Abstract
The preparation of cell membranes by ultracentrifugation of bacterial cell lysates, a pre-requisite for the purification of over-expressed membrane proteins, is both time-consuming and difficult to perform on a large scale. To overcome this bottleneck in the structural investigation of such proteins in the UK Membrane Protein Structure Initiative, we have investigated the alternative use of tangential flow filtration for preparation of membranes from Escherichia coli. This method proved to be superior to the conventional use of ultracentrifuges both in speed and in yield of membrane protein. Moreover, it could more readily be scaled up to process larger quantities of bacterial cells. Comparison of the purity and monodispersity of an over-expressed membrane protein purified from conventionally-prepared membranes and from membranes prepared by filtration revealed no substantial differences. The approach described should therefore be of general use for membrane protein preparation for a wide range of applications, including both structural and functional studies.
Introduction
Membrane proteins represent approximately 30% of the proteome in most organisms and play key roles in many aspects of cellular function, including nutrient uptake, energy transduction and cell-cell signalling. They also account for approximately 50% of current therapeutic drug targets Citation[1], and so gaining an understanding of their structure-function relationships is of both basic scientific interest and clinical importance. However, structural investigations of such proteins are hampered by their typically low natural abundance. While improvements in vector technology have rendered expression of prokaryote membrane proteins at levels up to 25% of the total inner membrane protein of Escherichia coli relatively routine Citation[2], inner membrane proteins in total account for only 200,000 of the 3,600,000 proteins in the bacterial cell Citation[3]. In order to purify the tens of mg quantities of protein typically required for crystallisation trials, it is therefore usually necessary to start with fermenter-scale cultures, even when the high-yielding auto-induction procedures introduced by Studier Citation[4] are employed. The downstream processing of large amounts of cell paste then represents a major bottleneck on the route to protein purification. Conventionally, the separation of membrane fractions from cytosolic and other components is performed by centrifugation Citation[5]. However, such procedures are time consuming, require expensive ultracentrifuges, and are limited by the rotor capacity: with a standard preparative ultracentrifuge typically no more than 60 g (wet weight) of cells can be processed at one time.
An alternative to centrifugation for the separation of particles of differing size is filtration through a membrane of defined pore-size, which may be accomplished using a technique known as Tangential Flow Filtration (TFF). In TFF, to avoid membrane fouling, the retained fraction (retentate), which contains molecules/particles too large to pass through the filter, is pumped tangentially across the membrane surface, whilst the filtrate, containing smaller particles/molecules, passes through the membrane. This process has been used for many years for the purification of water-soluble proteins Citation[6], inclusion bodies Citation[7], virus particles Citation[8] and nucleic acids Citation[9]. It has also been used as a means of preparing haemoglobin-free membranes from human erythrocytes Citation[10]. In addition to the relatively low capital costs of TFF, potential advantages of this approach include scalability and speed. In the present study we investigated whether TFF could be used as an alternative to traditional centrifugation methods for the purification of cell membranes from E. coli cells over-expressing membrane transport proteins.
Methods
Cell culture
Escherichia coli strain BL21 Star™ (DE3) (Invitrogen) cells harbouring a pTTQ18-based plasmid encoding a C-terminally RGSH6-tagged form of the adenine nucleotide exchanger Npt1 from Chlamydia trachomatis Citation[11] were used as a source of membranes. Auto-induction of expression Citation[4] was achieved by culturing the cells at 37°C in medium containing 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4 and 1 mM MgSO4, plus 1% (w/v) tryptone, 0.5% (w/v) yeast extract, 0.5% (w/v) glycerol, 0.05% (w/v) glucose, 0.2% (w/v) lactose and 0.1 mg/mL carbenicillin. Cells were cultured on a 30-l scale using a 40 l Applikon Bio Pilot Fermenter (Schiedam, The Netherlands). Dissolved oxygen was controlled at 40% (air saturation) by means of variable agitation between 200 and 400 rpm and the pH was controlled at approximately 7 with 2 M NaOH and 1 M phosphoric acid. Cells were harvested after 27 h and a yield of approximately 550 g (wet weight) was obtained.
Cell lysis
For cell lysis, samples were processed on ice. Cells (240 g wet weight) were resuspended and homogenized in 1.5 l of lysis buffer (20 mM Tris-HCl, pH 7.5 at 4°C, containing 0.5 mM EDTA) using a PowerGen 1000 shear mixer (Fisher Scientific) at 30,000 rpm for 5 min and then lysed by passing twice at 4°C through a cell disrupter (Constant Systems Ltd.) at 35 kpsi. Unbroken cells and other debris were then removed by centrifugation using a Beckman JLA 8.1000 Rotor at 14,000 gav for 45 min at 4°C. The supernatant was diluted to 1.6 l with lysis buffer.
Membrane preparation by centrifugation
To remove cytosolic proteins a sample (400 ml) of the supernatant produced as described above was centrifuged at 131,000 gav for 2 h at 4°C in a Beckman Type 45 Ti rotor in an Optima™ L-80 XP Ultracentrifuge. The supernatant was carefully decanted, the membrane pellets resuspended to a total volume of 200 ml in suspension buffer (20 mM Tris-HCl, pH 7.5 at 4°C), and centrifugation repeated as above for 1 h. Following a further round of resuspension and centrifugation, the final membrane pellet was resuspended in suspension buffer to yield a total volume of 26.5 ml and then drip-frozen in liquid nitrogen before storage at -70°C. Samples were taken of the starting material and of the supernatants and pellets from each centrifugation step.
Membrane preparation by TFF
As an alternative means of removing cytosolic proteins a sample (1200 ml) of the supernatant from cell lysis, produced as described above, was subjected to TFF at 4°C in a cold-room, using a Pellicon cassette TFF system (Millipore Corporation) equipped with a Pellicon microfiltration module of nominal pore size 0.5 µm. The system was assembled as illustrated in , using a 2 l side-arm Erlenmeyer flask as the retentate reservoir, a magnetic stirrer to keep the membranes in suspension, a 20-l container as a reservoir for the suspension buffer, a variable-speed peristaltic pump with a maximum flow rate of 1 l/min and pressure gauges in both the retentate and filtrate lines.
Figure 1. Schematic diagram showing assembly and operation of a tangential flow filtration apparatus for bacterial membrane preparation. The lower panel shows a magnified view of the filter, to illustrate how membrane vesicles are separated from smaller, water-soluble proteins.
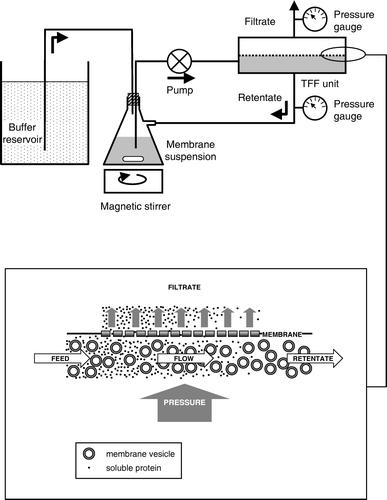
Filtration was initially performed using an empty buffer reservoir, allowing the volume of the membrane suspension to be reduced to approximately 400 ml. The suspension was then diluted with suspension buffer to a volume of 1.2 l, the buffer reservoir was filled with suspension buffer, the pump rate was adjusted to 1 l/min and the retentate line clamped sufficiently to maintain a pressure of between 0.6 and 0.8 bar. The air-tight seal provided by the silicone bung in the Erlenmeyer flask ensured liquid lost through filtration was replaced by buffer from the reservoir, thus keeping the volume of the retentate constant. After 20 l of buffer had passed through the system, the buffer reservoir was disconnected and the retentate was concentrated to the smallest volume possible without allowing the pressure to exceed 1 bar. Membranes remaining within the filtration module itself were displaced by passage of 50 ml suspension buffer through the system and combined with those in the Erlenmeyer flask, resulting in a final suspension volume of 210 ml, and then drip-frozen in liquid nitrogen before storage at -70°C. Samples of the retentate and of filtrate (from the filtrate line) were taken at regular intervals during filtration for subsequent analysis.
Protein purification
For purification of Npt1, membranes (20 mg protein) were solubilised by gentle mixing at 4°C in 5 ml of buffer A (50 mM HEPES-Cl, pH 7.4, containing 100 mM NaCl and 5% (w/v) glycerol), containing 5 mM imidazole and 1% (w/v) n-Dodecyl-β,D-maltoside (DDM; Anatrace). Insoluble material was removed by filtration through a 0.45 µm pore-size Acrodisc® 25 mm syringe filter (Pall Corporation). The filtrate was gently mixed for 2 h at 4°C with 0.5 ml Pierce HisPur Cobalt Resin (Perbio Science UK Ltd.) that had been pre-equilibrated with buffer A containing 5 mM imidazole. The resin was then washed in batch with 15 ml buffer A containing 5 mM imidazole and 0.05% (w/v) DDM, followed by 15 ml buffer A containing 20 mM imidazole and 0.05% (w/v) DDM. The bound protein was finally eluted in batch with 1 ml of buffer A containing 100 mM imidazole and 0.05% (w/v) DDM.
Membrane and protein characterization
Membrane protein concentrations were determined using the bicinchoninic acid (BCA) assay (Perbio Science UK Ltd). After SDS-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were detected using SimplyBlue™ Safestain (Invitrogen). Following electroblotting of gels, RGSH6-tagged Npt1 was detected by incubating with a horseradish peroxidase-labelled monoclonal antibody against oligohistidine tags (R&D Systems clone AD1.1.10) followed by SuperSignal® West Pico chemiluminescent substrate (Perbio Science UK Ltd). Signals were detected and quantified using a GeneGnome Detection system and GeneTools software respectively (Syngene Bio Imaging). Known amounts of hexahistidine-tagged Tobacco Etch Virus (TEV) protease Citation[12] were included on the gel to provide standards for this quantification.
Results
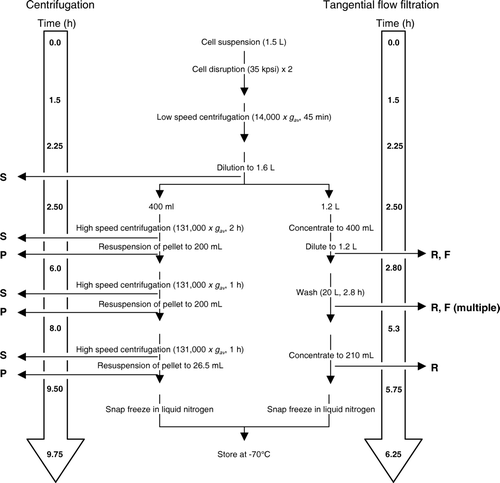
To compare the efficiencies of membrane preparation by centrifugation and tangential flow filtration, the two procedures were performed in parallel on samples of E. coli cell lysate from which unbroken cells and other debris had been removed by low-speed centrifugation, as described in Methods. Samples (1 ml) of resuspended membrane pellets and supernatants from the centrifugation steps, and of retentate and filtrate, were taken for analysis at the time points illustrated in .
Figure 2. Flow diagram comparing the approximate timings of steps in bacterial membrane preparation by centrifugation and by tangential flow filtration. The points at which samples were taken for analysis are also indicated: S = supernatant, P = pellet, R = retentate, F = filtrate. During the 20 l wash stage of the tangential flow filtration procedure, filtrate samples were taken at 10 or 15 min intervals, and retentate samples at 15 or 30 min intervals, as detailed in .
These were assayed for total protein by the BCA assay and by SimplyBlue™ Safestaining of SDS polyacrylamide gels. Membrane content was assessed by staining Western blots with horseradish peroxidase-labelled antibodies against the hexahistidine tag present in the membrane transporter Npt1.
Centrifugal preparation
The objective of centrifugation is to purify membranes by sedimentation, thereby separating them from water-soluble, cytosolic proteins. However, while, as expected, the supernatant following the first centrifugation step contained 62% of the total protein present in the initial lysate, reflecting the presence of cytosolic proteins, Western blotting unexpectedly showed that it also contained 20% of the membrane proteins. Similar losses of membrane protein in the supernatant were observed following the second and third centrifugation steps, such that the final yield of membrane protein was only 13% of that present in the initial cell lysate. It is possible that the high concentration of membranes in the suspension prevented their complete sedimentation, despite the use of an ultracentrifuge. Unfortunately, rotor capacity precluded use of more dilute suspensions, while time constraints did not allow the time of centrifugation to be increased – the overall process took almost 10 h to complete (). This highlights a key limitation when trying to process reasonable quantities of cell lysates for membrane protein purification by ultracentrifugation.
Tangential flow filtration preparation
In contrast to centrifugation, the objective of TFF was to purify membranes by passing them over a filtration membrane with a pore size sufficient to allow ready passage of cytosolic proteins but prevent passage of membrane vesicles and other membrane fragments. shows that this was achieved using a filter of nominal pore-size 0.5 µm: the histidine-tagged membrane protein Npt1 was detected only in the retentate and not in the filtrate produced by TFF of the E. coli cell lysate.
Figure 3. Time course of changes in the retentate and filtrate compositions during tangential flow filtration. (A) Protein concentration (•) in filtrate samples taken at the indicated times during the initial concentrating phase (0–15 min) and during the subsequent wash with 20 l buffer. (B) SDS-PAGE of filtrate samples (10 µl) taken at the indicated times, analysed for the presence of Npt1 by staining a western blot with antibody against the hexahistidine tag (upper panel) and for total protein by staining with SimplyBlue™ Safestain (lower panel). (C) Western blot of retentate samples (6.8 µg) taken at the indicated times, analysed for the presence of Npt1 by staining with antibody against the hexahistidine tag. The positions of marker proteins of known molecular mass, and of Npt1 (which migrates with an apparent molecular mass of 46 kDa), are shown on the left of B and C.
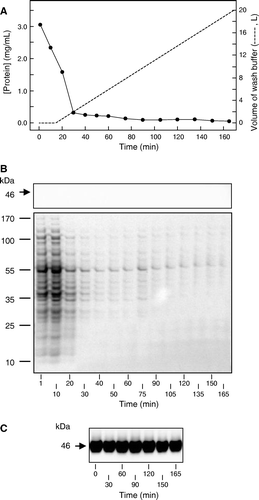
However, SDS-PAGE revealed that the filtrate did contain an abundance of other proteins; presumably originating from the bacterial cytosol (B). The major portion of this soluble protein was filtered during the initial 15 min step in which the volume of the retentate was reduced from 1.2 l to 400 ml, while most of the remainder was filtered during the first 15 min of the 20 l wash step (A). Comparison of the protein concentrations of the initial filtrate samples with those taken throughout the washing procedure indicated that approximately 98% of the total filterable protein had been removed during the 20 l wash, the entire process being completed in just over 6 h (). The overall yield of membrane protein, as assessed by quantitative western blotting, was 54% of that present in the initial lysate.
Comparison of membranes produced by centrifugation and TFF
SDS-PAGE followed by SimplyBlue™ Safestaining showed an indistinguishable pattern of proteins in the membranes produced by centrifugation and TFF (data not shown). Quantitative western blotting showed that Npt1 in each case comprised approximately 9% of the total protein, indicating similar degrees of membrane purity in the two preparations ().
Table I. Comparison of centrifugal and tangential flow filtration membrane preparations.
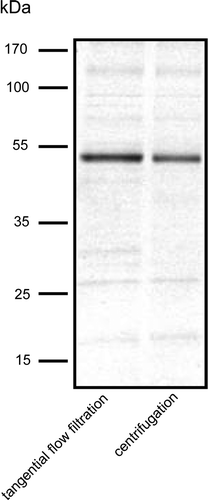
Given the approximately 4-fold greater yield of membranes obtained per unit weight of cells using the TFF procedure (), the latter would appear to be the method of choice for membrane protein production. However, before this conclusion could be definitively made, it was important to compare the suitability of the two preparations for downstream processing, and in particular for membrane protein purification. To this end, each was used for purification of His-tagged Npt1 by affinity chromatography as detailed in Methods. Membranes produced by centrifugation and TFF yielded 47 and 60 µg purified protein per mg membrane respectively. SDS-PAGE followed by SimplyBlue™ Safestaining revealed that the purity of the two preparations was similar, although slightly different patterns of contaminating minor bands were apparent ().
Figure 4. SDS-PAGE of Npt1 partially purified by Ni-NTA chromatography from DDM-solubilised membranes prepared either by centrifugation or tangential flow filtration, as indicated. Protein was detected by staining with SimplyBlue™ Safestain. The positions of marker proteins of known molecular mass are shown on the left.
Subsequent size exclusion chromatography (SEC) on a Superdex200 10/300GL column demonstrated monodispersity in both cases (data not shown). Although minor contaminants remained, this reflects the nature of Npt1 rather than of the starting material: in the case of several other transporters the purity and monodispersity of proteins purified from TFF membranes by affinity chromatography and SEC has been sufficient for crystallization trials. Examples are given in the accompanying paper in this special issue of Molecular Membrane Biology (Postis et al. this issue).
Discussion
The results obtained using the centrifugation and TFF methods for bacterial membrane preparation are compared in . Two advantages of the latter are clear. The first is that the TFF method results in an approximately 4-fold greater yield of membranes per gram of cell paste, because membrane losses resulting from incomplete pelleting upon centrifugation are avoided. The compositions of the resultant preparations are, however, similar and so there is also a 4-fold greater yield of any target membrane protein that has been over-expressed in the cells, in this case Npt1. The second advantage is that of time and labour: the TFF procedure takes only two thirds the time of the centrifugation procedure and involves fewer steps at which operator intervention is required. Moreover, larger quantities of cell paste can be processed at any one time. In the present study, 180 g cell paste were processed, but we have routinely processed up to 500 g of paste in the TFF apparatus by increasing the volumes of lysate and wash buffer employed. Such large amounts of cell paste are now routinely available from auto-induction expression procedures performed in fermenters, as detailed in the accompanying paper in this special issue of Molecular Membrane Biology (Deacon et al.). The ability to produce large amounts of membranes is particularly important for structural investigations of individual membrane proteins, not only because they typically represent ≤ 20% of the total membrane protein, but also because it overcomes batch-to-batch variability which can otherwise complicate crystallisation trials or other investigations. As described here for Npt1, the quality of membranes produced by TFF is comparable to that of membranes produced by centrifugation. Western blotting similarly revealed no significant differences in target concentration within membranes, produced by the two methods, from bacteria expressing a second transporter from the Membrane Protein Structure Initiative (MPSI) target list, the putative monovalent cation:proton antiporter designated target 0171 (data not shown). We have obtained preparations of similar quality for many other membrane transporters expressed in E. coli (please see the accompanying paper from Postis et al. (this issue), and therefore now routinely use this procedure for membrane protein production in the MPSI project.
Acknowledgements
This work was supported principally by the Biotechnology and Biological Sciences Research Council [grant numbers BBS/B/14418 (MPSI), 24/REI18440] and by the University of Leeds. We are grateful to Dr J. Bostock and Professor I. Chopra of the University of Leeds for provision of Chlamydia trachomatis genomic DNA encoding Npt1. Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov 2002; 1: 727–730
- Rahman M, Ismat F, McPherson MJ, Baldwin SA. Topology-informed strategies for the overexpression and purification of membrane proteins. Mol Membr Biol 2007; 24: 407–418
- Sundararaj S, Guo A, Habibi-Nazhad B, Rouani M, Stothard P, Ellison M, Wishart DS. The CyberCell Database (CCDB): a comprehensive, self-updating, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res 2004; 32: D293–295
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 2005; 41: 207–234
- Ward A, Sanderson NM, O'Reilly J, Rutherford NG, Poolman B, Henderson PJF. 2000. The amplified expression, identification, purification, assay and properties of histidine-tagged bacterial membrane transport proteins. In: Baldwin SA Membrane transport – a practical approach. Oxford: Oxford University Press. pp 141–166.
- van RR, Leonard LC, Hsu CC, Builder SE. Industrial scale harvest of proteins from mammalian cell culture by tangential flow filtration. Biotechnol Bioeng 1991; 38: 413–422
- Grimm KM, Trigona WL, Heidecker GJ, Joyce JG, Fu TM, Shiver JW, Keller PM, Cook JC. An enhanced and scalable process for the purification of SIV Gag-specific MHC tetramer. Protein Expr Purif 2001; 23: 270–281
- Morenweiser R. Downstream processing of viral vectors and vaccines. Gene Ther 2005; 12(Suppl. 1)S103–110
- Eon-Duval A, MacDuff RH, Fisher CA, Harris MJ, Brook C. Removal of RNA impurities by tangential flow filtration in an RNase-free plasmid DNA purification process. Anal Biochem 2003; 316: 66–73
- Rosenberry TL, Chen JF, Lee MM, Moulton TA, Onigman P. Large scale isolation of human erythrocyte membranes by high volume molecular filtration. J Biochem Biophys Methods 1981; 4: 39–48
- Tjaden J, Winkler HH, Schwoppe C, Van Der Laan M, Mohlmann T, Neuhaus HE. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J Bacteriol 1999; 181: 1196–1202
- van den Berg S, Löfdahl PA, Härd T, Berglund H. Improved solubility of TEV protease by directed evolution. J Biotechnol 2006; 121: 291–298