites chose to develop they own custom solutions instead of investing in a centralized LIMS.
The LIMS that does not match current work practices very well is hard to use and is unlikely to be successfully adopted. Furthermore, scientists should be prepared to spare some time defining their data management needs and communicating them to the LIMS administrator before introducing a centralized LIMS system.
For the MPSI PIMS is the centre of a federation of computational resources; there are a number of MPSI-integrated web tools, both local and remote, which allow members of the consortium to post further information related to protein targets. To act in a full compliance with the recent BBSRC's data sharing policy Citation[12], the majority of the information from MPSI PIMS database will be made publicly available after the MPSI grant has finished, providing detailed information on all experiments performed for the target structure determination and therefore forming an invaluable resource to identify the factors that contribute to successful structure determination of membrane proteins. On some targets the work will be carried out beyond the MPSI project and therefore the records relevant to these targets will not be made available immediately after MPSI project finishes. In our experience, using PIMS for the duration of the project is an effective way of ensuring that the experimental data is comprehensive and available in a form that is suitable for sharing.
The key feature of the PIMS is the ability of any registered member of the consortium to add and modify entries via an intuitive web interface which users can use with an ordinary web browser. Thus, PIMS can be accessed by scientists from any laboratory or from home with no need for initial set-up. It is possible to use the software using PCs, laptops, or handheld device accompanied by a barcode reader in any laboratory environment including a laboratory cold room.
Initial repository creation
It is important to introduce, agree and enforce the naming convention for common records within the project. The naming convention helps to reduce the need to get into the LIMS system to look up the details of the record. For instance, the constructs can be prefixed with the vector name and the target name. This eliminates the need to get into the LIMS when the tube with a sample is taken from the freezer. However, special care needs to be taken not to encode into the name information which may change in the future.
At the start of the project the MPSI steering board agreed on the targets name convention. According to the convention MPSI target names should include a two letter university abbreviation as a prefix, so for instance, university of Leeds target number 10 would be UL0010. This was useful before the PIMS became available, but as collaboration progressed and another university has taken a lead on some targets, the name of the target ceased to make sense.
Similar problems can arise for any common, shared data for example protocols, recipes, experiment types etc. The LIMS should support automatic naming of these types of records according to configurable patterns. PIMS does now support this feature for targets and experiments.
Initially MPSI target repository was populated by loading predefined targets from each of MPSI participating nodes. In MPSI some nodes have only a dozen targets while others namely the Leeds University group has their own web site with vital target information defined in hypertext files. Manchester University node defined their targets as Swissprot files with additional comments stored in a spreadsheet. The number of targets in both cases was in the order of several hundreds therefore, custom parsers had to be created for loading data into the structured PIMS database. Scientists from the labs which have only a few targets recorded these in the PIMS manually. To simplify target prioritization and scheduling and to provide some information on possible biological role of the target, all targets were classified according to the TCDB target classification Citation[13] upon loading. Target data uniformity helped to streamline communication within MPSI consortium thanks to the single underlying database.
Define access rights to allow the collaboration
LIMS store scientific results. They are a valuable asset which needs to be protected. Therefore LIMS have to implement some sort of mechanisms which restricts access to the data. PIMS allows fine grained access control rules to be set [unpublished work, Troshin PV, Griffiths SL, Diprose JM, Savitsky M, Niekerk CJ, Pajon A, Daniel EJ, Lin B, Morris C]. It is important that the scientists on the collaborative projects have equal access to the data and collaborative projects have to have key data accessible to everyone within the collaboration. On the other hand, the visibility of the data to external partners are a matter of local policies, and in extreme cases, access to anyone outside the collaboration can be prohibited.
At the same time, we found that some data do not help collaboration, and are better kept private to each of the collaborating nodes. These include scientifically unimportant data, for example chemicals, and the majority of the samples produced. This provides useful data separation and prevents scientists from accidental mistakes like using chemicals such as antibiotic solutions which belongs to the remotely collaborating partner. In the majority of cases, these data clutter the view and hinder access to more important data.
PIMS permits its administrator to define custom access rights for different users and groups. The access rights should be defined in such a way that collaboration is possible. For example it should be possible to perform experiments on the targets defined elsewhere, and use samples produced by the collaborator.
One of benefits of the LIMS, the accessibility of the data for collaborative projects, may also be seen as its greatest weakness. This is due to the fact that scientific results remain the property of the scientist before they are published. Trust among the collaborating partners is a prerequisite for a successful LIMS integration. Whatever the size of a lab, it should be able to look after the part of the LIMS for which it is responsible. Keeping the records in LIMS up to date is important for a collaboration to work. Making sure that recognition is always given to the right person, and never changed, can help with resolving the trust issues.
MPSI access right set-up
As the MPSI data is considered to be private for the consortium members for the period of the project, the access right management had to be in place for the very first PIMS release. The data access requirements were defined as follows: All MPSI scientists have to be able to record/update new data, and read but not change data recorded by the scientists from the collaborating node. Initially no one was allowed to delete the data, but later, this proved to be impractical, and at least one person in each location was granted delete privileges. In particular, much inconvenience was caused by mistakenly recorded data. However, the delete privileges were limited to the deletion of data imported by that institution. This set-up enabled collaboration between different sites and at the same time provided sufficient safeguards for the scientists to feel comfortable recording their precious results into PIMS. It is important to note that the access rights set-up permitted the recording of new constructs for a target defined by the collaborating node, as well as experiments performed on the sample by a collaborator.
Refining the data model
Initially PIMS utilized the ‘protein production model’ which is a logical representation of the processes and entities involved in all stages of the protein structure determination process derived from an earlier project Citation[14]. However, we soon discovered that modification was needed to accommodate new practices or to make useful generalizations. As a result of these efforts the initial data model was significantly modified, and the experiment and the protocol management were streamlined and generalized. The data model has been updated with the help of the special scripts. The PIMS core developer team conducted this work.
In general, our experience has shown that new needs are identified at each stage of the LIMS development. Therefore the data model needs to be easily adjustable. Performance requirements can also demand a data model change.
Data management outside PIMS
To satisfy MPSI data management needs we have used other resources in conjunction with PIMS. Such resources include a static website which has been secured and used to share the information for which it is impractical for PIMS to incorporate. For example an FTP server was used to share EM data from Sheffield, as well as to store the legacy data from Leeds University. The private area of the MPSI web server is open for MPSI members to contribute their data, ranging from the lists of genomic DNA libraries to PowerPoint presentations of results.
All the above are examples of the centralized approach to the data management. MPSI also has made significant use of de-centralized data repositories for publications, functional studies and crystallization data.
Usage statistics
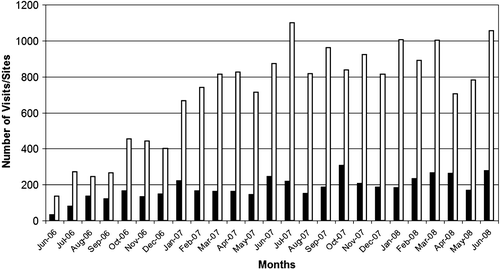
During the time of the project the usage statistics were collected and used among other things to steer the development. Due to the fact that the PIMS project was funded at the same time as MPSI project the usage did not reach high levels in the earlier years of the project ().
The benefits of PIMS as we have heard from MPSI scientists
Avoid repetition of unsuccessful experiments
During the period of project, PIMS can collect data on a large number of experiments. These will include both successful and, just as importantly, unsuccessful experiments. Without a LIMS system the only experiments that a scientist can normally find within published journals are the successful ones. Unsuccessful experiments are not normally published and are recorded to different degree of completeness in personal notebooks. Within PIMS the scientist can check whether similar condition was used by anyone else within the consortium and if so whether it yielded a successful result.
Never lose the samples
In a large laboratory it is common to find samples about which there is little or no information. This is not necessary caused by bad organization. It may be that someone, who knew what the sample is, left the laboratory or the information recorded cannot be found. Also, consider the situation when information is needed but the colleague responsible for creating the sample is temporarily unavailable. Such problems should not arise if PIMS is being used to record the details of a sample and its location.
Be confident in what you have been given
Find out the history of a sample you are about to use in your experiment, and make informed decisions about its quality. It is possible to display an image of the gel to decide on the quality of a sample. Other examples of important information are the amount of protein in the solution or the buffer/detergent conditions in which the sample is stored.
You choose where to work and when
With a LIMS the time and space restriction are lifted. It is possible to display data from any laboratory. Similarly laboratory data can be accessed with any web browser while outside the laboratory, for example at home or at a conference in another part of the world. Even in such remote locations it is possible to participate in the work of your laboratory as there are many reviewing, modifying and updating tasks that can be performed as well as simple passive monitoring task.
Case studies
Due to the degree of specialization of each of MPSI sites, each site uses PIMS in its own unique way. Therefore a description of a complete workflow would be unique for each of MPSI sites, and is outside the scope of this article. Instead, we would like to highlight several MPSI consortium data management tasks that have been successfully addressed within the PIMS framework.
Case study 1: Interfacing with legacy data management systems
The Leeds University group had a large amount of data on plasmids and primers in PDF files. These files were stored in the shared laboratory folder, and were hard to search or navigate. We extracted data and loaded them into PIMS, building intrinsic connections between the constructs, primers and target information. This way of recording data worked very well and the MPSI researchers rapidly moved completely to PIMS. Based on this positive experience other laboratories from Leeds University are now adopting PIMS. This system is constantly being improved with enhanced and new construct are being added with workflows and protocols continuing to be streamlined.
Case study 2: Invitrogen and Sigma-Aldrich primer order loading
The Leeds laboratory orders primers from Invitrogen or Sigma-Aldrich by completing MS Excel forms which contain information about the primers. Design of primers is done using Vector NTI software Citation[15] followed by the insertion of primer specification into either Invitrogen or Sigma-Aldrich spreadsheets. The supplier makes the primers and adds supplementary data to the spreadsheet, such as melting temperatures, primers’ concentration and primer samples optical density. Finally MPSI scientists add the information on the plasmids and the targets to the same spreadsheet. To avoid additional effort when recording these data into the PIMS system, we have developed a tool which loads information on primers from the excel spreadsheets. The reverse can be performed namely PIMS can create primer order spreadsheets from information stored within the database.
Case study 3: MPSI specific protocol development, experiments interlinking
MPSI developed a number of protocols which are common for all sites. The protocols were defined in PIMS and therefore shared across the whole consortium and serve the role of templates for experiments.
The PIMS protocol consists of 6 parts: protocol details, method, inputs, parameters, outputs and a files section. The details section contains the protocol name, objective, details, experiment type, and a ‘copy’ button, which provides a convenient way of making a new protocol based on this. The method section holds textual version of the protocol. The new protocol can also be designed from scratch and input and outputs samples can be added by using ‘add new’ option. The parameters can be added in the same way; the only difference is that there are three parameter types, which are the number, the text and Yes/No parameter. Input samples are the reagents in an experiment, (for example, antibiotics in bacterial culture), output samples are the product, (for example membranes in a cell fractionation experiment), whereas parameters relate to the conditions of experiments, such as temperature or centrifuge rotation speed. The default units and amounts can be specified for input and output samples, but this, similarly to the parameter defaults, can be overridden later with the actual experimental values.
To put theory into practice let us look at the MPSI transformation protocol in detail (). The antibiotics and the media samples are the standard lab chemicals recorded in PIMS. Competent cells could come prepared from external supplier or alternatively be produced by the PIMS competent cells preparation experiment. The output of this transformation experiment is recombinant cells. They can be used as inputs in any other experiment which takes recombinant cells as input, such as protein expression.
Figure 2. The PIMS protocol view. The screenshot of the PIMS protocol displaying the transformation protocol. Input, output samples and parameters are shown. Please note that due to lack of space not all the parameters of this protocol are shown. The lacking parameters are the (a) temperature of heat shock, (b) time of pre-incubation on ice, (c) time of regeneration after shock on ice, (d) whether culture was shaken (Yes/No). The dustbin icon permits deleting of the parameters and samples. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
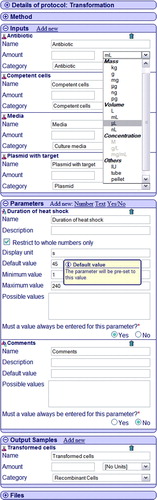
Finally the files section contains the list of files which have been linked to the protocol. This can be documents with the original protocol description or images which may be helpful in the experiment result interpretation and any other files which the protocol designer decided to provide.
PIMS protocols help to ensure that the values supplied in the experiments are valid. The protocol designer should make use of these facilities wherever possible. For instance it is easy to require the value of the parameter by setting the ‘is required’ flag to yes. For the number parameters the range of valid values can be specified so that PIMS can check whether the value set is within the range. Also the list of possible values for the parameter can be defined by the protocol designer. Finally, the default values if defined help to avoid the need to complete the field as PIMS will do it automatically. All the fields of the protocol should be self explanatory but if something is unclear by pointing the mouse on the field the context help can be accessed.
PIMS protocols offer a large degree of flexibility. Therefore the design of the protocol is largely down to the scientist judgement. It is possible to define the protocol with many or no inputs, outputs or parameters. When designing the protocol it is central to identify the data that is important to capture, some obvious inputs or parameters which are not important can be omitted. Ideally the protocol should contain sufficient details for a successful reproduction of the experiment. There are no benefits in recording protocols which do not contain sufficient data for reproducing the experiment. Recording every detail is not good either. Recording experiments where too many parameters and inputs have to be specified may be time consuming and will not yield any additional benefits. For example consider whether it is important to record an amount of the competent cells or the amount of media on the Petri dish in the transformation experiment, perhaps not. Such amounts are not usually measured precisely. At the same time it is important to know what the media was and what competent cells were used. PIMS is quite flexible in these questions and permits leaving amounts fields empty.
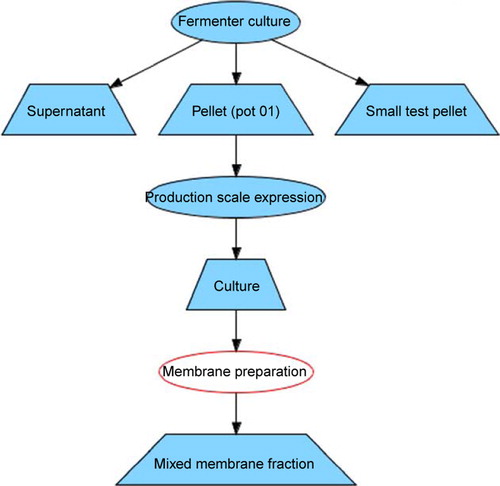
Experiments in PIMS can be chained by means of defining output sample(s) which are the input sample(s) for the next experiment in the chain. For instance, a fermentation experiment produces cells, which are used as input samples in a production scale experiment to make a larger amount of cells or culture. The membranes are extracted from cultured cells and these three experiments form a chain shown in .
The history of each sample produced can be conveniently tracked from the very first experiment in the chain to the last one. In PIMS the experiment chains with inputs and outputs can be visualized as a directed acyclic graph. Experiments on the graph are displayed as ellipses with the experiment type, the samples, which link the experiments, are represented by a trapezium displaying the sample name; arrows are used to indicate the order of the experiments in the series. The detailed information on each node on the graph can be obtained by clicking on the corresponding shape.
PIMS supports sample bar-coding and can generate barcode labels. In conjunction with location tracking functionality PIMS records where a container originates and what it contains. These protocols are therefore more than database entries they also hold information about physical location and histories of the samples that arise from the pipeline of experiments.
Case studies conclusion
As can be seen, we had to tailor PIMS to MPSI scientists’ needs which involved importing data from legacy systems as well as providing unified representation for the diverse data sets coming from the different nodes. Furthermore, the change of suppliers can also mean a need for a change in the LIMS. For example in case study 2 we describe that PIMS can generate an order for external supplier. Different suppliers have different order formats requiring a change to the LIMS component responsible for order generation. We have had to do it at least once therefore, we conclude that a large protein research laboratory needs some IT support for bespoke work, however good the LIMS and other tools that it uses.
PIMS for MPSI – the data mining perspectives
We foresee that the following analysis of the PIMS data will produce interesting results in the near future once a critical mass of experimental data is recorded.
Conditions which lead to the highest expression levels of a target protein (shown in ).
Optimized purification protocols for membrane proteins
Plasmid and strains combinations which are best for membrane protein expression (i.e. produce soluble, stable and mono disperse proteins)
Detergents which are the best at solubilization and stabilization of the membrane proteins.
Detergents which are best for crystallizing a target protein.
Figure 3. Chain of experiments in the PIMS, starting from fermenter culture preparation through production scale expression to membrane extraction. Experiments are displayed as ellipses with the experiment type, the samples, are represented by a trapezium displaying the sample name; arrows indicate the order of the experiments in the series. For simplicity and due to space limitations some input and output samples were not defined. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
This list is only a small part of what could be mined, and once the data are more extensive we can expect to find answers to even more interesting questions.
Towards universal LIMS for structural proteomics
LIMS can turn a laboratory into a round the clock working environment, where researchers deposit their data during the working hours and the supervisors review the results in the evenings, the environment where the need for face-to-face meetings diminishes or sufficiently reduces. But are there many LIMS that can perform tasks required of a structural genomics programme?
Despite a large number of ongoing efforts, by and large, they have enjoyed only limited success, have only been accepted by one or two groups they were initially developed for, and none has been widely adopted Citation[16], Citation[17]. Several sites have been able to develop a LIMS that encodes their current workflow Citation[18]. Such LIMS software has not proved usable at other locations, and is not future proof. This is largely due to the immense scale of the problem and quickly emerging new tools, databases and methods of work. Most commercial LIMS are better suited for well established specialized processes, and are not a viable alternative in the ever changing practice of an academic environment Citation[19].
Even the largest efforts in protein structure determination, such as the US funded Protein Structure Initiative (PSI) has different solutions for their data management needs at the different centres Citation[20–22]. PSI uses several separate databases for core data sharing. One of such core components is the PSI Material Repository Citation[23]. Another is a Protein Expression Purification and Crystallization Database (PepcDB) Citation[24].
However, as the content of PepcDB is assumed to be one of the enduring legacies of the PSI, the data are not comprehensive as some vital information like reagents are not stored while other information like protocols are stored as unstructured text Citation[25]. Experimental data is only really useful if there is enough information for an experimenter at another laboratory to reproduce the result. The scientist will naturally want to record only the information he or she needs to reproduce the experiment. For this reason, it is essential that the consistency of data can be checked and enforced upon the deposition. Typical LIMS would have stored this information in a structured manner, thus enabling easier data mining.
We believe that PIMS is avoiding similar problems by being flexible enough to accommodate very diverse practices at different laboratories. This flexibility lies in the reference data mechanism which allows scientists to specify new types of records in PIMS. In particular, experiments types, target milestones, sample categories, protocols etc.
Truly flexible LIMS needs to have well defined extension interfaces, so that new modules can be easily developed and plugged into the main system. The ability to extend the system would facilitate community development and would also enable robotic manufactures to provide pluggable interfaces for their robots, directly within the LIMS, so that data conversion would not be required. The work is ongoing to ensure that the PIMS meet all the criteria stated above.
Is it right to assume that once the LIMS is provided the scientists will be keen on using it? What else does it take for a LIMS to take up? The benefits of LIMS come with a cost. There are a number of requirements which have to be satisfied to get benefits from the LIMS system. These are discussed below.
PIMS is not one person LIMS
In contrast to a laboratory notebook where no restriction on data format is required, PIMS provides a structured LIMS and implies the means to extract and compare many records with a single key-stroke, however in many cases PIMS will inevitably be less flexible than a document based environment. A single researcher could use PIMS but it is likely that more time would be required to record the data than in a free-format application. Additionally the PIMS system needs to be configured and maintained, which is not a trivial undertaking. In our experience, the benefit from PIMS arises when several people are recording their results in a collaborative project. PIMS allows access to all of the data accumulated by a team of researchers as and when needed.
Finally, as a data format is enforced by PIMS, an agreed standard between collaborators is a natural outcome of PIMS usage: This allows the streamlining of data input and aids communication.
Using PIMS on collaborative projects
LIMS greatly facilitates collaboration. However, collaborators should be prepared to share the majority of their results with each other. The consequence of this is that publication policies have to be in place and agreed by all collaborating partners.
There is a potential for an individual contribution to be lost in a large collaborative projects especially ones that use centralized LIMS like the PIMS. If LIMS does not permit saving individual credentials, scientists will be reluctant to contribute the data to such a LIMS, and the collaboration may suffer. Therefore the individuals’ contribution has to be protected at all costs and it is essential that the LIMS retain the individual's credentials, and is implementing the strict policies to control the change of an ownership of a record. It should cater for the situations where the person who records the data is not the one who did an experiment. On a similar note, the person who enters the target may not be the one who works on it, and not the one who is in charge of defining it as a scientific objective to work towards. Regardless of the size of the contribution all individuals involved in the work need to be properly credited for their contribution.
PIMS as web-based LIMS (with web browser interface) is not suitable for massive data exchange
Web based LIMS are useful if only an average amount of data needs to be passed from client to server and the other way around. If gigabytes of data routinely have to be transmitted, then web based LIMS would be unlikely to be a viable solution. Although, if the server is located at the depositing location, so that the data can be stored directly into the local database, and users need only a subset of the data, then, web-based LIMS can still applied. In our experience, the transfer of several gigabytes of data in a month in a relatively small portion is perfectly feasible with the web-based LIMS. The top data transfer for MPSI PIMS reached 1.5 gigabyte per month.
Interface as many instruments as possible
Recording the data into the LIMS is not the most exciting nor is it the most rewarding task for scientist. Therefore all efforts should be made to capture the data from instruments used in the laboratory to reduce the amount of work the scientist need to do manually.
Furthermore, capturing the data directly from the instrument helps to ensure that a complete dataset and all the activities which were performed irrespective of whether the result has been achieved are recorded. This is especially important as negative results are often neglected and are not recorded into the LIMS. This is understandable as for the bench scientists the negative results does not contain any value. While this may be true for one experiment, it is certainly is not true for many. Once the critical mass of the experiments is recorded, it may be possible to mine them and to determine the cause of a failure. At the same time capturing negative results are as important as recording positive as this will help to avoid them in the future.
PIMS, through its extension XtalPIMS, supports crystallization imaging robots. Interfacing with other robotic equipment is high on the PIMS developer team agenda.
Change in work practice
LIMS provides structured ways of recording information. Before the LIMS era, scientists could record their data in an unstructured way; however, this is not possible with LIMS.
Introduction of a data management system in scientific practice implies a change in working practice. The nature of the data management systems is to provide structured, easily searchable data storage. There are two major benefits of the LIMS. The first is automation and robotic integration, when there is an instrument which produces a lot of data which cannot be efficiently handled by human operator. The second is ease of data sharing, which is especially crucial for distributed collaborations.
Structured LIMS requires a systematic approach to data entry, and the researcher needs to make a distinction between the sample and the experiment. Also if the data entry is sparse and incomplete, the LIMS will not yield full benefits, and data analysis on an incomplete dataset can produce misleading results.
A successful LIMS needs many users who are eager to share the data. Interestingly, we found that to get the benefits of the LIMS distributed collaboration is not required. It is sufficient to have a big group of scientists on site. In the case of MPSI, the biggest is in the University of Leeds, therefore, not surprisingly, they were among the first who started to appreciate the benefits of PIMS.
On the contrary, for weakly interacting partners, whether distributed or not, the LIMS does not provide many benefits. A case example would be an MPSI Oxford node, which due to the nature of its duties – computational modelling studies – does not contribute the same amount of data as other nodes. However in the long term even a small group could benefit from future data mining or information on samples. Therefore close collaboration seems to be the key for successful LIMS adaptation.
Simple vs. complete records
Apart from the sound architecture, successful LIMS has to be easy to use. When building or configuring a LIMS, conflicting requirements can often lead to the production of unusable solutions and inconsistencies within the LIMS. This can happen even when the data storage is consistent.
LIMS are used by different people with very different needs and this can be a source for conflicting requirements. One person can be a data miner, and from his/her point of view the greater the amount of data the better. From the researcher's point of view, the easier it is to record the information, the better. Scientists can tolerate much more ambiguity than the data miners, without losing their grip on what actually was recorded. For scientists, ideally the data should be recorded automatically, with minimum intervention.
This is not to mention the difference between different scientists’ practices and habits. Some are used to recording everything in detail, others just a bare minimum. To satisfy everyone, ideally different interfaces should be developed. Without them providing excessive amounts of details can lead to the system which satisfies no one truly. PIMS offered at least two different views for each record, one more advanced with all the details, another simpler without related record details. However, even this proved to be insufficient in some cases and more specialized views which show various bits of information belonging to different entities are required. Providing such views is the ongoing work of PIMS developers at the moment.
We found that providing the right level of detail is absolutely crucial. If there is too much information, to record some may be reluctant to record anything at all. If it is not possible to record enough details some crucial information can go missing, which renders the whole chunk of data useless.
Furthermore, additional details often detract from the important information and programming unwanted functionality uses up the limited resource such as the developers’ time. LIMS that is able to record a vast number of parameters is great as long as the robotic system populates it automatically and the scientists are shielded from the problems of data entry.
In practice, many scientists wanted to be able to see additional data, but few were willing to enter manually this extra detail.
The balance between complexity and simplicity is not easy. The best combination is to have a LIMS which is highly modular and allow a site administrator or the individual user to pick and choose the parts of the system they would like to use and type of records they would like to see. On the other hand not every laboratory has highly computer literate researchers willing to engage with LIMS support or enough funds to employ a dedicated LIMS administrator. To address this problem a completely different approach has to be employed such as providing an infrastructure for protein production laboratories where the experts who are involved in the constant development of the system keep it running.
Not only record the data, but mine it later
Recording the data, in an increasingly high-throughput environment is a big issue, but a bigger issue is to record data uniformly, so that the same data recorded through the different interfaces remains the same. This seems obvious, but this proved to be complicated for scientific data in our experience. During the PIMS development the data model has been changed 34 times despite the fact that it was highly developed at the start of the project. Furthermore, we learned that minimal data requirements have to be defined with great care as firstly they tend to differ hugely from one laboratory to another, and secondly if they are defined inconsistently and are incomplete, data mining can be problematic. Most importantly the key, rarely changed data has to be defined correctly and modified very carefully by a dedicated person. In PIMS, such data is called reference data, and is provided in the form of xml files which can be modified by the PIMS administrator. This data includes experiment types, chemical and biochemical suppliers, hazard phrases, sample categories, target milestones and many others. This guarantees that data from different PIMS deployments is interoperable, and can be mined in the same database if necessary. Furthermore, changes in the reference data can be rolled out to a number of production databases at once, and all the modifications can be kept in one reference data repository. Finally, PIMS can update the reference data on live databases without restarting.
Export from PIMS
PIMS supports exporting to XML, PDF, CSV, and Excel formats. It is possible to export the majority of the data recorded into the PIMS. In particular information on the single and plate experiments, samples, recipes, targets, constructs, primers, people (person whose details are recorded), organizations (i.e., suppliers, or universities), chemicals (i.e., NaCl), hazard phrases, locations (i.e., freezer 1), holders (i.e., box A). All reports and lists can be exported to either of these formats – XML, PDF, CSV, MS Excel (). For PDF export PIMS support WYSIWYG (what-you-see-what-you-get) paradigm Citation[26], therefore the information one see on the screen will look exactly the same once exported. Prior to the exporting it is often possible to choose what columns to hide, and sort the report as required. The columns chosen and the sorting order are preserved during the export. In this way it is possible to limit the number of records with precisely defined search criteria.
Figure 4. List of an entry clones (gateway vectors). List of entry clones containing information on the MPSI entry clones, whom they were designed by, where they can be found, when they were made, which restriction sites can be used for re-cloning the gene of interests to the expression vector. The magnifying glass icon in the left column leads to the detailed view of the record. The ‘Map’ link leads to the genbank formatted vector with inserted gene map file. Genbank file contains comments embedded by Vector NTI software used by MPSI laboratories, so that the map of the vector with inserted gene of interest can be readily visualized. The records in the columns can be sorted by Name, Designed By, Location, Box, Flanking site, Date, Position in the box. The sequence link leads to the sequence of the vector. Four exporting options are available: CVS, MS Excel, XML and PDF. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
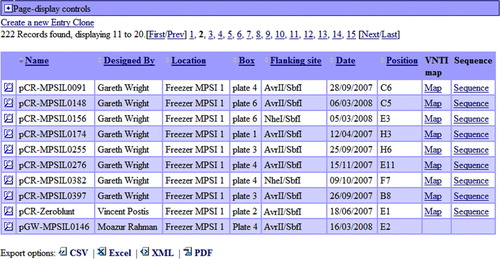
The options listed above are limited to one type of record. By means of this functionality it is not possible to export the information on several types of record at once. For instance to export all targets which have reached the stage of protein expression would need a more specific report. Luckily PIMS is supplied with a number of pre-built reports like this. The targets scoreboard can be used to get the list of the targets which reached an expressed stage. For the experiments with the same protocol it is also possible to compare experiment parameters. PIMS also has list of constructs with the targets, active targets report, samples reports. One of the most comprehensive reports PIMS has to date is the ‘target to crystal report’. It graphically displays the chain of experiments which led from the first experiments performed on the targets, to the last experiment which produced a crystal.
Various structure diagrams of the PIMS data model can be invaluable. The PIMS data model is open access therefore for the qualified programmer it would not be difficult to extract any data from PIMS.
Acknowledgements
We would like to thank all MPSI scientists for providing the wealth of information on their working practices and for the active participation in the PIMS development. The PIMS team, namely Dr Susanne Griffiths, Mr Bill Lin, Mr Ed Daniel, Mr Marc Savitsky, Mrs Anne Pajon and Dr Jon Diprose. Also, special thanks to MPSI scientists who have being willing to change their working practices, despite some inconveniences at the beginning. Namely Prof. Steve Baldwin, Dr Vincent Postis, Mr Gareth Wright, Dr Peter Roach, Dr Robert Cleverley, Dr Sarah Deacon, Dr Xiaobing Xia, Mr Frank Kroner, Mrs Isobel Black, Dr Karen McLuskey, Mrs June Southall, Dr Mads Gabrielsen, Mr James Kean, Dr Muhammad Imran, Dr Svetomir Tzokov and Dr Moazur Rahman. Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- BBSRC SPoRT grant details [Internet]. Available from the website: http://oasis.bbsrc.ac.uk/netans-bin/gate.exe?f=toc&p_lang=english&p_d=GABI&p_L=25&p_search=searchgabi&a_search=&p_s_SPRO=SPORT.
- Hibernate [Internet]. Available from the website: http://www.hibernate.org/.
- PostgreSQL database [Internet]. Available from the website: http://www.postgresql.org/.
- Oracle database [Internet]. Available from the website: http://www.oracle.com/technology/products/database/oracle11g/index.html.
- Sun java servlet and JSP specifications [Internet]. Available from the website: http://java.sun.com/products/servlet/reference/api/index.html.
- Apache Tomcat web application server [Internet]. Available from the website: http://tomcat.apache.org/.
- Protein Data Bank, An Information Portal to Biological Macromolecular Structures, statistics [Internet]. Available from the website: http://www.rcsb.org/pdb/statistics/contentGrowthChart.do?content=total&seqid=100.
- Burley SK. An overview of structural genomics. Nature Struct Biol 2000; 7: 932–934
- Protein Information Management System. STFC Daresbury Laboratory, Warrington, UK. [Internet]. Available from the website: http://www.pims-lims.org.
- Huia R, Edwards A. High-throughput protein crystallization. J Structural Biol 2003; 142: 154–161
- LabVantage SAPPHIRE LIMS [Internet]. Available from the website: http://www.labvantage.com/products/sapphire/index.html.
- BBSRC Data Sharing Policy [Internet]. Available from the website: http://www.bbsrc.ac.uk/publications/policy/data_sharing_policy.pdf.
- Membrane Transport Proteins Transporter Classification Database [Internet]. Division of Biological Sciences, University of California San Diego, US. Available from the website: http://www.tcdb.org/.
- Pajon A, Ionides J, Diprose J, Fillon J1, Fogh R, Ashton AW, Berman H, Boucher W, Cygler M, Deleury E, Esnouf R, Janin J1, Kim R, Krimm I, Lawson CL, Oeuillet E, Poupon A, Raymond S, Stevens T, Tilbeurgh H, Westbrook J, Wood P, Ulrich E, Vranken W, Xueli L, Laue E, Stuart DI, Henrick K. Design of a data model for developing laboratory information management and analysis systems for protein production. Proteins 2005; 58: 278–284
- Lu G, Moriyama EN. Vector NTI. A balanced all-in-one sequence analysis suite. Brief Bioinformat 2004; 5: 378–388
- Fulton KF, Ervine S, Faux N, Forster R, Jodun RA, Ly W, Robilliard L, Sonsini J, Whelan D, Whisstocka JC, Bucklea AM. CLIMS: Crystallography Laboratory Information Management System. Short communications Acta Crystallogr D Biol Crystallogr 2004; 60: 1691–1693
- Haquin S, Oeuillet E, Pajon A, Harris M, Jones AT, van Tilbeurgh H, Markley JL, Zolnai Z, Poupon A. Data management in structural genomics: An overview. Methods Mol Biol 2008; 426: 49–79
- Kats A, Prilusky J, Peleg Y, Dym O, Unger T, Albeck S, Sussman JL. 2008. PLUMS, a Proteomics Laboratory Unabridged Management System. The Israeli Bioinformatics annual Symposium (IBS). Abstract.
- Albeck S, Alzari P, Andreini C, Banci L, Berry IM, Bertini I, Cambillau C, Canard B, Carter L, Cohen SX, et al. SPINE bioinformatics and data-management aspects of high-throughput structural biology. Acta Crystallogr D Biol Crystallogr Oct 2006; 62: 1184–1195
- Zolnai Z, Lee PT, Li J, Chapman MR, Newman CS, Phillips GN, Rayment I, Ulrich EL, Volkman BF, Markley JL. Project management system for structural and functional proteomics: Sesame. JSFG 2003; 4: 11–23
- 2007 Protein Structure Initiative ‘Bottlenecks’ Workshop [Internet]. Available from the website: http://www.nigms.nih.gov/News/Reports/Bottlenecks032007.htm.
- Bonanno JB, Almo SC, Bresnick A, Chance MR, Fiser A, Swaminathan S, Jiang J, Studier FW, Shapiro L, Lima CD, et al. New York-Structural GenomiX Research Consortium (NYSGXRC): A Large Scale Center for the Protein Structure Initiative. J Struct Functional Genomics 2005; 6: 225–232
- Protein Structure Initiative. The PSI Materials Repository (PSI-MR) [Internet]. Available from the website: http://www.hip.harvard.edu/PSIMR/index.htm.
- Protein Structure Initiative, an Information Portal to Biological Macromolecular Structures [Internet]. PepcDB – Protein Expression Purification and Crystallization Database Available from the website: http://pepcdb.pdb.org/.
- Protein Structure Initiative, an Information Portal to Biological Macromolecular Structures [Internet]. PepcDB Schema: Clearly explained. Content and Report Formats Available from the website: http://pepcdb.pdb.org/help/PepcDB_help.html.
- Chamberlin D. Document convergence in an interactive formatting system. IBM J Res Develop 1987; 31(1)59