Abstract
The sodium solute symporters (SSS) and neurotransmitter sodium symporters (NSS) are two families of secondary transporters that are not related in amino acid sequence. Nonetheless, recent crystal structures showed that the Na+/galactose (SSS) and Na+/leucine (NSS) transporters have similar core structures. The structural relatedness highlights the need for classification methods for membrane protein structures based on other criteria than amino acid similarity. Here, we demonstrate that a method based on hydropathy profile alignments convincingly identifies structural similarity between the NSS and SSS families. Most importantly, the method shows that one of the largest transporter families for which a crystal structure is elusive (the amino acid/polyamine/organocation or APC superfamily), also shares the similar core structure observed for the Na+/galactose and Na+/leucine transporters. The APC superfamily contains the major amino acid transporter families that are found throughout life. Insight into their structure will significantly facilitate the studies of this important group of transporters.
Introduction
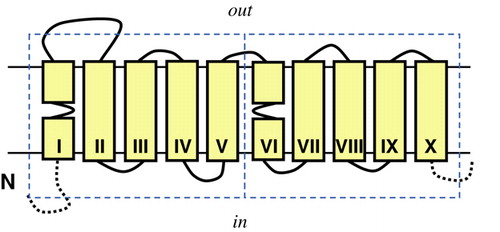
Very recently, the crystal structure of a Na+/galactose symporter vSGLT was reported at a resolution of ∼ 3 ? Citation[1]. The transporter found in Vibrio parahaemolyticus is the first member of the sodium solute symporter (SSS) family for which a crystal structure has been solved. The SSS family is a ubiquitous family of Na+-coupled transporters including the well studied mammalian transporters for glucose (SGLT) and iodide (NIS) and the bacterial proline transporters (PutP) Citation[2]. The Na+/galactose symporter has 14 transmembrane segments (TMSs) with a core of two structurally similar domains of five TMSs each. The two domains have opposite orientations in the membrane and bind the galactose molecule at their interface. The domains are further characterized by a break in the first transmembrane α-helix (). Surprisingly, the structural arrangement of the core of the galactose transporter had been seen before in LeuT Citation[3], a leucine transporter of the neurotransmitter sodium symporter (NSS) family, a family unrelated to the SSS family in amino acid sequence. The observation stresses the importance of methods other than amino acid sequence analysis to structurally classify membrane proteins.
Figure 1. Membrane topology model of the core of 10 TMSs shared by the Na+-galactose transporter vSGLT of Vibrio heamatolyticus and the Na+-leucine transporter LeuT of Aquifex aeolicus. The core consists of two domains (dashed boxes) of five TMSs each that have the same fold but opposite orientation in the membrane (inverted topology), a structural motif that is observed frequently in membrane proteins. vSGLT contains one additional TMS at the N-terminal side of the core (N) and 3 at the C-terminal side (C). LeuT contains two additional TMSs at the C-terminal side. Solid yellow boxes represent transmembrane segments. This Figure is reproduced in colour in Molecular Membrane Biology online.
The MemGen method uses hydropathy profile alignments to predict structural similarity of membrane proteins not related in sequence Citation[4]. Although the method predicted the structural similarity between the transporters of the SSS and NSS families Citation[5], the limited data set available at the time (1998) made the method prone to the identification of false positives. Here, we show that using the vast number of sequences in current databases, the MemGen method convincingly demonstrates structural similarity of the SSS and NSS families. In fact, TMSs II-XI of the SSS transporters corresponding to TMSs I-X of the NSS transporters are identified correctly as the core of the proteins. More importantly, based on the larger dataset, the method also decidedly predicts the same structural core in the transporters of the amino acid-polyamine-organocation (APC) superfamily Citation[6], one of the largest remaining families of secondary transporters for which no crystal structure is available. The APC superfamily is the main family of amino acid transporters found in all domains of life.
Methods
Computational methods
Members of the SSS, NSS and AAT families were collected by BLAST searches Citation[7] of a database of proteins coded on the genomes of 649 available microbes. vSGLT of Vibrio heamatolyticus (SSS), LeuT of Aquifex aeolicus (NSS) and the aromatic amino acid transporter AroP of Escherichia coli were used as queries. AroP is a member of the AAT family, one of the families in the APC superfamily. Sequences were selected from each set that shared between ∼ 20 and 60% sequence identity by pairwise alignment. Family hydropathy profiles were calculated following multiple sequence alignment by CLUSTALW2.0 Citation[8] as described Citation[4]. The structure divergence scores of the family profiles (SDS, see Citation[4]) were 0.123, 0.125, and 0.125 for the SSS, NSS and AAT families, respectively. The algorithm to find the optimal alignment of the family profiles and calculation of the S-factor to discriminate between similar and dissimilar structures have been described as well Citation[4].
Results and discussion
The hydropathy profile of the amino acid sequence of a membrane protein shows a characteristic peak pattern due to the clustering of hydrophobic residues in the parts of the polypeptide chain that are embedded in the membrane. MemGen uses the hydropathy profile as a ‘reporter’ or ‘fingerprint’ of the global folding of the protein to compare the structures of different proteins, i.e., for structural classification. First, an average ‘family hydropathy profile’ is calculated based on the multiple sequence alignment of a set of homologous proteins (e.g. from the SSS family), which show unambiguous similarity at the amino acid level. Then, in a way similar to comparing amino acid sequences, MemGen compares averaged hydropathy profiles of different families of membrane proteins (e.g. SSS and NSS) by finding the optimal alignment of the profiles, without using the amino acid sequences, and allowing for insertions and deletions. The divergence of the hydropathy profiles within a family (e.g. the variation in the single protein profiles in the SSS family) is then compared to the divergence between the averaged hydropathy profiles of different families (e.g. between NSS and SSS) to provide a numerical criterion for structural similarity. Similarity is expressed in the similarity score S that takes values of 1 or below for similar structures. When the method was introduced, and only a very limited set of sequences was available, 13 families of secondary transporters were classified into 4 structural classes, termed ST[1], ST[2], ST[3], and ST[4] Citation[4]. Later, by screening the available sequence databases, many more families were assigned to these structural classes Citation[5], Citation[9]. All the families in one particular class are predicted to have the same global fold. The SSS, NSS and APC families are all found in structural class ST[2].
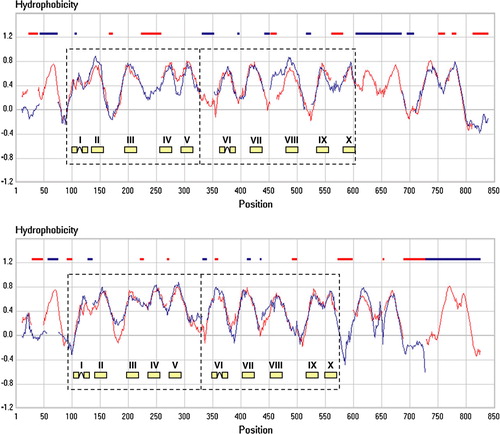
The limited data set used before made the method prone to errors because of poorly defined average family hydropathy profiles. We now reevaluate the predicted structural similarity of the SSS and NSS families, for which crystal structures of representatives are now available, and APC families, for which no crystal structure has been determined. The sequences of members of the SSS, NSS, and APC families were extracted from a database of proteins coded on 649 bacterial and archaeal genomes. The families contained 36, 113 and 133 sequences, respectively, with pair wise sequence identities in each family ranging between 20 and 60%. Optimal alignment of the SSS and NSS profiles resulted in an S (similarity) score of 0.879, which is strongly indicative of similar structures (, top). Similarly, optimal alignments of the SSS and APC profiles (, bottom) and of the NSS and APC profiles (not shown) result in similarity scores of 0.900 and 0.944, respectively, showing that the APC superfamily belongs to the same structural class. The section marked as ‘core’ is present in all three profiles and can be found in all other families in structural class ST[2], while regions outside the core may be present or not (not shown). Clearly, the SSS family contains an extra TMS at the N-terminus that is not present in the NSS and APC families and the number TMSs at the C-terminal side of the core is variable even within the families.
Figure 2. Hydropathy profile alignment of the SSS and NSS (top) and the SSS and AAT (bottom) families. The family hydropathy profile of the SSS family is indicated in red, those of the NSS (top) and AAT (bottom) families in blue. The AAT family is one of the families in the APC superfamily. Red and blue bars at the top indicate positions where gaps were introduced in the profiles by the alignment algorithm. Dashed boxes show the two domains in the core structure. The position of the 2 times five TMS is indicated. This Figure is reproduced in colour in Molecular Membrane Biology online.
Structural class ST[1] in the MemGen classification system largely correspond to the Major Facilitator Superfamily (MFS) from sequence homology Citation[4], Citation[5]. ST[2] contains the APC superfamily and the SSS and NSS families discussed here plus a number of smaller families of transporters, mainly for amino acids. ST[3] has been analyzed most extensively and contains a total of 36 families among which the families of the Ion Transporter superfamily (IT) and 2-hydroxycarboxylate transporter family (2HCT) and Na+-coupled glutamate transporters (ESS) Citation[9], Citation[10]. Finally, ST[4] is unique in that it contains a single family of glutamate and neutral amino acid transporters (DAACS) Citation[10]. Until now, available crystal structures of secondary transporter proteins supported the MemGen classification in the sense that proteins from different classes (LacY and GlpT in ST[1] Citation[11], Citation[12], LeuT in ST[2] Citation[3] and GltPh in ST[4] Citation[13] clearly represented different structures (see also reference Citation[14]). The high resolution structures of vSGLT and LeuT now also provide support for the MemGen classification of families within a structural class: two proteins in the same MemGen class, but unrelated in sequence, share the same global fold. Before, this had been demonstrated only at the low resolution level of the membrane topology for families in class ST[3] Citation[15].
In the MemGen classification, the APC superfamily is in the same structural class (ST[2]) as the SSS and NSS families, meaning that they share the core structure observed in the vSGLT and LeuT transporters. This should significantly facilitate the characterization of this large and important group of transporters.
Acknowledgements
This work was supported by The Netherlands Organisation for Scientific research (NWO, Vidi grant to DJS, ECHO grant to JSL) and the European Community's Seventh Framework Programme, grant agreement no. 211441-BIAMFOOD to JSL.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Faham S, Watanabe A, Mercado Besserer G, Cascio D, Specht A, Hirayama BA, Wright EM, Abramson J. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 2008; 321: 810–814
- Jung H. The sodium/substrate symporter family: structural and functional features FEBS Letters 2002; 529: 73–77
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of the Na+/Cl−-dependent neurotransmitter transporters. Nature 2005; 437: 215–223
- Lolkema JS, Slotboom DJ. Estimation of structural similarity of membrane proteins by hydropathy profile alignment. Molec Mem Biol 1998; 15: 33–42
- Lolkema JS, Slotboom DJ. Hydropathy profile alignment. A tool to search for structural homologues of membrane proteins. FEMS Microbiol Rev 1998; 22: 305–322
- Jack DL, Paulsen IT, Saier MH, Jr. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 2000; 146: 1797–1814
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search. Nucleic Acids Res 1997; 25: 3389–3402
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23: 2947–2948
- Lolkema JS, Slotboom DJ. Classification of 29 families of secondary transport proteins into a single structural class using hydropathy profile analysis. J Molec Biol 2003; 327: 901–909
- Lolkema JS, Slotboom DJ. Sequence and hydropathy profile analysis of two classes of secondary transporters. Molec Membr Biol 2005; 22: 177–189
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science 2003; 301: 610–615
- Huang Y, Lemieux MJ, Song J, Auer M, Wang D-N. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 2003; 301: 616–620
- Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 2004; 431: 811–818
- Lolkema JS. Domain structure and pore-loops in the 2-hydroxycarboxylate transporter family. J Molec Microbiol Biotechnol 2006; 11: 318–325
- Dobrowolski AJ, Sobczak I, Lolkema JS. Experimental validation of membrane topology prediction by hydropathy profile alignment: membrane topology of the Na+-glutamate transporter of Escherichia coli. Biochemistry 2007; 46: 2326–2332