Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) envelope spike (S) glycoprotein is responsible for the fusion between the membranes of the virus and the target cell. In the case of the S2 domain of protein S, it has been found a highly hydrophobic and interfacial domain flanked by the heptad repeat 1 and 2 regions; significantly, different peptides pertaining to this domain have shown a significant leakage effect and an important plaque formation inhibition, which, similarly to HIV-1 gp41, support the role of this region in the fusion process. Therefore, we have carried out a study of the binding and interaction with model membranes of a peptide corresponding to segment 1073–1095 of the SARS-CoV S glycoprotein, peptide SARSL in the presence of different membrane model systems, as well as the structural changes taking place in both the lipid and the peptide induced by the binding of the peptide to the membrane. Our results show that SARSL strongly partitions into phospholipid membranes and organizes differently in lipid environments, displaying membrane activity modulated by the lipid composition of the membrane. These data would support its role in SARS-CoV mediated membrane fusion and suggest that the region where this peptide resides could be involved in the merging of the viral and target cell membranes.
| Abbreviations | ||
| 16NS | = | 16-Doxyl-stearic acid |
| 5NS | = | 5-Doxyl-stearic acid |
| BPS | = | Bovine brain L-α-phosphatidylserine |
| CF | = | 5-Carboxyfluorescein |
| Chol | = | Cholesterol |
| CoV | = | Coronavirus |
| DMPG | = | 1,2-Dimyristoyl-sn-glycero- phosphoglycerol |
| DPH | = | 1,6-Diphenyl-1,3,5-hexatriene |
| EPA | = | Egg L-α-phosphatidic acid |
| EPC | = | Egg L-α-phosphatidylcholine |
| EPG | = | Egg L-α-phosphatidylglycerol |
| ESM | = | Egg sphingomyelin |
| FP | = | Fusion peptide |
| FTIR | = | Fourier-transform infrared spectroscopy |
| HIV-1 | = | Human immunodeficiency virus type 1 |
| HR | = | Heptad repeat region |
| LUV | = | Large unilamellar vesicles |
| MLV | = | Multilamellar vesicles |
| POPC | = | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| POPE | = | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine |
| POPG | = | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, POPS, 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine |
| PTM | = | Pre-transmembrane domain |
| SARS | = | Severe acute respiratory syndrome |
| SARSL | = | Loop peptide of SARS |
| SIV | = | Simian immunodeficiency virus |
| TFE | = | Trifluoroethanol |
| Tm | = | Main transition temperature |
| TM | = | Transmembrane domain |
Introduction
In late 2002, the emergence of a form of pneumonia called severe acute respiratory syndrome (SARS) was attributed to a previously unknown coronavirus termed SARS-CoV Citation[1–4]. Similar to other coronavirus, SARS-CoV membrane fusion is mediated by the viral spike glycoprotein located on the viral envelope. Trimers of the spike protein form the peplomers that protrude from the virion giving these viruses their distinctive crown-like appearance. SARS-CoV S glycoprotein seems to be cleaved into S1 and S2 subunits in Vero-E6 infected cells Citation[5], but cleavage is not an absolute requirement for the mechanism of fusion Citation[6]. Additionally, infection mediated by the SARS-CoV protein can be inhibited by specific inhibitors of the endosomal protease cathepsin L Citation[7] suggesting that protein cleavage might play an important role activating spike protein for membrane fusion.
SARS-CoV infection, similarly to other envelope viruses, is achieved through fusion of the lipid bilayer of the viral envelope with the host cell membrane Citation[8], although recent studies have shown that the entry of SARS-CoV into the cell may be through pH- and receptor-dependent endocytosis Citation[9]. Interestingly, raft microdomains have been also shown to be involved in virus entry Citation[9]. The S1 subunit forms a globular head and is responsible for recognition of the host cell receptors Citation[10], which for the SARS-CoV has been identified as angiotensin converting enzyme 2 (ACE2) and CD209L Citation[11], Citation[12]. The S2 subunit is responsible for the fusion between viral and cellular membranes Citation[13]. S2 sequence is highly conserved in coronavirus and contains different functional regions critical for membrane fusion as occurs in other class I viral fusion proteins (see ): a stretch of about 20 residues situated downstream of the minimum putative cleavage site and considered the fusion peptide Citation[14], Citation[15], a second fusion peptide (helper?) for the fusion peptide located immediately upstream of the HR1 regionCitation[16], Citation[17], two heptad repeat regions HR1 and HR2 Citation[18] and a stretch of about 13–18 residues that precedes the TM domain at the C terminus, i.e., the pretransmembrane domain Citation[16], Citation[19], Citation[20].
Figure 1. Hydrophobic moment, hydrophobicity, interfaciality distribution, and relative position of the SARSL peptide used in this study along the SARS-CoV spike S2 domain, assuming it forms an α-helical wheel Citation[16]. Only positive bilayer-to-water transfer free-energy values are depicted (the darker, the higher). The scheme of the structure of SARS-CoV spike glycoprotein S2 (amino acid residues 758–761 to 1255 for the full-length) according to literature consensus is also shown. The relevant functional regions are highlighted: the putative fusion peptide (FP), the internal fusion peptide (IFP), the predicted heptad repeat regions pertaining HR1, HR2, and the pre-transmembrane (PTM), transmembrane (TM) domains Citation[14], Citation[16], Citation[19], Citation[20], Citation[34]. The sequence of the peptide used in this work is also shown.
![Figure 1. Hydrophobic moment, hydrophobicity, interfaciality distribution, and relative position of the SARSL peptide used in this study along the SARS-CoV spike S2 domain, assuming it forms an α-helical wheel Citation[16]. Only positive bilayer-to-water transfer free-energy values are depicted (the darker, the higher). The scheme of the structure of SARS-CoV spike glycoprotein S2 (amino acid residues 758–761 to 1255 for the full-length) according to literature consensus is also shown. The relevant functional regions are highlighted: the putative fusion peptide (FP), the internal fusion peptide (IFP), the predicted heptad repeat regions pertaining HR1, HR2, and the pre-transmembrane (PTM), transmembrane (TM) domains Citation[14], Citation[16], Citation[19], Citation[20], Citation[34]. The sequence of the peptide used in this work is also shown.](/cms/asset/98c28d25-da45-4207-8442-aa5619cac96d/imbc_a_392792_f0001_b.gif)
Membrane fusion proteins share common motifs functioning as an integrated unit. In the case of the HIV gp41 envelope protein, as well as in other fusion proteins of other retroviruses, it has been shown that the presence of a conserved immunodominant domain in the region of the loop separating the two heptad repeats that reverse the polypeptide chain Citation[21–24]. This region seems important in the stabilization of the trimeric helical hairpin either in aqueous solution or in the presence of membranes Citation[25]. Moreover, it has been shown that mutations in the loop region of gp41 affect the formation and dilation of fusion pores that are permeable to relatively small soluble molecules suggesting a role of this region in late steps of the membrane fusion process Citation[26]. Finally, different peptides corresponding to this region have shown membrane binding and membrane perturbing capabilities Citation[27–29], and therefore the interaction of this domain with the target cell membrane may be of key importance in the viral fusion mechanism. In the case of the SARS-CoV S glycoprotein, it was found a highly hydrophobic and interfacial domain flanked by the HR1 and HR2 regions Citation[16] (), and different peptides pertaining to this domain have shown a significant leakage effect Citation[16] and an important SARS-CoV plaque formation inhibition Citation[30]. These results support a role of this region in the fusion process such as it has been shown for the loop domain of HIV-1 gp41. Apart from that, while much progress has been made in understanding the membrane fusion mechanism of different viruses, available data concerning coronaviruses, particularly in the case of SARS-CoV, are scarce. Therefore, the purpose of the present work was to gain an insight into the implication of the SARS-CoV loop domain in the membrane interaction.
We have studied a 23-amino acid sequence, peptide SARSL, pertaining to this domain, both in aqueous solution and in the presence of different membrane model systems, to evaluate its incorporation and location in membranes, determine its secondary structure and to study its effect on the integrity of the membrane. The structural and dynamical changes which take place in both the SARSL peptide and the phospholipid molecules induced by its interaction with the lipid bilayer have been characterized using monolayer penetration and fluorescence and infrared spectroscopy. Our results show that SARSL strongly partitions into phospholipid membranes and organizes differently in lipid environments, displaying membrane-activity modulated by the lipid composition of the membrane. These data suggest that the regions where the peptide SARSL resides could be involved in the merging of the viral and target cell membranes.
Materials and methods
Materials and reagents
A 23-residue peptide, SARSL, pertaining to the S2 domain of SARS-CoV (1073REGVFVFNGTSWFITQRNFFSPQ1095, with N-terminal acetylation and C-terminal amidation) was obtained from Genemed Synthesis (San Francisco, CA, USA). The peptide was purified by reversed-phase HPLC (Vydac C-8 column, 250×4.6 mm, flow rate 1 ml/min, solvent A, 0.1% trifluoroacetic acid, solvent B, 99.9 acetonitrile and 0.1% trifluoroacetic acid) to better than 95% purity, and its composition and molecular mass were confirmed by amino acid analysis and mass spectroscopy. Since trifluoroacetate has a strong infrared absorbance at approximately 1,673 cm−1, which interferes with the characterization of the peptide amide I band Citation[28], residual trifluoroacetic acid, used both in peptide synthesis and in the high-performance liquid chromatography mobile phase, was removed by several lyophilization-solubilization cycles in 10 mM HCl. Egg phosphatidylCholine (EPC), egg phosphatidylglycerol (EPG), egg phosphatidic acid (EPA), bovine brain phosphatidylserine (BPS), Cholesterol (Chol), 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1,2-Dimiristoyl-sn-glycero-3- phosphatidylglycerol (DMPG), 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylserine (POPS), 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG) and 1-Palmitoyl-2-oleoyl-sn-glycero-3- phosphatidylcholine (POPC) were obtained from Avanti Polar Lipids (Alabaster, AL, USA). 5-Carboxyfluorescein (CF), (>95% by HPLC), deuterium oxide (99.9% by atom), Triton X-100, EDTA, and HEPES were purchased from Sigma-Aldrich (Madrid, ES, EUR). All other reagents used were of analytical grade from Merck (Darmstad, Germany, GER, EUR). Water was deionized, twice-distilled and passed through a Milli-Q equipment (Millipore Ibérica, Madrid, ES, EUR) to a resistivity higher than 18 MΩ cm.
Liposome preparation
Aliquots containing the appropriate amount of lipid in chloroform-methanol (2:1 vol/vol) were placed in a test tube, the solvents were removed by evaporation under a stream of O2-free nitrogen, and finally, traces of solvents were eliminated under vacuum in the dark for >3 h. The lipid films were resuspended in an appropriate buffer and incubated either at 25°C or 10°C above the phase transition temperature (Tm) with intermittent vortexing for 30 min to hydrate the samples and obtain multilamellar vesicles (MLV). The samples were frozen and thawed five times to ensure complete homogenization and maximization of peptide/lipid contacts with occasional vortexing. Large unilamellar vesicles (LUV) with a mean diameter of 0.1 µm (leakage measurements) were prepared from multilamellar vesicles by the extrusion method Citation[31] using polycarbonate filters with a pore size of 0.1 µm (Nuclepore Corp., Cambridge, CA, USA). The phospholipid and peptide concentration was measured by methods described previously Citation[32], Citation[33].
Peptide binding
Peptide partitioning into membranes was evaluated by the change of the tryptophan fluorescence by successive additions of small volumes of LUVs to the peptide sample (2.67×10−5 M). Fluorescence spectra were recorded in a SLM Aminco 8000 spectrofluorimeter with excitation and emission wavelengths of 290 and 348 nm, respectively, and 4 nm spectral bandwiths. Measurements were carried out in 20 mM HEPES, 50 mM NaCl, EDTA 0.1 mM, pH 7.4. Intensity values were corrected for dilution, and the scatter contribution was derived from lipid titration of a vesicle blank. Partition coefficients Kp and fluorescence anisotropies <r> were obtained as described previously Citation[19], Citation[28], Citation[34].
Fluorescence quenching
Fluorescence quenching of the Trp emission by acrylamide and fluorescence quenching of the lipophilic probes 5NS or 16NS were performed as described previously Citation[19], Citation[34–36].
Leakage measurement
LUVs with a mean diameter of 0.1 µm were prepared as indicated above in buffer containing 10 mM Tris, 20 mM NaCl, 0.1 mM EDTA, pH 7.4, and CF at a concentration of 40 mM. Non-encapsulated CF was separated from the vesicle suspension through a Sephadex G-75 filtration column (Pharmacia, Uppsala, Sweden) eluted with buffer containing 10 mM Tris, 0.1 M NaCl, 1 mM EDTA, pH 7.4. Leakage of intraliposomal CF was assayed by treating the probe-loaded liposomes (final lipid concentration, 0.125 mM) with the appropriate amounts of peptide on microtiter plates using a microplate reader (FLUOstar; BMG Labtech, Offenburg, Germany), each well containing a final volume of 170 µl stabilized at 25°C. The medium in the microtiter plates was continuously stirred to allow the rapid mixing of peptide and vesicles. One hundred percent release was achieved by adding Triton X-100 to the microtiter plates to a final concentration of 0.5% (wt/wt). Fluorescence measurements were made initially with probe-loaded liposomes, then by adding peptide solution, and finally 100% release was achieved by adding Triton X-100 to the microtiter plates (final concentration of 0.5% (wt/wt)). Leakage was quantified on a percentage basis as previously described Citation[37].
Insertion of peptides into lipid monolayers
Insertion of peptides into lipid monolayers, residing on an air/water interface, was measured using magnetically stirred circular Teflon wells (Multiwell plate, subphase volume 3 ml, Kibron Inc., Helsinki, FIN). Surface pressure (π) was monitored with a Wilhelmy wire attached to a microbalance (DeltaPi, Kibron Inc., Helsinki, FIN, EUR) interfaced to a computer. Lipids were mixed at the indicated molar ratios in chloroform (approximately 1 mM) and then spread onto the air-buffer interface (20 mM HEPES, 0.1 mM EDTA, pH 7.4). The lipid monolayers were allowed to equilibrate for approximately 15 min at different initial surface pressures (π0) before the injection of the peptides into the subphase. The increment in π after peptide addition was complete in approximately 30 min and the difference between the initial surface pressure (π0) and the value observed after the penetration of peptide into the films was taken as Δπ. The data shown represent the average from triplicate measurements and are represented as Δπ vs. π0. These graphs yield the critical surface pressure πc corresponding to the lipid lateral packing density preventing the intercalation of the peptides into the lipid films. All measurements were performed at ambient temperature (25°C).
Infrared spectroscopy
Aliquots containing the appropriate amount of lipid in chloroform/methanol (2:1, v/v) were placed in a test tube containing 200 µg of dried lyophilized peptide. After vortexing, the solvents were removed by evaporation under a stream of O2-free nitrogen, and finally, traces of solvents were eliminated under vacuum in the dark for more than 3 h. The samples were hydrated in 200 µl of D2O buffer containing 20 mM HEPES, 50 mM NaCl, 0.1 mM EDTA, pH 7.4 and incubated at 10°C above the phase transition temperature (Tm) of the phospholipid mixture with intermittent vortexing for 45 min to hydrate the samples and obtain multilamellar vesicles (MLV). The samples were frozen and thawed five times to ensure complete homogenization and maximization of peptide/lipid contacts with occasional vortexing. Finally the suspensions were centrifuged at 15000 rpm at 25°C for 15 min to remove the possibly peptide unbound to the membranes. The pellet was resuspended in 25 µl of D2O buffer and incubated for 45 min at 10°C above the Tm of the lipid mixture, unless stated otherwise. Approximately 20 µl of the pelleted sample were placed between two CaF2 windows separated by a 56-µm-thick Teflon spacer in a liquid demountable cell (Harrick, Ossining, NY). The spectra were obtained in a Bruker IFS66s FTIR spectrometer using a deuterated triglycine sulfate detector. Each spectrum was obtained by collecting 200 scans with a triangular function at a resolution of 2 cm−1. The spectrometer was continuously purged with dry air at a dew point of −40°C to remove atmospheric water vapor from the bands of interest. Once the sample was mounted in the holder of the instrument, it was equilibrated at the lowest temperature used for at least 20 min before acquisition. An external bath circulator, connected to the infrared spectrometer, controlled the sample temperature.
Time-resolved Fluorescence Spectroscopy
Fluorescence intensity and anisotropy decays were obtained by the single-photon timing technique. For Trp excitation at λ = 295 nm, a cavity-dumped frequency doubled dye laser of Rhodamine 6G was described elsewhere Citation[38]. For DPH excitation at 375 nm, a solid state Ti:sapphire laser in a setup previously described Citation[39] was employed. The emission wavelength (350 nm for Trp and 430 nm for DPH) was selected by a Jobin-Yvon H329 monochromator (Horiba Jobin-Yvon, Longjumeau, FR-EU). For other details see references above. For the anisotropy decays, the parallel (vertically polarized) and the perpendicular (horizontally polarized) components of the fluorescence decay were separately collected with the same acquisition time. Background decays were obtained and subtracted, although the average count rate was always less than 3% of the sample. The time-scales ranged from 7.5–14.0 ps/channel for Trp and from 20.0–57.6 ps/channel for DPH. For other details on the time-resolved anisotropy measurements and analysis see Citation[40].
Hydrophobic moment, hydrophobicity, and interfaciality
The hydrophobic moment calculations were carried out according to Eisenberg Citation[41] and the scale for calculating hydrophobic moments was taken from Engelman Citation[42]. Hydrophobicity and interfacial values, i.e., whole residue scales for the transfer of an amino acid of an unfolded chain into the membrane hydrocarbon palisade and the membrane interface respectively, have been obtained from Citation[43]. Each specific value in the two-dimensional plot represents the mean of the values pertaining to the hydrophobic moment, hydrophobicity and interfaciality of the amino acid at that position and its neighbours Citation[16]. Positive values correspond to positive bilayer-to water transfer free energy values and therefore, the higher the value, the greater the probability to interact with the membrane surface and/or the hydrophobic core.
Results
Binding of SARSL peptide to lipid vesicles
The ability of the SARSL peptide to interact with membranes was determined from fluorescence studies of the peptide intrinsic Trp in presence of model membranes containing different phospholipid compositions at different lipid/peptide ratios. The Trp fluorescence emission intensity of the SARSL peptide decreased upon increasing the lipid/peptide ratio, indicating a significant change in the environment of the Trp moieties of the peptide (A). The change on the Trp fluorescence of the peptide allows obtaining its partition coefficient, Kp, for different phospholipids (summarized in ). Kp values in the range 106–107 were obtained, indicating that the peptide was bound to the membrane surface with high affinity. Slightly higher Kp values were obtained for negatively charged phospholipid-containing bilayers, EPG, EPA and BPS, than liposomes containing the neutral phospholipid EPC. These results were further corroborated by the displacement in the maximum emission wavelength of Trp in the presence of phospholipids LUVs. In buffer, the peptide had an emission maximum at 345 nm, typical for Trp in a polar environment, whereas, in the presence of increasing concentrations of membranes, the emission maximum presented a shift of about 4–6 nm to shorter wavelengths in negatively charged liposomes, but only 1 nm for zwitterionic ones. Differences in the fluorescence anisotropy values of the SARSL peptide were also observed when in the presence of phospholipid model membranes. It was found that addition of LUVs to the peptide increased significantly the anisotropy values (), indicating an eventual decrease in the mobility of the Trp residue. The increase in anisotropy was lower in the zwitterionic phospholipid EPC than in the presence of the negatively charged phospholipids BPS, EPG and EPA. The SARSL peptide has a positive net formal charge of +1, so that an electrostatic effect might be the reason to observe relatively high binding and anisotropy values for compositions containing negatively charged phospholipids. However, it is not only the electrostatic attraction that is playing a role, since, as shown below, the peptide affects more significantly membranes containing zwitterionic phospholipids rather than those containing negatively-charged ones. The peptide mobility will be addressed in more detail later in the article, in the framework of the time-resolved data.
Figure 2. (A) Change on the Trp fluorescence intensity of the SARSL peptide in the presence of increasing lipid concentration, (B) Stern-Volmer plots of the quenching of the Trp fluorescence emission of SARSL by acrylamide in aqueous buffer (∇), and in the presence of LUVs, (C) Doxyl NS quenching (5NS, open symbols, and 16NS, black symbols) of the Trp fluorescence of the SARSL peptide in the presence of LUVs at different lipid compositions. The lipid compositions used were EPC (▪), BPS (▴), EPG (•), EPA (♦). The spin label concentration is the effective quencher concentration calculated as stated in the Material and Methods section.
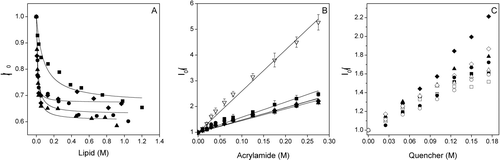
Table I. Partition coefficient (Kp), spectral shift (Δλ), anisotropy value, acrylamide Stern-Volmer (KSV) quenching constant obtained from the fluorescence of the Trp residue of the SARSL peptide in buffer, in the presence of different model membranes.
Location and penetration of SARSL in the bilayer
To assess the accessibility of the Trp residue of the SARSL peptide to the aqueous environment before and after binding to membranes having different phospholipid compositions, the water-soluble quencher acrylamide, an efficient neutral quencher probe Citation[34] was used. Stern-Volmer plots for the quenching of Trp by acrylamide, recorded in the absence and presence of lipid vesicles, are shown in B and the resultant Stern-Volmer constants are presented in . The data reveal a weaker decrease in the fluorescence intensity in the presence of liposomes, indicating that the Trp residue was less accessible to the quencher in the presence of LUVs. The KSV value for the zwitterionic phospholipid EPC was slightly higher than those found in the presence of negatively charged phospholipids. As observed in B, the plots are linear with a unitary intercept showing that the Stern-Volmer dynamic quenching formalism describes accurately the data (it will be shown later that the variations in the peptide fluorescence lifetime do not affect this result). The transverse location (penetration) of the SARSL peptide in the lipid bilayer was evaluated by monitoring the relative quenching of the fluorescence of the Trp residue by the lipophylic spin probes 5NS and 16NS when the peptide was incorporated in the fluid phase of vesicles having different phospholipid compositions (C). It can be seen that, in general, and for each one of the different membrane compositions studied, the SARSL peptide was quenched more efficiently by 16NS, quencher for molecules near or at the interface, than by 5NS, quencher for molecules buried deeply in the membrane allowing to conclude that SARSL remained close to the lipid/water interface. It can be seen that in general, and for each one of the different membrane compositions studied, although the SARSL peptide was quenched slightly more efficiently by 16NS, the difference for the two probes is not very high. This allows the conclusion that Trp resides in between these probes.
Fluorescence intensity decay of the peptide
All the parameters describing the peptide fluorescence intensity decay are given in . As expected for Trp Citation[34], the decays are described by the sum of three exponentials, with typical values for the components lifetimes. The mean fluorescence lifetime at 25°C is around 2 ns, also a typical behavior for Trp residues. However, the lifetime-weighted quantum yield had an unusual behavior. In the absence of static quenching, this parameter should be proportional to the steady-state fluorescence intensity (to the quantum yield), and it is expected to decrease monotonically with raising temperature. As shown in , this is not the case, because the amplitude averaged lifetime increases from 25–50°C. This means that in addition to thermal deactivation of the excited state, there should be another factor contributing to the lower quantum yield at 25°C. For the peptide in solution, this suggests that the peptide conformation and/or aggregation state changes between 25 and 50°C. Either the peptide becomes denatured, or an aggregate is destroyed at the higher temperature, taking a Trp quencher side-chain away from the vicinity of the indole ring. Upon interacting with lipid bilayers of different compositions, the parameters describing the fluorescence intensity decays of the peptide tend to change, but not dramatically Citation[27], Citation[28], Citation[35]. The general trend observed here is a slight increase of both the amplitude-weighted and the intensity-weighted average lifetimes, except for EPC, for which the interaction with the peptide is weaker and the changes on peptide/structure/dynamics are smaller. The slight increase observed for the amplitude-weighted lifetime in all the anionic membranes studied is at variance with the marked decrease observed for the steady-state fluorescence intensity. This effect should be due to a strong conformational change and/or extensive aggregation of the peptide in the membrane. The fact that the amplitudes and the lifetime components change only mildly, and also that the order of the relative amplitudes was kept, suggests that a strong conformational change is not taking place, and favors an interpretation of the data as an extensive aggregation upon interaction with the membrane.
Table II. Fluorescence lifetime components τi, normalized amplitudes αι, and fluorescence mean lifetime (amplitude weighted, , and intensity weighted, <τ > ) of the Trp residue of the SARSL peptide in buffer, in the presence of phospholipid model membranes at 25°C. The reduced χ2 values of the fits to the experimental fluorescence decays are also given. The peptide/lipid ratio was 1:30.
Time-resolved fluorescence anisotropy of SARSL peptide
In order to gain more insight into the dynamic behavior of SARSL, as well as information on its aggregation state in water, the fluorescence anisotropy decay of the peptide was obtained in different systems. From visual inspection of the experimental fluorescence anisotropy decays (not shown), it could be observed that the fluorescence anisotropy at very long times tended to zero in buffer solution, and to a small value in zwitterionic membrane (EPC). In addition, the dynamic component was slower for the peptide interacting with membranes than in water solution, and in the case of anionic membranes, even at very long times, the fluorescence anisotropy had a significant positive value. The decay of the anisotropy starts in general at a value lower than the fundamental anisotropy of Trp at the excitation wavelength used, which is r0~0.2 Citation[44]. This is probably due to a very fast wobbling motion of the indole moiety of the Trp residue that is unresolved, especially in the case of the lipid containing samples, due to stronger scattering. For the peptide in buffer, the results of the analyses of the anisotropy decays are shown in . A very fast component (<100 ps) had to be included in the analysis in order to obtain a low chi-square and random distribution of residuals, but because it could not be accurately determined, it is not presented in the Table. The rotation corresponding to the longer rotational correlation time was responsible for the remaining depolarization. In order to determine if this was due to some kind of segmental motion or due to the rotation of the whole peptide, the volume of the peptide was calculated from its molecular mass Citation[45]. In this way, a volume of 3402 Å3/molecule was obtained. In the predicted rotational correlation time for a sphere of this volume rotating in water is also shown. It is clear that both at 15°C and 25°C, the experimental value of the rotational correlation time corresponds to a peptide dimer. At these two temperatures the peptide has probably the same molecular organization. This result correlates well with the tendency of the peptide to form aggregates in which a quenching of the Trp residue fluorescence takes place, and that those aggregates are partially or totally destroyed when reaching 50°C. The fluorescence anisotropy decay in membranes is clearly different from those in water. The parameters retrieved from the analysis of those decays are given in . It is clear from the data displayed in that the rotational dynamics of the peptide is different in the several systems. However, the most significant differences (considering the error associated to the parameters majored by 15%, not shown) are a smaller value of r∞ for EPC, and a longer rotational correlation time for EPA (in the case of EPG, there is a broad surface minimum for which r∞ could be slightly higher and φ slightly shorter, which would be effectively closer to the φ of BPS/EPC and r∞ closer to BPS; in the EPG system the average lifetime is longer than in BPS and this could also allow a better definition of the dynamic part, and the retrieval of a smaller r∞). Focusing on the two major differences mentioned, the anisotropy decay in EPC can be interpreted in a similar way to the one observed in water, but in a medium of higher viscosity (slower rotational correlation time), and also with some hindering (non-zero anisotropy for long times), but the peptide seems to be loosely associated to the membrane. For the other lipids, this is not the case. The rotational dynamics of the Trp residue is severely hindered. Taking the fact that no strong structural alterations in the vicinity of the Trp residue are concluded from the fluorescence intensity decay parameters (), together with the anisotropy decay data, this point to the formation of large aggregates that have only limited motion in the time scale of the fluorescence emission. In the case of EPA, the rotational correlation time is significantly longer. This may be due to the establishment of strong interactions at the level of the small and the less hydrated EPA headgroup that slow down the Trp residue rotational motions as it anchors the peptide at the membrane/water interface. This is fully consistent with the larger spectral shift (low hydration) observed for the peptide in the presence of EPA, but also larger water accessibility (less protection by the small phospholipid headgroup). The KSV value is actually the product of the bimolecular quenching rate constant times the average fluorescence lifetime of the fluorophore in the absence of quenching. In this way, it is important to check if there are significant variations in that parameter in order to compare quencher accessibility from KSV values. By dividing KSV () by <τ> (), it is verified that the water accessibility decreases in the order EPC > EPA > BPS~EPG.
Table III. Long rotational correlation time φ from the anisotropy decay of the peptide SARSL in buffer solution at different temperatures. The global chi-square is also given. For comparison, the rotational correlation time for different aggregates of the SARSL peptide at those temperatures is also indicated. The inclusion of a very short rotational correlation time (<100 ps) was necessary to account for the initial depolarization, but its value could not be accurately determined.
Table IV. Fitting parameters for the anisotropy decay of the peptide SARSL (rotational correlation time φ, associated depolarization β, and limiting anisotropy r∞) in lipid membranes of several compositions at 25°C. The global chi-square is also given.
Membrane leakage
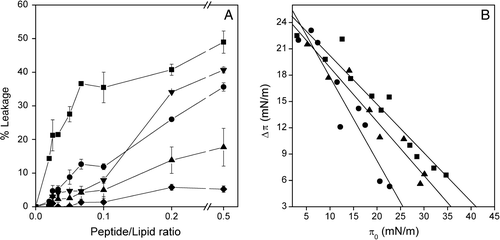
In order to further explore the possible interaction of the SARSL peptide with phospholipid model membranes, we studied the effect of the peptide on the release of encapsulated fluorophores trapped inside LUVs. The extent of leakage observed at different lipid-to-peptide ratios and the effect of lipid composition is shown in A, where it can be seen that the peptide was able to induce the release of the internal contents of the liposomes in a dose-dependent manner. In the presence of liposomes containing zwitterionic phospholipids, the SARSL peptide induced a significant leakage value, whereas in liposomes containing negatively-charged phospholipids much lower leakage values were found. At the lowest lipid/peptide ratio used, the highest percentages of leakage (49%, 41% and 36%) were observed for EPC, EPC/EPG/Chol (5:3:1) and EPG, respectively. Lower leakage values were found in the presence of EPC/BPS/Chol (5:3:1) (18%) and EPC/EPA/Chol (5:3:1) (5%) (A). Interestingly, at the lower peptide/lipid ratio used and more usual in the normal context of protein-membrane scenario, the membrane containing only zwitterionic phospholipid had a significant effect in the probe release (A). These results indicate that the peptide affects more significantly membranes containing zwitterionic phospholipids rather than those containing negatively-charged ones.
Figure 3. (A) Effect of the SARSL peptide on membrane rupture of LUVs at different lipid compositions, at different lipid-to-peptide molar ratios. The lipid compositions used were EPC (▪), EPC/BPS/CHOL at a molar ratio of 5:3:1 (▴), EPG (•), EPC/EPG/CHOL at a molar ratio of 5:3:1 (▾), EPC/EPA/Chol at a molar ratio of 5:3:1 (♦). (B) Insertion of the SARSL peptide into lipid monolayers. Increments in surface pressure (Δπ) of lipid monolayers due to the addition of SARSL peptide into the subphase is illustrated as a function of the initial pressure (π0). The lipid compositions used were POPC (•), POPS (▴), POPG (▪).
Monolayer insertion of peptide
The intercalation of the SARSL peptide in lipid monolayers with different initial pressures π0 was observed by measuring the increment in surface pressure (Δπ) following the addition of the peptides into the subphase. As it can be seen in B, SARSL readily intercalated into the POPC film with a πc of 41 mN/m, but POPS or POPG attenuated the peptide insertion with πc values observed at approximately 35 and 25 mN/m, respectively.
Perturbation induced by the peptide in the lipid palisade
The fluorescence intensity decay of the membrane probe DPH was obtained in order to evaluate the perturbation induced by the peptide in the membrane hydrophobic core. It was previously verified for other membrane active peptides that a superficial location is also compatible with strong perturbation in the lipid core (e.g., Citation[34]). If the environment of the DPH chromophore is more hydrated this will readily lead to a concomitant decrease of the amplitude-averaged and/or the intensity averaged fluorescence lifetimes. In , the parameters describing the fluorescence intensity decay of DPH are shown for a zwitterionic lipid (POPE), and an anionic one (DMPG). The values were obtained both below (15°C) and above (35°C) the main transition temperature of the phospholipids (25°C for POPE Citation[46] and 23°C for DMPG Citation[36]), i.e., in the gel and in the fluid phase, respectively, and in the absence and in the presence of the SARSL peptide. In the absence of the peptide, the fluorescence decay of DPH is always bi-exponential, except for DMPG at 15°C, where a longer component of ~12 ns is the major one, giving rise to a longer lifetime-weighted quantum yield and average lifetime. This corresponds to the gel phase of the saturated phospholipid, for which the order and rigidity is kept deeper along the acyl chains. In the presence of the SARSL peptide, the decay becomes in general more complex. An intermediate component either appears or becomes more important to the total decay, with a consequent decrease of the intensity and amplitude averaged fluorescence lifetimes of DPH. This shows that in all cases, the peptide perturbs the membrane in such a way that it is sensed in the hydrophobic core. The effect of the peptide in the average lifetimes is similar (though lesser) to the effect of the fusion peptide of the SARS-CoV in the same or similar lipids Citation[34]. However, the effect on the lifetime components is different. In the presence of the SARS-CoV fusion peptide the decay was more complex, originating a short (sub-nanosecond) component with an amplitude of more than ~30%, and, e.g., the amplitude averaged lifetime becomes in every case <3 ns. In the case of the SARSL peptide, the stronger effect is felt on the intermediate component, and although this is related to an increase in the polarity of the environment of the DPH fluorophore, there is no direct contact with water (that would lead to a component < 0.5 ns), and in fact the SARSL peptide does not perturb the bilayer structure in a way as drastic as the fusion peptide does, as expected from the proposed function of the fusion and the loop peptides. Another interesting feature is that in the case of the fusion peptide, the relative effect on the average lifetimes was stronger in the anionic lipid both below and above Tm. In the case of the SARSL peptide, it is clear that below Tm, the effect is much more pronounced in DMPG, whereas above Tm, it is in POPE membranes that the effect is more marked.
Table V. Fluorescence lifetime components τι, normalized amplitudes αι, and fluorescence mean lifetimes (amplitude weighted, , and intensity weighted, <τ > ) of DPH in phospholipid model membranes in the absence, in the presence of the SARSL peptide below, above the phospholipid Tm. * From ref. Citation[34].
Secondary structure of SARSL peptide
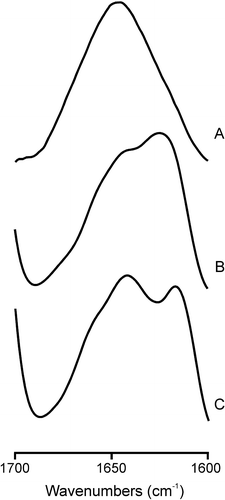
The existence of structural changes on the SARSL peptide induced by membrane binding was studied by infrared spectroscopy. For the peptide in solution, the Amide I’ band was quite symmetric with a maximum at about 1646 cm−1, implying that the peptide consisted mainly of a mixture of helical and unordered structures Citation[47] (A). However, a significant change in the Amide I’ envelope was observed in the presence of model membranes of EPC (B) and EPG (C). In the presence of EPC, the Amide I’ band consisted of a main band at about 1625 cm−1 and a shoulder at about 1642 cm−1, whereas in the presence of EPG two bands of similar intensity appearing at about 1641 and 1618 cm−1 were observed. Since bands at about 1625 cm−1 can be assigned to β-sheet structures, whereas bands at about 1618 cm−1 correspond to aggregated ones, these data would imply that the SARSL peptide structure changed from a mixture of helical and unordered structures to a mixture of helical, unordered and β-sheet structures in the presence of EPC and a mixture of helical, unordered and aggregated structures in the presence of EPG. These results imply that the secondary structure of the SARSL peptide was affected by the phospholipid composition of the membrane.
Discussion
Enveloped viruses use membrane fusion proteins in order to juxtapose and merge the viral and cellular membranes. The viral fusion protein of SARS-CoV is the envelope Spike glycoprotein and the S2 domain is the responsible for fusion. Recent studies point to the fact that there are several regions within Class I and Class II membrane fusion proteins which are involved in the interaction with the membrane to accomplish fusion Citation[48–51]. Although in other Class I fusion proteins plentiful data have been obtained to understand the implication of these membrane-interacting segments in the fusion mechanism, available information concerning the possible membrane-active regions in coronavirus, and particularly in the case of SARS-CoV, is scarce. Many studies performed to examine the interaction of synthetic peptides mimicking functional regions have been shown to be very valuable in the understanding of membrane viral fusion. Several peptides have been identified in the SARS-CoV spike glycoprotein as belonging to membrane interacting regions Citation[14], Citation[16], Citation[19], Citation[20], Citation[34]. In this work we have focused on the possible roles of the loop region in the membrane fusion process. We have selected a specific segment from the SARS CoV sequence which corresponds to the loop region, SARSL, to carry out an in-depth biophysical study aimed at elucidating the capacity of this region to interact and disrupt membranes, as well as to study the structural and dynamic features which might be relevant for that disruption.
From the methodological point of view, it is noteworthy that the screening of the protein sequence taking into account hydrophobic moment, hydrophobicity and interfaciality allowed a precise identification of the membrane-active sequences, and in addition provided insight into their functions. In general, the experimental biophysical studies conducted corroborate the putative functions of those sequences, at the same time revealing new and less expected features that contributed to the better understanding of the molecular mechanism of infection by SARS-CoV ().
The SARSL peptide binds with high affinity to model membranes, although it showed a higher affinity for anionic-phospholipid compositions than those containing zwitterionic phospholipids. These results were further corroborated by the study of the change in the maximum emission wavelength of the Trp residue, as well as the increase in anisotropy in the presence of model membranes. The greater affinity for anionic phospholipids rather than for zwitterionic ones was already expected, due to the positive net charge of the peptide. The interaction with membrane vesicles decreased the Trp quenching by acrylamide as illustrated by the low KSV values obtained, indicating a low accessibility of the quencher to the peptide. The lipophylic probe quenching results suggest a shallow-to-intermediate location of the peptide in the membrane, but the extent of quenching is relatively low compared to other peptides Citation[19], Citation[52], Citation[53]. This fact and the decrease in fluorescence suggest a possible aggregation of the peptide when bound to the membrane. The fluorescence intensity decay of the peptide supports the experiments and conclusions commented above.
Time-resolved fluorescence anisotropy indicates the formation of a peptide dimer in solution and the formation of large aggregates in the presence of membrane vesicles. Recently, it has been found that the loop region of the gp41 from SIV increases the stability of the trimeric helical hairpin, both in solution and in the presence of membranes Citation[25]; interestingly, mutations in this region almost abolished the formation of syncytia between transfected HEK293T cells and target HeLa-P4 cells Citation[26]. Therefore, the loop region might mediate the interactions between different gp41 trimers facilitating the completion of later steps of the membrane fusion process. The formation of a dimer in solution and larger aggregates in the presence of membranes suggests a similar role of the loop region of the spike glycoprotein of SARS-CoV. Moreover, the formation of aggregates by the SARSL peptide is supported by the FTIR data, being observed an increase of aggregated structures at the expense of the α-helical structure content in the presence of either phospholipids or the lipid mimetic TFE (not shown).
We have also shown that SARSL peptide is capable of affecting the membrane integrity shown by the release of encapsulated probes as well as by perturbation of the environment of the DPH fluorophore in LUVs, being the highest effect observed for neutral phospholipids. These experiments are in agreement with monolayer experiments, because the higher increase in πc is obtained in the presence of neutral phospholipids, indicating a higher change in the superficial tension of the monolayer.
Recently, it has been shown the inhibition of SARS-CoV plaque formation by loop pertaining peptides and on this basis it has been hypothesized that these peptides might hamper the loop by a steric interaction, preventing either the initial extension of the fusion protein, the transition to the six-helix bundle state, or the interaction of the S2 subunit with the cellular membrane Citation[30]. Our results would support this notion and add new information about how this region can contribute to the interaction with the membrane. Moreover, the binding of the peptide to the membrane and the modulation of the lipid biophysical properties could be related to the conformational changes which might occur during the activity of the SARS-CoV S protein. Although the peptide is not deeply buried in the membrane, it is able to affect the lipid milieu from the membrane surface down to the hydrophobic core. It is known that, in the case of Class I membrane fusion proteins, several fragments pertaining to different trimers can promote the formation of local nipples in the cell membrane leading to the formation of local bends which could induce zones of non-lamellar phases on the outer leaflet Citation[54]. Therefore, the region where the SARS-CoV SARSL peptide is located could interact with the negatively-charged leaflet favoring the fusion process, and leading to the fusogenic pore formation, similarly to other Class I proteins Citation[28].
The present data would suggest that disassembly of the membrane core could possible happen only after exposure to negatively charged membranes, either by flip-flop or when in contact with the inner leaflet, i.e., by making small holes in the outer leaflet, as shown by the very short component induced in the fluorescence decay of DPH by the SARS-CoV fusion peptide Citation[34]. The SARSL would thus have a more primordial role because importantly it interacts strongly and specifically with zwitterionic lipids whereas the SARSFP interacts only weakly and non-specifically (e.g., the Kp of SARSL to EPC is ~106 () and for SARSFP the interaction was too weak to allow determination of a Kp value Citation[34]. In this way the transverse asymmetry of the target cell membrane, which is challenged by the perturbation induced by the SARSL peptide in the outer leaflet, becomes of major importance for the process. In addition, the perturbation of the membrane is somewhat controlled because the membrane core becomes less hydrophobic () but not as much as with the SARSFP. This may be important to prevent fusion to occur randomly. SARSL would have a more facilitator and controller role, whereas SARSFP would have a more effector role in the fusion process.
Acknowledgements
This work was supported by grant BFU2005-00186-BMC (Ministerio de Ciencia y Tecnología, Spain) to J.V. and PTDC/QUI/68151/2006 (Fundação para a Ciência e a Tecnologia, Portugal). J.G. is recipient of pre-doctoral fellowship from the Autonomous Government of the Valencian Community, Spain and a FEBS short-term fellowship. R.F.M. de A. and M.P. acknowledge research grants (POCTI/QUI/57123/2004 and POCTI/QUI/68151/2006) from FCT, Portugal. We are especially grateful to Prof. Paavo Kinnunen and Dr Alexander Fedorov for their collaboration in the monolayer and time-resolved experiments, respectively. Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003; 300: 1394–1399
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1953–1966
- Kunstman KJ, Puffer B, Korber BT, Kuiken C, Smith UR, Kunstman J, Stanton J, Agy M, Shibata R, Yoder AD, et al. Structure and function of CC-chemokine receptor 5 homologues derived from representative primate species and subspecies of the taxonomic suborders Prosimii and Anthropoidea. J Virol 2003; 77: 12310–12318
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003; 361: 1319–1325
- Wu XD, Shang B, Yang RF, Yu H, Ma ZH, Shen X, Ji YY, Lin Y, Wu YD, Lin GM, et al. The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells. Cell Res 2004; 14: 400–406
- Follis KE, York J, Nunberg JH. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology 2006; 350: 358–369
- Huang IC, Bosch BJ, Li F, Li W, Lee KH, Ghiran S, Vasilieva N, Dermody TS, Harrison SC, Dormitzer PR, et al. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem 2006; 281: 3198–3203
- Qinfen Z, Jinming C, Xiaojun H, Huanying Z, Jicheng H, Ling F, Kunpeng L, Jingqiang Z. The life cycle of SARS coronavirus in Vero E6 cells. J Med Virol 2004; 73: 332–337
- Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res 2008; 18: 290–301
- Taguchi F. The S2 subunit of the murine coronavirus spike protein is not involved in receptor binding. J Virol 1995; 69: 7260–7263
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne SA, Somasundaran M, Sullivan JL, Luzuriaga KL, Greenough TC, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003; 426: 450–454
- Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD, Jr, Thackray LB, Young MD, Mason RJ, et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA 2004; 101: 15748–15753
- Xiao X, Chakraborti S, Dimitrov AS, Gramatikoff K, Dimitrov DS. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem Biophys Res Commun 2003; 312: 1159–1164
- Sainz B, Jr, Rausch JM, Gallaher WR, Garry RF, Wimley WC. Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J Virol 2005; 79: 7195–7206
- Guillen J, de Almeida RF, Prieto M, Villalain J. Structural and dynamic characterization of the interaction of the putative fusion peptide of the S2 SARS-CoV virus protein with lipid membranes. J Phys Chem B 2008; 112: 6997–7007
- Guillen J, Perez-Berna AJ, Moreno MR, Villalain J. Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16/18-mer peptide scan: implications for the viral fusion mechanism. J Virol 2005; 79: 1743–1752
- Guillén J, Pérez-Berná AJ, Moreno MR, Villalaín J. A second SARS-CoV S2 glycoprotein internal membrane-active peptide. Biophysical Characterization and Membrane Interaction. Biochemistry 2008; 47: 8214–8224
- Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 2003; 77: 8801–8811
- Guillen J, Moreno MR, Perez-Berna AJ, Bernabeu A, Villalain J. Interaction of a peptide from the pre-transmembrane domain of the severe acute respiratory syndrome coronavirus spike protein with phospholipid membranes. J Phys Chem B 2007; 111: 13714–13725
- Sainz B, Jr, Rausch JM, Gallaher WR, Garry RF, Wimley WC. The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: putative role during viral entry. Biochemistry 2005; 44: 947–958
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell 1997; 89: 263–273
- Caffrey M, Cai M, Kaufman J, Stahl SJ, Wingfield PT, Covell DG, Gronenborn AM, Clore GM. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. Embo J 1998; 17: 4572–4584
- Wang JJ, Steel S, Wisniewolski R, Wang CY. Detection of antibodies to human T-lymphotropic virus type III by using a synthetic peptide of 21 amino acid residues corresponding to a highly antigenic segment of gp41 envelope protein. Proc Natl Acad Sci USA 1986; 83: 6159–6163
- Gallaher WR, Ball JM, Garry RF, Griffin MC, Montelaro WC. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses 1989; 5: 431–440
- Peisajovich SG, Blank L, Epand RF, Epand RM, Shai Y. On the interaction between gp41 and membranes: the immunodominant loop stabilizes gp41 helical hairpin conformation. J Mol Biol 2003; 326: 1489–1501
- Bar S, Alizon M. Role of the ectodomain of the gp41 transmembrane envelope protein of human immunodeficiency virus type 1 in late steps of the membrane fusion process. J Virol 2004; 78: 811–820
- Santos NC, Prieto M, Castanho MA. Interaction of the major epitope region of HIV protein gp41 with membrane model systems. A fluorescence spectroscopy study. Biochemistry 1998; 37: 8674–8682
- Contreras LM, Aranda FJ, Gavilanes F, Gonzalez-Ros JM, Villalain J. Structure and interaction with membrane model systems of a peptide derived from the major epitope region of HIV protein gp41: implications on viral fusion mechanism. Biochemistry 2001; 40: 3196–3207
- Pascual R, Moreno MR, Villalain J. A peptide pertaining to the loop segment of human immunodeficiency virus gp41 binds and interacts with model biomembranes: implications for the fusion mechanism. J Virol 2005; 79: 5142–5152
- Sainz B, Jr, Mossel EC, Gallaher WR, Wimley WC, Peters CJ, Wilson RB, Garry RF. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res 2006; 120: 146–155
- Mayer LD, Hope MJ, Cullis PR. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim Biophys Acta 1986; 858: 161–168
- Böttcher CSF, Van Gent CM, Fries C. A rapid and sensitive sub-micro phosphorus determination. Anal Chim Acta 1961; 1061: 203–204
- Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 1967; 6: 1948–1954
- Guillén J, de Almeida RF, Prieto M, Villalaín J. Structural and dynamic characterization of the interaction of the putative fusion peptide of the S2 SARS-CoV virus protein with lipid membranes. J Phys Chem B 2008; 112: 6997–7007
- Pascual R, Contreras M, Fedorov A, Prieto M, Villalain J. Interaction of a peptide derived from the N-heptad repeat region of gp41 Env ectodomain with model membranes. Modulation of phospholipid phase behavior. Biochemistry 2005; 44: 14275–14288
- Contreras LM, de Almeida RM, Villalain J, Fedorov A, Prieto M. Interaction of alpha-melanocyte stimulating hormone with binary phospholipid membranes: structural changes and relevance of phase behavior. Biophys J 2001; 80: 2273–2283
- Perez-Berna AJ, Guillen J, Moreno MR, Bernabeu A, Pabst G, Laggner P, Villalain J. Identification of the membrane-active regions of hepatitis C virus p7 protein: Biophysical characterization of the loop region. J Biol Chem 2008; 283: 8089–8101
- de Almeida RF, Borst J, Fedorov A, Prieto M, Visser AJ. Complexity of lipid domains and rafts in giant unilamellar vesicles revealed by combining imaging and microscopic and macroscopic time-resolved fluorescence. Biophys J 2007; 93: 539–553
- de Almeida RF, Loura LM, Fedorov A, Prieto M. Lipid rafts have different sizes depending on membrane composition: a time-resolved fluorescence resonance energy transfer study. J Mol Biol 2005; 346: 1109–1120
- De Almeida RF, Loura LM, Prieto M, Watts A, Fedorov A, Barrantes FJ. Structure and dynamics of the gammaM4 transmembrane domain of the acetylcholine receptor in lipid bilayers: insights into receptor assembly and function. Mol Membr Biol 2006; 23: 305–315
- Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol 1984; 179: 125–142
- Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem 1986; 15: 321–353
- Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol 1996; 3: 842–848
- Valeur B, Weber G. Resolution of the fluorescence excitation spectrum of indole into the 1La and 1Lb excitation bands. Photochem Photobiol 1977; 25: 441–444
- Kuntz ID, Jr, Brassfield TS. Hydration of macromolecules. II. Effects of urea on protein hydration. Arch Biochem Biophys 1971; 142: 660–664
- Epand RM, Bottega R. Determination of the phase behaviour of phosphatidylethanolamine admixed with other lipids and the effects of calcium chloride: implications for protein kinase C regulation. Biochim Biophys Acta 1988; 944: 144–154
- Arrondo JL, Goni FM. Structure and dynamics of membrane proteins as studied by infrared spectroscopy. Prog Biophys Mol Biol 1999; 72: 367–405
- Moreno MR, Guillen J, Perez-Berna AJ, Amoros D, Gomez AI, Bernabeu A, Villalain J. Characterization of the interaction of two peptides from the N terminus of the NHR domain of HIV-1 gp41 with phospholipid membranes. Biochemistry 2007; 46: 10572–10584
- Peisajovich SG, Samuel O, Shai Y. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J Mol Biol 2000; 296: 1353–1365
- Peisajovich SG, Epand RF, Epand RM, Shai Y. Sendai virus N-terminal fusion peptide consists of two similar repeats, both of which contribute to membrane fusion. Eur J Biochem 2002; 269: 4342–4350
- Moreno MR, Giudici M, Villalain J. The membranotropic regions of the endo and ecto domains of HIV gp41 envelope glycoprotein. Biochim Biophys Acta 2006; 1758: 111–123
- Moreno MR, Pérez-Berná AJ, Guillén J, Villalaín J. Biophysical characterization and membrane interaction of the most membranotropic region of the HIV-1 gp41 endodomain. Biochim Biophys Acta 2008; 1778: 1298–1307
- Pérez-Berná AJ, Guillén J, Moreno MR, Gómez-Sánchez AI, Pabst G, Laggner P, Villalaín J. Interaction of the most membranotropic region of the HCV E2 envelope glycoprotein with membranes. Biophysical characterization. Biophys J. 2008; 94: 4737–4750
- Dimitrov AS, Xiao X, Dimitrov DS, Blumenthal R. Early intermediates in HIV-1 envelope glycoprotein-mediated fusion triggered by CD4 and co-receptor complexes. J Biol Chem 2001; 276: 30335–30341