Abstract
Notch is a conserved cell signalling receptor regulating many aspects of development and tissue homeostasis. Notch is activated by ligand-induced proteolytic cleavages that release the Notch intracellular domain, which relocates to the nucleus to regulate gene transcription. Proteolytic activation first requires mechanical force to be applied to the Notch extracellular domain through an endocytic pulling mechanism transmitted through the ligand/receptor interface. This exposes the proteolytic cleavage site allowing the signal to be initiated following removal of the Notch extracellular domain. Ligands can also act, when expressed in the same cell, through non-productive cis-interactions to inhibit Notch activity. Furthermore, ligand selectivity and Notch activation are regulated by numerous post-translational modifications of the extracellular domain. Additional non-canonical trans and cis interactions with other regulatory proteins may modulate alternative mechanisms of Notch activation that depend on endocytic trafficking of the full-length receptor and proteolytic release of the intracellular domain from endo-lysosomal surface. Mutations of Notch, located in different regions of the protein, are associated with a spectrum of different loss and gain of function phenotypes and offer the possibility to dissect distinct regulatory interactions and mechanisms, particularly when combined with detailed structural analysis of Notch in complex with various regulatory partners.
Introduction
Notch is a cell surface membrane protein acting as a receptor that mediates a vitally important developmental cell-cell signal (Hori et al., Citation2013; Wharton et al., Citation1985). It has been just over a century since the first description of the Notch gene, detected as a dominantly inherited mutation that results in notching of the adult Drosophila wing. Study of the Notch mutation played an important part in Morgan's work to establish his theory of the gene (Morgan, Citation1917), and it was studies by Poulson of Notch deficiency mutations that linked a gene mutation to a specific defect in embryonic morphogenesis for the first time (Poulson, Citation1937). Complete loss of function of Notch gives rise to the classical lateral inhibition phenotype resulting in a grossly expanded nervous system at the expense of epidermal cell fates (Lehmann et al., Citation1981). Continuing genetic analysis of the Notch locus over subsequent decades has uncovered a multitude of other less severe loss and gain of function alleles with diverse phenotypic consequences in different tissues, indicating the pleiotropic developmental roles of Notch (Foster, Citation1975; Kelley et al., Citation1987; Ramain et al., Citation2001; Welshons & Von Hale, Citation1962; Welshons, Citation1971; Yamamoto et al., Citation2012). Notch continues to play an important role in the adult organism, orchestrating many cell fate decisions regulating adult tissue homeostasis (Chiba, Citation2006; Conboy & Rando, Citation2005; Lampreia et al., Citation2017; Ohlstein & Spradling, Citation2007). Notch is highly conserved across the metazoans and is represented by four homologues in the human genome (Weinmaster, Citation1997). Not surprisingly the disease associations linked to human Notch misregulation are many and varied (Aster et al., Citation2017; Mašek and Andersson, Citation2017; Siebel & Lehndahl, Citation2017). Understanding mechanistic links between mutations affecting Notch structure and the resulting loss and gain of function genotypes is an important and challenging focus of research today. Exploring these questions requires a multidisciplinary approach spanning application of physical techniques and detailed atomic structure through to whole organism genetic studies.
Regional specialisation within the Notch gene locus was already evident from fine scale recombination genetic mapping of its diverse allele phenotypes. Cloning of the Drosophila Notch gene in the mid 1980s (Wharton et al., Citation1985) began a journey to place this intriguing genetic architecture within a protein/structure function context. Notch was revealed as a highly modular membrane protein (), notably comprised, in its extracellular domain, of 36 tandem-repeated modules related to epidermal growth factor (EGF). In its intracellular domain a highly conserved feature is a region of seven tandem-repeats of an Ankyrin-domain motif, flanked by two nuclear localisation sequences. The two canonical ligands for Notch, Delta and Serrate (or Jagged in vertebrates), are also characterised by multiple repeats of EGF-like modules, together with a conserved N-terminal region including Delta, Serrate, Lag2 (DSL) domain (Lag2 being a C.elegans homologue) (). The ligands themselves are membrane bound proteins and thus the signal mediated by Notch is a short range one between adjacent cells which is deployed in many developmental contexts (Bray, Citation1998, Citation2016).
Figure 1. Relationship between Notch modular structure and its activation mechanism. (A) The modular structure of Drosophila Notch and its ligands Serrate and Delta. Vertebrate Notch and ligand homologues have similar domain organisation with some minor differences in number of EGF-like modules in some cases. The seven ankyrin (Ank) domain repeats in the intracellular domain are a highly conserved feature. The heterodimer region (HD) contains a Furin cleavage site known as S1 and processing of Notch during its secretory trafficking results in a heterodimeric molecule expressed at the cell surface, held together through non-covalent interactions in the HD region. S1 processing may not be significant for Drosophila Notch however (Kidd & Lieber, Citation2002). Locations of sequenced Drosophila classical Notch alleles are indicated. Arrows indicate missense mutations, * indicates frame shift or stop codon. Red arrows and red * indicate loss of function, blue arrows and blue * indicate gain of function. Green arrow indicates mutant with complex interpretation. Orange labelled mutants are recessive lethal, others are homozygous viable with variety of adult phenotypes. TS1 is a temperature sensitive allele. Further details of Notch allele phenotypes can be obtained from FlyBase (http://flybase.org). (B) Ligand-induced Notch activation mechanism. Notch is activated by binding of membrane bound ligands expressed on adjacent cells through the EG8-12 region of the receptor and the C2 to EGF3 region of the ligands. Endocytosis of the ligand applies tension on Notch extracellular domain, which causes a local unwinding of the Negative Regulatory Region (NRR) structure. Engagement of Notch with the endocytic machinery of the signal receiving cell may also contribute to this mechano-transduction mechanism. This structural change exposes a proteolytic cleavage site (S2), which lies in the HD region, for processing by Adam10/Kuzbanian metalloprotease. After removal of the bulk of the extracellular domain, the remaining membrane tethered Notch intracellular domain (NICD) is released by γ-secretase processing and NICD is transported to the nucleus through the action of two nuclear localisation sequences (nls), which flank the Ank-domain region. The PEST region (Proline Serine Glutamate Threonine rich) regulates NICD turnover and limits the signal activation. C) Crystal structure of the NRR and HD region from human Notch1 (Gordon et al., Citation2009). LNR (Lin12 Notch repeats) and HD region indicated by colour coded labels. Blue side chain V1721 indicates S2 cleavage site. In the inactive confirmation this site is protected in the core of the structure through interactions with hydrophobic side chains L1481 and F1483 indicated in red. Grey spheres indicate location of Calcium ions. D) Crystal structure (Choi et al., 2009) of human Notch1 Ram-Ank region of NICD complex (green) with the CSL transcriptional activator (yellow) and recruited coactivator protein Mastermind (Mam, pink), bound together as a complex to a DNA helix (Refer online version for colour figure).
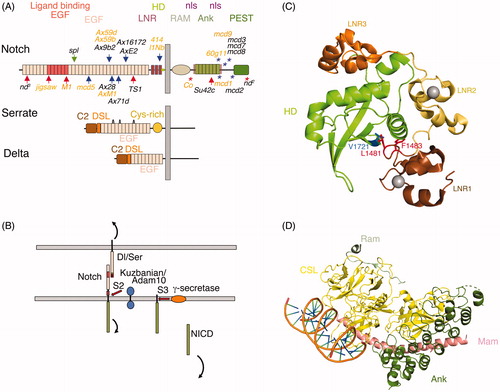
Genetic studies in the Drosophila and C.elegans model organisms together with further biochemical studies in mammalian cells uncovered a conserved signalling pathway dependent on proteolytic activation of Notch (Greenwald, Citation1998; Mumm and Kopan, Citation2000) (). In this pathway, ligand binding to Notch, coupled with engagement of both receptor and ligand with the endocytic machinery of the cell, results in an applied tension in the picoNewton range that exposes an Adam10 metalloprotease cleavage site that is normally hidden within the Negative Regulatory Region (NRR) () (Gordon et al., Citation2015; Kopan & Ilagan, Citation2009; Wang & Ha, Citation2013; Seo et al., Citation2016). Exposure of this cryptic cleavage site by this mechano-transduction mechanism results in the shedding of the bulk of the extracellular domain of Notch which, still bound to ligand, is transendocytosed into the signal donor cell (Lai et al., Citation2005; Langridge & Struhl, Citation2017; Parks et al., Citation2000). The remaining membrane tethered intracellular domain is subsequently cleaved by the gamma-secretase complex within the membrane spanning region to release the Notch intracellular domain as a soluble entity allowing it to traffic to the nucleus, whereupon it associates in a complex with a transcription factor CSL (denoted CBF in mammals, Suppressor of hairless in Drosophila and Lag1 in C. elegans). Other cofactors such as Mastermind are simultaneously assembled into this complex displacing co-repressor proteins from CSL and switching it from a repressor of gene transcription to an activator complex (Morel et al., Citation2001; Mumm & Kopan, Citation2000). The ankyrin domain region together with the membrane proximal RAM domain together act as a platform for binding to CSL and the other cofactors involved in assembling this activating complex (). Recent work supports a dynamic model in which NICD binding to CSL increases the latters residency time by opening chromatin structure to increase accessibility (Gomez-Lamarca et al., Citation2018). Considerable detailed 3 D structural information is available giving us atomic-resolution overview of the structure/function relationships of intracellular domain functions (Arnett et al., Citation2010; Nam et al., Citation2006; Wilson & Koval, Citation2006).
At first site this appears to represent a relatively simple pathway lacking any amplification step, however first impressions are deceptive. Many layers of regulation, including O-glycosylation of the extracellular domain, mechanical force, membrane lipids and intracellular domain posttranslational modifications ensure the setting of Notch signalling levels is deeply embedded into the physiology of the cell and integrated with other cell signalling networks in pattern formation (Bray, Citation2016). A fundamental question begged by the modular nature of the Notch structure is the relationship between structure and function and how this is reflected in the phenotypic diversity arising from Notch mutations located in different regions of the structure. In Drosophila many missense mutant alleles have been uncovered with diverse loss or gain of function phenotypes arising respectively from decreased or increased Notch signalling activity. These varied alleles phenotypes display a range of different tissue specificities hinting at different underlying regulatory mechanisms that may be disrupted (). For example mutations in a region encompassing EGF24-29 of Drosophila Notch produce gain of function phenotypes, which prevents formation of regions of wing veins (Kelley et al., Citation1987), while a mutation in the EGF18 produces a gain of function specifically associated with sensory microchaetae loss (Ramain et al., Citation2001).
Much progress has been made in building and understanding of Notch structure through a piecemeal approach to different sections. These structural advances will form a solid basis to begin to explain the mechanistic links between genotype to phenotype for many classes of Notch allele. In this review I will discuss recent progress in understanding of Notch structure-function relationships in particular focussing on the roles of the Notch extracellular domain.
Dissecting notch/ligand interactions
The earliest assignment of Notch function to a specific sub-region of the extracellular domain came from cell-cell aggregation studies, which aimed to discern the nature of the ligand-binding site on Notch (Rebay et al., Citation1991). Stable cell contacts form when Notch and ligand expressing cells are co-cultured, providing a simple assay to identify regions involved in Notch recruitment by ligands. Drosophila Notch comprises 36 tandemly repeated motifs related by homology to Epidermal Growth Factor, a conserved modular protein building block found in many extracellular and cell surface proteins. Using nested deletion constructs, cell aggregation assays identified just two of these modules, EGF11 and 12 as being necessary and sufficient to allow Notch mediated cell aggregation, focussing attention on this region (Rebay et al., Citation1991). The first structural insights into EGF module structure came from NMR studies of an expressed fragment of the blood clotting protein Factor IX, which confirmed the similarity of the 3 D structure of EGF-like modules to that of Epidermal growth factor (Baron et al., 1992). Mutagenesis studies followed which revealed the presence of a calcium-binding site within the N-terminal region formed by certain conserved acidic amino acid residue side chains (Handford et al., Citation1991). This calcium binding signature sequence was found to denote a subset of calcium-binding EGF modules (cb-EGF) found also in Notch, including the ligand binding EGF11 and 12. Structural investigations by NMR and crystallography of an expressed human Notch1 fragment encompassing EGF11-13 and similar EGF module arrays from the matrix protein Fibrillin gave further insight into domain-domain linkages and their influence on Notch/ligand interactions (Cordle et al., Citation2008; Downing et al., Citation1996; Hambleton et al., Citation2004; Rao et al., Citation1995). Calcium binding to the N-terminus of the EGF domain, together with a conserved hydrophobic packing interaction, stabilises the domain interface of calcium binding-EGF pairs. (Downing et al., Citation1996; Hambleton et al., Citation2004) (). The importance of Calcium ions to ligand binding was confirmed through in vitro binding studies with isolated protein fragments (Cordle et al., Citation2008). Interestingly physiological fluctuations in extracellular and intracellular calcium may regulate Notch activity in some developmental contexts such as the establishment of left-right asymmetry in chick embryos (Raya et al., Citation2004) and during mechano-sensing control of progenitor cell differentiation in the Drosophila gut (He et al., Citation2018). In Drosophila the importance of the Calcium ion binding site was reinforced through identification of a loss of function Drosophila developmental mutant NotchM1 (de Celis et al., Citation1993). This allele has a missense mutation in the Calcium-binding consensus of EGF12, an elegant early example of complementary data arising from structural and genetic approaches. Indeed missense mutations in the ligand-binding region of Notch, now known to encompass EGF8-12 (see below), have been identified in cancers where wild type Notch normally acts as a tumour suppressor protein (Agrawal et al., Citation2011; Stransky et al., Citation2011; Wang et al., Citation2011).
Figure 2. The structure of the Notch/ligand complex. (A) EGF11-13 from human Notch1 (Cordle et al., Citation2008) forms an extended rigid structure with EGF-EGF module interfaces stabilised by Calcium (grey sphere) binding sites (purple side chains) and hydrophobic packing interactions (green side chains). Side chains L468, I477 labelled in blue indicate residues whose mutation reduces Notch/ligand binding and signalling (Whiteman et al., Citation2013). (B) Crystal structure of human Jag-1 C2 to EGF3 (Chillakuri et al., Citation2013). Magenta labelled side chain indicates a conserved F207 residue whose mutation strongly affects binding and Notch signal activation when altered in Drosophila Serrate. Yellow side chains F199 and R203 indicate conserved positions, which when mutated in Drosophila Serrate strongly reduce signalling but have weak affects on binding. They further can act in a dominant negative manner when expressed in vivo in Drosophila wing discs (Cordle et al., Citation2008). (C) A crystal structure of a complex of rat Notch1 EGF8-12 and Jag1 C2-EGF3. Arrow shows approximate position of hinge bending conformational change compared to Jag1 crystallised alone. This conformational change is proposed to facilitate catch bond behaviour (Luca et al., Citation2017). (D) Detail of the EGF11/12 interface with ligand C2/DSL region. Side chains highlighted in (A) and (B) are coloured as above. Additional side chains highlighted are labelled. Jag1 F207 is deeply buried in a hydrophobic pocket supplied by Notch EGF11 comprising P422 and F436 (dark blue) that are a conserved part of the general EGF-module consensus sequence together with the R448 side chain (cyan), which is conserved within EGF11. The Jag1 F199 and R203 engage with conserved EGF11 residues E423 and H424 (Red). The former forming a salt bridge with Arg203 and also a hydrogen bond with conserved DSL domain Y210. In EGF12 side chains L468 and I477 (blue) form hydrophobic interactions surface with F94 and F126 in the C2 domain (side chains shown in orange). O-fucose attached to conserved threonine in EGF 12 (black) also contributes to this interface. (E) Detail of Notch EGF8-10 interface with Jag1 EGF1-3. Hydrophobic interactions mediated by conserved residues from Notch (dark blue) and from Jagged EGF1-3 (green). Black side chain represents an O-fucosylated T311, which contributes to binding affinity, making a H-bond with N298 in EGF3 of Jag-1. In purple is V324, which is the location of the Drosophila Njigsaw allele, whose mutation affects Serrate signalling but not activation through Delta (Yamamoto et al., Citation2012) (Refer online version for colour figure).
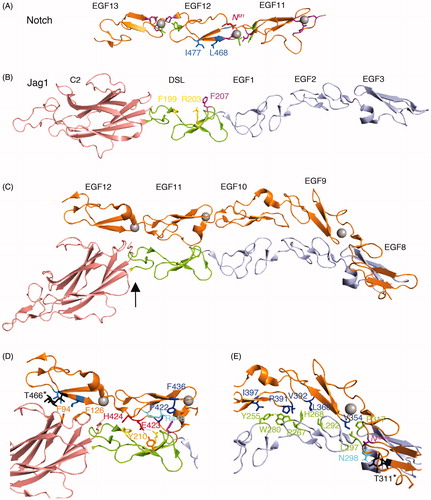
Genetic analyses combined with domain deletion studies and in vitro binding and signalling assays have also defined important regions of the ligands involved in binding and activating Notch to include an N-terminal region structurally similar to lipid-binding C2 domains, the DSL domain and the EGF1-3 modules (Andrawes et al., Citation2013; Chillakuri et al., Citation2013; Cordle et al., Citation2008; Komatsu et al., Citation2008; Liu et al., Citation2017; Parks et al., Citation2006; Schuster-Gossler et al., Citation2016). More detailed insights into Notch/ligand complex formation have been provided through structural studies involving NMR and crystallography. The first Jagged structure in a non-complexed form was obtained for the N-terminal region of Jagged-1 (Jag-1) encompassing the DSL domain and EGF1-3, a fragment which retained Ca-dependent in vitro binding to Notch, albeit at low affinity (Cordle et al., Citation2008). The structure revealed a surface of the DSL domain with several highly conserved, surface-exposed residues (). Mutagenesis of the Drosophila ligand Serrate confirmed the function of this region in Notch binding and signalling. Interestingly mutagenesis revealed a mutation class that severely disrupted both Notch/Serrate mediated cell aggregation and signalling, and a mutation class, which strongly affected signalling but had only mild effects on Notch/Serrate interactions. Thus complex formation alone was insufficient in itself to efficiently activate Notch in some cases. Recent work, which obtained a crystal structure of a Jag-1 N-term to EGF3 fragment complexed with Notch1 EGF8-12, has suggested a possible explanation for such genetic dissection of binding from activation (Luca et al., Citation2017). To obtain crystal structures of ligand/Notch complexes an elegant directed evolution approach has been utilised in which a PCR generated mutant library of ligand fragments encompassing the Notch binding region was expressed on the surface of yeast cells by fusion to a cell wall protein. Several rounds of affinity selection for Notch1 binding were applied resulting in the identification of ligands bearing a combination of mutations that together resulted in sufficient affinity for successful co-crystallisation. Importantly, compared to the unbound structure, Jagged-1 in the bound form had undergone a hinge bending conformational change with the linking region between the C2 and DSL domain acting as a pivot (Luca et al., Citation2017) (). The hinge bending motion thus allows the extended interface to form involving contacts through both the C2-DSL and EGF1-3 regions. Further insights were obtained using assays to measure the affinity of the Notch/ligand complex when different tensions were applied (Luca et al., Citation2017). Intriguingly the affinity of wild type Jag-1 increased when higher tensions were applied exhibiting so called catch-bond behaviour, whereas the mutant high affinity Jag-1 protein bound with lower affinity at higher applied tensions (a slip bond). It is possible that the mutant ligand construct arising from the directed evolution approach already favoured the high affinity bent conformation found in the complex and hence did not display catch bond behaviour on application of tension.
Activation of Notch depends on the application of tension to reveal the cryptic proteolytic cleavage site in the NRR domain. As tension is generated due to engagement of endocytic machinery of both Notch and ligand then the resulting increased affinity of the complex may allow longer lifetime of the complex. In this case the cryptic proteolytic activation site can become sufficiently exposed to allow cleavage by the Adam10 enzyme (Luca et al., Citation2017). Notch/ligand complex formation can therefore be separated into two stages, before and after tension is applied, and this two step process might therefore underlie the apparent genetic dissection of binding and activation steps that was discussed above (Cordle et al., Citation2008). It is therefore informative to map onto the structure of the complex, the DSL domain residues previously proposed to be involved in binding and activation (Cordle et al., Citation2008; Whiteman et al., Citation2013). Mutation of Jag1 Phe207 (Phe257 in Drosophila Serrate) has both a strong affect on Notch ligand interactions and strongly reduces signalling. In the complex this residue occupies a hydrophobic pocket created by conserved Pro422 and Phe436 in Notch EGF11 and thus appears to play a central role in complex formation (). The conserved Jag1 residues Arg201 and Asp205 (Arg251 and Asp255 in Drosophila Serrate) when mutated had weak and proportional affects on both binding and signalling and lie peripheral to the main binding surface. On the other hand Jag1 Phe199 and Arg203 (Phe249 and Arg253 in Drosophila Serrate) form an additional interaction interface with His424 and Glu423 in Notch EGF11 respectively, including a salt bridge formed between the Glu423 and Arg203 residues and a Hydrogen bond to Jag1 Tyr210 (Luca et al., Citation2017). Mutation of equivalent Phe249 and Arg253 residues in Drosophila Serrate also strongly reduces Notch signalling but has only weak effects on ligand/receptor complex formation. It is interesting therefore to speculate whether these mutant ligand proteins can form the initial low affinity complex but the catch bond cannot be initiated to facilitate signal activation. Interestingly when Drosophila Serrate mutants at Phe249 or Arg253 are overexpressed in Drosophila tissue they can act dominant negatively on endogenous Notch activity perhaps by competing with endogenous ligand to form non-productive complexes (Cordle et al., Citation2008).
The EGF12/Jag-1 interface comprises a largely hydrophobic interaction comprised of conserved residues. In Notch EGF12, mutation of Leu468 and Ile477 (Leu504 and Val513 in Drosophila Notch) has shown these residues to be important for Notch ligand complex formation and signalling (Cordle et al., Citation2008). Consistent with these identified roles, inspection of the co-crystal structure shows that both residues form a hydrophobic interface with two conserved Phenylalanines in the Jag-1/Serrate C2 domain (). Side chain and backbone interactions with the conserved O-fucosylated Threonine 466 residue in Notch EGF12 also play a part in this C2 domain/EGF12 interface () (Luca et al., Citation2017), consistent with increased ligand affinity observed when this residue is fucosylated in vitro (Taylor et al., Citation2014).
An additional extensive interaction interface is formed through bulky hydrophobic residues of Notch EGF9 and 10 and Jag-1 EGF1 and 2 (), which fill the cavity that lies between the peptide backbones for respective receptor and ligand structures. A fucosylated residue, Threonine 311 in Notch EGF8 additionally interacts with EGF3 in Jag1. Thus two O-fucose modifications in EGF12 and EGF8 of Notch are directly involved in ligand complex formation and were found to contribute to binding affinity, although in this study the EGF8 O-fucosylation was found to be the more significant for signal activation for the particular Notch1/Jag-1 combination investigated (Luca et al., Citation2017).
It is interesting to compare the Jagged1/Notch complex with that of Delta-like4/Notch complex that has also been published (Luca et al., Citation2015). This complex comprised an Nterm-EGF2 fragment of Delta-like 4 (Dll4) and the EGF11-13 of Notch1. It revealed a similar antiparallel arrangement of Notch and ligand, also involving DSL/EGF11 and C2 domain/EGF12 interfaces (). However there were differences in detail resulting in less buried surface area in this region for the Jag-1 complex compared to Dll4 (Luca et al., Citation2017). This reduced interface for Jag-1 Notch may be compensated for by the additional interactions between Notch EGF8 to 10 and EGF1-3 of Jag-1, facilitated by the curvature of the elongated Notch EGF array, centred around the EGF9/EGF10 module junction. However this region of Notch was not present in the Dll4 complex and so this supposition has not been tested directly. However a missense mutant allele, known as jigsaw, in EGF8 of Drosophila Notch has previously been found to affect Serrate but not Delta induced signalling, suggesting the importance of this region in contributing to ligand specificity (Yamamoto et al., Citation2012). Reproducing this mutation in a conserved residue in mouse Notch2 produced a similar discrimination, selectively reducing activation by Jag-1 but not Dll1. Nevertheless the contribution EGF8 O-fucosylation to Dll-1 binding affinity suggests that specific contacts with the Delta ligand in this region are waiting to be uncovered (Kakuda & Haltiwanger, Citation2017).
Figure 3. The Notch Dll4 complex. Crystal structure of the Dll4 C2 to EGF1 region in complex with Notch1 EGF11-13 (Luca et al., Citation2015). The C2 domain makes similar contacts with EGF12 of Notch1 to those observed for Jag1 ligand, involving conserved Phenylalanine residues (F76, F109). The DSL domain residue F195 (conserved with Jag1 F207) is similarly buried in the EGF11/Jag1 interface and Leu187, a conserved substitution for Jag1 F199, is similarly involved. Differences in detail can be discerned between the Jag1 and Dll4 complexes. The EGF11 conserved R448 forms a salt bridge with DLL4 D218 (orange) and its side chain is buried by the DSL domain residue Y216 (grey), side chain interactions which do not exist for the Jag-1 complex.
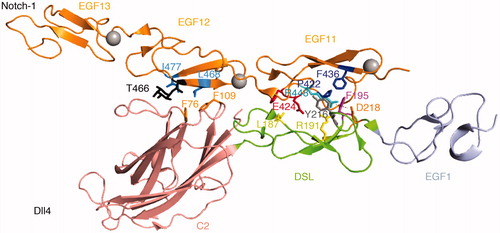
Differences in the detail of the contacts at the level of individual amino acid side chains can also be discerned by comparing the different structures. In the EGF12/C2 domain interface, similar contacts to the Jag1 complex are made involving Notch EGF12 residues Leu468, Ile477 and Thr466, with the two conserved Phe76 and Phe109 residues in the Dll-4 C2 domain (). In the EGF11/DSL interface, as with Jag1 Phe207, the conserved Dll4 Phe195 appears to play a pivotal role in the complex through conserved contacts with P422 and F436 in Notch EGF11. In Dll4 the equivalent position in the DSL domain of Jagged1 Phe199 is occupied by Leu187, although it still protrudes similarly between the Notch1 Glu424 and His425 residues. However in the Dll4 complex, the salt bridge between Dll4 Arg191 (Arg203 in Jag1) and Glu424 in EGF11 does not form. Instead the Notch EGF11 Arginine 448 protrudes into a hydrophobic pocket between Dll4 Phe 195 and Tyr216. Notch 1 Arg448 further forms a salt bridge with the Dll4 Asp218 residue that lies at the end of the DSL domain. Neither the Tyr216 nor Asp218 are conserved in Jag1. It is likely that further structures of different combinations of Notch and ligand partners will eventually reveal additional subtle differences, which may contribute to partner-preferences in ligand/Notch interactions. For example Delta-like 4 is reported to bind with a ten fold higher affinity to Notch1 than Delta-like 1 (Andrawes et al., Citation2013; Shimizu et al., Citation2000). However it must be born in mind that these detailed comparisons are derived from complexes containing different size fragments and may need to be further validated with additional structures.
Whatever detailed differences in amino acid side chain contacts there are between different Notch ligand interfaces it is important to consider post-translational modifications of Notch to understand the molecular basis of differential ligand preferences. As discussed above O-fucose modifications of Notch in EGF8 and 12 are known to be directly involved in the Notch/ligand interface and Fringe mediated extension of O-Fucose residues is further known to alter the affinity for ligands (Taylor et al., Citation2014). In total around 20 EGF modules in Notch have the consensus sequence for O-fucose modification and most of these are efficiently modified in cells during secretory transport through the endoplasmic reticulum (Kakuda & Haltiwanger, Citation2017). Importantly O-fucose residues can be extended by the activity of Fringe proteins, which are B3-N-acetylgucosaminyl transferases that covalently link GlcNAc units to the O-fucose moiety. This activity was first identified in Drosophila and leads to enhanced Notch signalling by Delta but decreased activation by Serrate, a feature important in establishing developmental boundaries (Domínguez & de Celis, Citation1998; Fleming et al., Citation1997; Panin et al., Citation1997; Papayannopoulos et al., Citation1998). In mammals, there are three fringe homologues, Lunatic Fringe (Lfng), Manic Fringe (Mfng) and Radical Fringe (Rfng) whose activity also differentially regulates Notch activation by different ligands; however, the situation is more complex (Johnston et al., Citation1997). For example Lfng and Mfng both activate Dll1 signalling through Notch1 and down-regulate Notch1 activation by Jag-1. However Rfng activates both Dll1 and Jag-1 activation of Notch1. Recently published elegant work utilising mass spectrometry has provided important understanding of the molecular basis for such differences and highlighted which EGF domain modifications are important in ligand discrimination (Kakuda & Haltiwanger, Citation2017). Whereas most available sites for O-fucosylation were utilised, the activity of the Fringe proteins in further extending this modification were more selective. LFng and Mfng modification of O-fucose in EGF8 and 12 increased the activity of Dll1 while modification of EGF6 and 36 was responsible for down-regulating Jag-1 activation. Rfng modified EGF8 and EGF9 to activate both Dll1 and Jag-1 signalling and consistent with this positive effect on Jag-1 it did not modify EGF6 or 36. Interesting the Fringe modifications led to increased binding of ligand whether or not the outcome was an increase or decrease in Notch activation (Kakuda & Haltiwanger, Citation2017; Taylor et al., Citation2014). This suggests that the inhibitory affect on Jag-1 signalling occurs after the initial binding. Whether this paradox is linked to catch-bond initiation after binding or some other process remains to be seen. Because of the above described complexity caution is needed when comparing the literature on ligand binding affinities because much will depend on the specific post-translational modifications added by the particular cell lines used.
As well as Notch ligand preferences being regulated, there is also reported bias in outputs driven by different ligand/receptor couplings. Despite a common core proteolytic activation pathway, different ligands drive different phenotypic outcomes on cell fate. For example several reports illustrate the non-equivalence of Dll1 and Dll4 in controlling cell fates in different developmental contexts (Mohtashami et al., Citation2010; Preusze et al., Citation2015; Sörensen et al., Citation2009). Recent work has suggested that different dynamics of the signalling output may contribute to ligand specific outcomes on target gene expression (Nandagopal et al., Citation2018). Dll1 was found to produce a pulsed signalling output, which was modulated by frequency, whereas Dll4 produced a more sustained signalling output, which was modulated by amplitude. These different signalling modulations activated expression of distinct target genes and had opposing outcomes on myogenic cell differentiation. It was proposed that pulsatile signalling behaviour may be favoured where ligand receptor clustering is required for Notch activation with transendocytosis resulting in simultaneous activation of many Notch molecules at a time, whereas the sustained signalling of the Dll4 mode may reflect a lack of any need for clustering of complexes to occur. In this case signals can be generated from individual ligand receptor pairs favouring a continuous signal generation.
A further role for the ligand has been proposed based on the lipid binding properties of its N-terminal domain. Crystallographic studies revealed the strong structural homology between the N-terminal region of Notch ligands with calcium-dependent, phospholipid-binding, C2 domains (Chillakuri et al., Citation2013). Binding assays to liposomes of carefully controlled composition showed that the Jag1 C2 domain indeed mediates phospholipid binding in a calcium-dependent manner (Suckling et al., Citation2017). Mutation of residues involved in calcium coordination within the C2 domain did not affect ligand binding to Notch but did prevent signalling. Intriguingly Jag-1 C2 domain mutants found in the inherited disease extrahepatic biliary atresia also reduce signalling but not binding (Suckling et al., Citation2017). This mutation data therefore suggests the Notch/ligand complex may further require a membrane interface with the C2 domain for efficient Notch activation. This proposal is further supported by the fact that by adding Notch1 EGF11-13 to Jag1, the latters binding binding to lipid was stimulated, suggesting a three-component complex involving membrane association (Suckling et al., Citation2017). Interestingly there is much variability between different Notch ligands in the surface loops that comprise the lipid-binding site and it is possible that altered membrane composition might tune activation of Notch by different ligand receptor pairings. The nature of the lipids that bind to the C2 domain in vivo is not known, but the involvement of sphingomyelin and gangliosides in promoting ligand discrimination was implicated in liposome studies. It is worth to note that alternative but not mutually exclusive roles have been reported for glycosphingolipid interactions to regulate Notch ligand endocytosis in C.elegans, Drosophila and mammalian models (Hamel et al., Citation2010; Heuss et al., Citation2013; Katic et al., Citation2005). Glycosphingolipids are also suggested to regulate endocytosis of Drosophila Notch along with its ligand-independent activation (Shimizu et al., Citation2014).
Cis-inhibitory notch/ligand complexes
A notable feature of Notch regulation is that ligands can also act as down regulators of Notch signalling when expressed in the same cell as Notch (cis-inhibition) (de Celis et al., Citation1997; Klein et al., Citation1997; Micchelli et al., Citation1997). This is a reciprocal interaction since cis-association with Notch also inhibits the ability of ligand to signal out to adjacent cells (Becam et al., Citation2010). Thus cis-inhibition can reinforce the directionality of Notch signalling during pattern formation. The nature of the cis-complex compared to the trans-binding configuration, and how it may be affected by different mutations is at present unclear. To date structural studies of 3 D complexes have not provided any clear insights as to whether there is a single mode of binding for cis or trans or if there are alternate modes of interaction. However studies using mutational analysis have indicated some of the binding surfaces are in common. In Drosophila cis-inhibition can be readily observed when Serrate is expressed in the developing wing imaginal disc in a pattern that crosses the dorsal ventral boundary of the wing. Notch activation is induced only in the cells adjacent to the Serrate-expression domain. Within the Serrate expressing cells Notch activity is repressed by cis-inhibition including the endogenous Notch activity within the dorsal-ventral boundary. Mutation of Phe257 in the DSL domain of Drosophila Serrate abrogates both transactivation and cis-inhibition actions of Serrate, implicating the DSL domain in both trans and cis-outcomes of Serrate on Notch (Cordle et al., Citation2008). Furthermore the ability of Notch to cis-inhibit the Serrate in vivo is reduced by the EGF12 NotchM1 mutation, also implicating this region of Notch in both trans and cis-inhibition (Becam et al., 2012). However, other work has suggested that the requirements for the cis and trans-interaction differ in some respects. Domain deletion experiments have identified EGF-modules 4-6 of Serrate as being necessary for cis-inhibition by Serrate but which are not required for trans-activation (Fleming et al., Citation2013). Whether the requirement EGF4-6 region is one of a spacer region to allow complex formation in the cis-presented condition, or whether further binding interfaces remain to be uncovered remains to be determined. In this regard it is interesting that Dll3, a mammalian Notch ligand lacking certain conserved DSL domain residues normally involved in trans-binding complexes, does not activate Notch but preserves cis-inhibition activity, which may therefore occur through other Notch ligand/interfaces (Chapman et al., Citation2011; Ladi et al., Citation2005). Furthermore covalent modification of Drosophila Notch by addition of xylose to O-glucosylated residues in the Notch EGF16 to EGF20 region has been reported to reduce trans-binding to Delta but not cis-inhibitory interactions, indicating some distinctiveness in the respective complexes (Lee et al., Citation2017). Further understanding of the cis-ligand interaction interface and identification of the reciprocal interaction surfaces on Notch will likely help to interpret further genotype/phenotype connections for different Notch mutant alleles.
Extracellular and intracellular interactions modulate non-canonical notch activation mechanisms
Despite the large extracellular domain of Notch only a small region, EGF8-12 has been implicated in canonical DSL-domain ligand binding needed to initiate signalling. The roles of the remaining portions of the EGF-module tandem array are unclear. Indeed a signalling-competent synthetic receptor/ligand interaction system has been devised that replaces the entire EGF-module region of Notch and can still be activated (Gordon et al., Citation2015). In this case the only extracellular domain requirements to activate the signal are the ability to form a ligand/receptor complex and to be able to withstand the application of tension up to the point at which the NRR unwinds to expose the metalloprotease cleavage site. Given these simple mechanical requirements we can rule out an essential role for an allosteric conformational change transmitted by ligand binding to the NRR. It is necessary therefore to look beyond the core signal activation mechanism to understand the different regulatory roles fulfilled by regions outside the ligand-binding interface. In this regard it is useful to consider that Notch can also be activated independently of DSL-domain ligands following its endocytosis (Hori et al., Citation2011; Mukherjee et al., Citation2011; Palmer et al., Citation2014; Sakata et al., Citation2004; Schneider et al., Citation2013; Shimizu et al., Citation2014; Wilkin et al., Citation2004, Citation2008), (), by a mechanism best characterised in Drosophila models but which may also be relevant to human Notch in some contexts (Choy et al., Citation2017; Gómez-del Arco et al., Citation2010; Zheng et al., Citation2013). Signal activation by this mechanism critically depends on Notch being retained in the endosomal membrane and not being transferred off the endosomal surface by incorporation into the intra-luminal vesicles (ILVs) (Hori et al., Citation2011; Shimizu et al., Citation2014; Wilkin et al., Citation2008). The importance of this critical regulatory node is highlighted by the consequences of loss of function mutations of components of the ESCRT complexes, which mediate cargo recruitment into the ILVs. Loss of ESCRT function thus leads to potent ectopic Notch activation (Thompson et al., Citation2005; Vaccari & Bilder, Citation2005; Vaccari et al., Citation2009). When retained on the endosomal surface the Notch intracellular domain remains accessible to the cytoplasm with its extracellular domain exposed to the endosomal lumen. Proteolytic removal of the extracellular domain likely occurs after endosome/lysosome fusion and the Notch intracellular domain can be released in the normal manner by the gamma-secretase complex which is present in the endo/lysosomal membranes (Schneider et al., Citation2013; Shimizu et al., Citation2014; Wilkin et al., Citation2008). This mechanism of action is promoted by Deltex, a ring finger ubiquitin ligase that binds to the Notch ankyrin repeats within its intracellular domain. Deltex has two functions, firstly to promote Notch endocytosis through a Clathrin-mediated mechanism, and secondly to retain Notch on the endosome, suppressing its entry into ILVs (Shimizu et al., Citation2014) (). This mechanism of action depends on fusion of the late endosome and lysosome and may occur on the lysosomal membrane. The mechanism is independent of the Adam10 metalloprotease and may depend instead on lysosomal proteases for initial removal of the Notch extracellular domain (Schneider et al., Citation2013; Shimizu et al. Citation2014). Interestingly Notch can also be internalised by a Clathrin-independent endocytic pathway that is dependent on membrane cholesterol and the synthesis of membrane glycosphingolipids, components of membrane microdomains associated with lipid rafts (Shimizu et al., Citation2014). Entry of Notch into this pathway is regulated by a HECT domain ubiquitin ligase, Suppressor of deltex (Su(dx)), which also promotes Notch transfer into ILVs sequestering it from cytoplasmic access (), and hence promotes its down-regulation. The latter can however be prevented when the ubiquitin ligase activity of Su(dx) is inactive, in which case retention of Notch on the endosomal membrane is associated with a basal level of ligand-independent Notch signalling (). In Drosophila this occurs at low temperature when basal ligand-independent activity is thought to maintain Notch signalling above critical thresholds when ligand-dependent signalling is reduced. This provides a mechanism of temperature compensation through altered balance of trafficking fluxes (Shimizu et al., Citation2014). It is important to note that depending on the context, Deltex can act in flies also to down-regulate Notch activity by reducing its access to ligand-dependent activation, an outcome for which it cooperates with Su(dx) (Shimizu et al., Citation2014).
Figure 4. DSL-ligand-independent activation of Notch in the endo/lysosomal pathway. Notch can be endocytosed through Clathrin-dependent and independent routes (Shimizu et al., Citation2014). Clathrin-mediated endocytosis is promoted through interaction with the E3 ubiquitin ligase protein Deltex (Dx). The latter also acts to promote Notch retention on the endosomal membrane rather than being transferred into intraluminal vesicles of the maturing endosome. On lysosome fusion, mediated by Rab7 and the HOPS complex, the extracellular domain of Notch is removed, independently of the activity of Adam10, and NICD is released by γ-secretase activity. Suppressor of deltex (Su(dx)) also interacts with NICD and promotes Notch endocytosis through a lipid raft-dependent and Clathrin-independent route (Shimizu et al., Citation2014). Su(dx) also acts via Notch ubiquitination to promote Notch internalisation into intraluminal vesicles where it is sequestered from activation and is degraded on lysosomal fusion. If the ubiquitin ligase of Su(dx) is inactive then Notch is retained on the endosomal membrane and can be activated in this membrane domain by an Adam10-dependent activity (Shimizu et al., Citation2014). Additional interactions at the cell surface through Crumbs and ZO-1 regulate these alternative mechanisms of activation by limiting Notch Dx-dependent endocytic trafficking of Notch (Nemetschke & Knust, Citation2016; Shimizu et al., Citation2017).
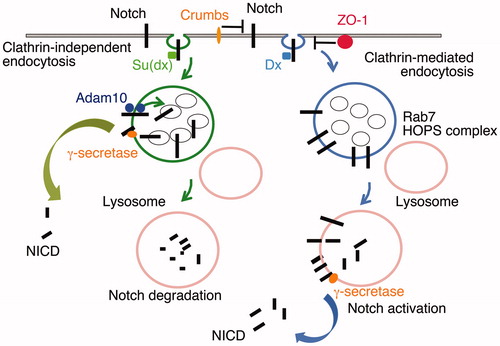
Mutations of Drosophila deltex and Su(dx) have wing phenotypes respectively similar to Notch loss and gain of function alleles (Fostier et al., Citation1998; Xu & Artavanis-Tsakonas, Citation1990). It is therefore interesting to consider whether some Notch mutant alleles may also produce phenotypes by perturbing the flux balance within the associated endosomal trafficking network. Indeed the gain of function phenotypes of certain Abruptex class Notch mutations are critically dependent on the presence of a functional deltex gene (Gorman & Girton, Citation1992; Xu & Artavanis-Tsakonas, Citation1990;). The Abruptex phenotypes may arise from disruption of novel interactions of Notch that regulate its endocytic trafficking. Indeed it is becoming clearer that endocytic Notch activation mechanisms are themselves subject to further levels of regulation from both extracellular and intracellular components. Recent papers have shown by in vitro and in vivo studies that two cell junctional proteins ZO-1 and Crumbs interact with Notch respectively through the latters intracellular and extracellular domains to suppress Deltex-dependent signalling. In both cases this appears to be due to suppression of Notch endocytosis (Das and Knust, Citation2018; Nemetschke & Knust, Citation2016; Shimizu et al., Citation2017). Interestingly, the loss of either Crumbs or ZO-1 activity produces Notch gain of function phenotypes in different tissues. Thus, alternative interactions with Notch may provide specificity to the tuning of Notch signalling capacity in different developmental contexts.
In mammals, ZO-1 is a tight junction component and also present in some specialised adherens junctions. In Drosophila ZO-1 is mainly localised to the adherens junctions and its loss results a Deltex-dependent gain of Notch activity reflected by an increase in the size and capacity of the germ line stem cell niche of the Drosophila ovary, a tissue in which ZO-1 is strongly expressed (Djiane et al., Citation2011; Shimizu et al., Citation2017). Recent work has shown how stretch-dependent regulation of its protein interactions allows ZO-1 to act as a tension sensor (Spadaro et al., Citation2017). It is plausible that similar mechanisms may control ZO-1 action on Notch in flies in coordination with tissue assembly, and may also underlie the observed cell density-dependent regulatory outcomes on Notch (Shimizu et al., Citation2017).
Crumbs is an apically localised cell membrane protein which, like Notch, has an extracellular domain substantially comprised of EGF-like modules. In the developing Drosophila embryo Crumbs regulates apical-basal polarity of the epithelial cells, a function that only requires the Crumbs intracellular domain. In the larval wing imaginal discs there is no requirement for this activity of Crumbs but instead its loss of function results in increased Notch endocytosis, independently of ligands, and increased Notch signalling activity. Unlike its role in apical basal cell polarity, the activity of Crumbs to suppress Notch endocytosis is a function that requires only its extracellular domain. The extra Notch activity arising from Crumbs loss of function is suppressed by simultaneous removal of Deltex and also mutants in the HOPS complex. The latter is required for late endosomal/lysosomal fusion and is only required for Dx-dependent activation of Notch and not for the ligand-induced or the alternative lipid raft-associated endocytic Notch activation mechanisms (Shimizu et al., Citation2014; Wilkin et al., Citation2008). Crumbs action on Notch is itself regulated by an extracellular interaction of Crumbs with the WD40 protein Ebi which promotes Crumbs ubiquitination and downregulation (Nguyen et al., Citation2016). Crumbs can also be regulated through intracellular domain interactions. In zebrafish brain development, the FERM domain protein Mosaic Eyes down-regulates Crumbs to maintain Notch signalling and regulate proliferation of neuroepithelial stem cells (Ohata et al., Citation2011). It is therefore likely that both ZO-1 and Crumbs provide hubs allowing cross talk with other physiological processes to tune Notch activity.
It is interesting that other extracellular proteins have been shown to bind and activate Notch through either canonical or Deltex-dependent activities. F3/Contactin is a glycosyl phosphatidylinositol (GPI)-anchored immunoglobulin domain superfamily protein, which acts as a neuronal cell adhesion protein. It has been found to act as an alternative ligand for Notch, regulating vertebrate oligodendrocyte maturation in the developing brain, and hence has an important role in axon myelination (Hu et al., Citation2003). Its activity was found to produce nuclear localised NICD, dependent on gamma-secretase activity. Deltex-1, one of four human Deltex homologues was found to be required for Contactin-induced oligodendrocyte maturation (Hu et al., Citation2003). Interestingly loss of this signal may have relevance for the progression of multiple sclerosis, which results from loss of axon myelination (Aparicio et al., Citation2013; Brosnan et al., Citation2009). A Contactin-related protein NB-3 also acts as a Notch ligand, likely at an earlier step in the oligodendrocyte lineage, through a similar Deltex1-dependent mechanism (Cui et al., Citation2004). NB3 also acts through Notch to promote proper cerebellum development, a process regulated by an additional extracellular interactor, Adropin (Wong et al., Citation2014). The precise stage at which mammalian Deltex interacts with and facilitates this process is at present unclear, as different activities of mammalian Deltex on Notch have been reported. Given that there are four mammalian Notch homologues and four Deltex homologues some diversity and specialisation of function may be expected. Some previous work has suggested that mammalian Deltex1 can bind to the Notch1 NICD and regulate its interactions and activity in the nucleus, acting to inhibit CSL-signalling (Yamamoto et al., Citation2001). Additionally recent work suggests that in some contexts mammalian Deltex4 can promote ligand-induced signalling by increasing Notch endocytosis of Notch1 in the trans-ligand complex, contributing to endocytic-dependent unwinding of the NRR domain in the activation step (Chastagner et al., Citation2017). On the other hand Deltex1 has also been proposed to down-regulate Notch activity by acting to prevent Notch1 recycling to the cell surface via a Rab4 dependent trafficking route (Zheng & Conner, Citation2018). Deltex1 and Deltex3 acting in a heterodimeric complex are also reported to down-regulate Notch activity and promote vessel sprouting of the lymphatic system (Choi et al., Citation2017).
An alternative ligand has also been identified for human Notch3 protein (Rauen et al., Citation2009). Y Box protein-1 (YB-1) is a cold shock protein that is involved in transcriptional regulation, but is also secreted from the cell by a non-classical secretion pathway involving its ubiquitination and recruitment by the ESCRT complex component TSG101. It furthermore binds to the Notch extracellular domain and activates gamma-secretase-dependent signalling by Notch3 to upregulate the expression of the Notch target HES2, and this may be relevant to the progression of mesangioproliferative nephritis, an inflammatory disease of the kidney (Rauen et al., Citation2009; Raffetseder et al., Citation2011). It will be interesting to determine whether this alternative ligand also acts through a Deltex-mediated process.
It remains to be seen whether some of the diverse mutant allele phenotypes associated with missense mutations at different locations in the Notch extracellular domain may be explained by disruption of such alternative extracellular interactions. Both Contactin and NB-3 bind to Notch within the EGF22-34 which lies outside of the normal ligand binding domain but encompassing a region within which, in Drosophila Notch, several gain of function Notch alleles are located (Cui et al., Citation2004; Hu et al., Citation2003). YB-1 was, like NB-3, found to bind to Notch via EGF-module region that is C-terminal to the DSL-ligand binding site, in this case encompassing EGF13-33 of Notch3 (Rauen et al., Citation2009). The EGF18-36 region of Drosophila Notch has also been found to interact, using molecular crosslinking studies with Wingless, a Drosophila Wnt protein (Wesley, Citation1999; Wesley & Saez, Citation2000). There is a close interrelationship between Notch and Wingless/Wnt signalling components and signal cross talk between the respective signals has been documented at many levels. Recent work indicated the downstream Wingless pathway component Dishevelled can bind to and activate the ubiquitin ligase Su(dx) providing a potential route of cross talk at the level of Notch trafficking, however whether this intracellular interaction is in anyway linked to a potential interaction between Wingless and the Notch extracellular domain is at this point unclear (Mund et al., Citation2015).
When considering the potential alternative interactions of the Notch extracellular domain it is also necessary to consider whether there are dimeric interactions between different Notch molecules and what roles these might play in Notch regulation. Dimeric interactions of the intracellular domain mediated through a contact interface within the ankyrin domain are implicated in activating a subset of Notch responsive enhancers that contain paired CSL binding sites (Arnett et al., Citation2010). It is also possible that regions of the extracellular domain contribute to Notch/Notch dimeric contacts. A number of studies have suggested such dimeric complexes exist and are mediated through the EGF region of the Notch extracellular domain, however the physiological significance is not clear (Kelly et al., Citation2010; Sakamoto et al., Citation2005; Vooijs et al., Citation2004). On possibility is that inter or intra-molecular interactions between different EGF-module regions of Notch may modulate receptor/ligand interactions. Using binding assays with isolated fragments the EGF modules 22-27 were found to bind to EGF11-14 region and to compete with the ligand Delta for binding to this region (Pei & Baker, Citation2008; Sharma et al., Citation2013). It was suggested that gain of function mutations that map to this Abruptex region might disrupt this Notch/Notch interaction and make ligand binding more accessible, although this was not tested directly (Pei & Baker, Citation2008). It is also not clear if such an interaction occurs intramolecularly or if the interaction might result in dimerisation of Notch proteins. While calcium binding EGF domains form an extended rigid conformation, non-calcium binding inter-domain interfaces can produce a bent configuration or may be flexible (Weisshuhn et al., 2016), which might favour either intra or intermolecular contacts.
The N-terminal region of the notch extracellular domain
If the EGF8-12 region forms a platform for canonical ligand activation and the EGF13-36 region mediates a number of alternative ligand interactions, then is there a distinct function for the EGF1-7 domains that lie N-terminal to the DSL ligand binding sites? There is little direct mechanistic information but genetic data suggest that this region also carries a distinctive regulatory activity. Mutations in the human Notch3 gene are associated with a dominantly inherited disorder CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy), which is a disease whose pathology is associated with vascular smooth muscle misfunction and frequent ischaemic strokes and dementia. The distribution of these disease-associated mutations shows an interesting enrichment within the N-terminal EGF repeat region of Notch, in particular within EGF modules 1-5 (Joutel et al., Citation1997). Whether the CADASIL phenotype reflects a specific mis-regulation of Notch through disrupting an N-terminal specific function is however unclear. CADASIL mutations are most often ones that remove or add cysteine residues, disrupting the disulphide bond formation, which is a core feature of the EGF module structure. Misfolding and protein aggregation might lead to the observed accumulation of Notch3 extracellular domain in granular osmiophilic material, which is a hallmark of the CADASIL disease phenotype (Yamamoto. et al., Citation2013). However this does not provide a straightforward explanation for the observed clustering of CADASIL mutants in the N-terminal EGF-modules and the consequences on Notch3 trafficking and causes of Notch3 extracellular domain accumulation are not well understood. Indeed mutant analysis of Drosophila Notch has shown that cysteine mutations located in different EGF repeat regions can have quite different tissue specific phenotypic outcomes which may be loss or gain of Notch activity suggesting disruption of more specific regulatory functions may contribute to their outcomes. Cysteine mutations in the Abruptex region of Notch are linked to gain of function activity that suppresses wing vein differentiation (Kelley et al., Citation1987). In contrast a Drosophila Notch cysteine replacement mutant in the EGF2 module, a location often mutated in CADASIL, is associated with reduced Notch activity affecting wing development, and in cell culture has been found to increase cell surface localisation through reduced endocytosis (Bardot et al., Citation2005). Therefore there may yet be interacting partners to be identified that modulate Notch localisation through this N-terminal region. A similar diversity of mutant outcomes also arises from Cysteine mutants in different EGF domain regions of the extracellular matrix protein Fibrillin, whose disruption leads to the connective tissue disorder Marfan's syndrome (Schrijver et al. Citation1999). Intriguingly structural studies have identified an unusual inter-module linkage around the EGF5-6 interface of Notch 1 that produces a significant bend in the extended Notch structure (Weisshuhn et al., Citation2016). Whether this structural feature has functional significance and whether this feature is relevant to the clustering of CADASIL mutants in this region of Notch3 remains an interesting avenue for further exploration.
Conclusions and future perspectives
Biophysical approaches to the study of Notch are allowing its key regulatory complexes to be understood at a highly detailed level. These approaches are at their most powerful however when combined with mutational data to probe Notch mechanism and regulation. Intensive studies of Notch over 100 years has provided a rich source of function-perturbing mutations from both classical genetic approaches and more recent genome-wide sequencing efforts which link Notch to various diseases and inherited disorders. Exploiting this mutational database alongside structural investigations will likely reveal important new information about the normal and pathological regulation of Notch activity. With gene editing techniques in model organisms becoming routine, there is ample scope for understanding and integrating knowledge of protein structure/function across multiple scales from atomic resolution up to changes in the organismal phenotype. The Drosophila model offers the capacity and speed to feasibly allow the introduction of germline mutations at a medium throughput. Over a century since the initial identification of the Notch locus by Morgan lab, the fruit fly is set to continue to make important contributions to the study of Notch protein structure/function relationships in the modern era.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. 2011. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333:1154–1157.
- Andrawes MB, Xu X, Liu H, Ficarro SB, Marto JA, Aster JC, Blacklow SC. 2013. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J Biol Chem 288:25477–25489.
- Aparicio E, Mathieu P, Pereira Luppi M, Almeira Gubiani MF, Adamo AM. 2013. The Notch signaling pathway: its role in focal CNS demyelination and apotransferrin-induced remyelination. J Neurochem 127:819–836.
- Arnett KL, Hass M, McArthur DG, Ilagan MX, Aster JC, Kopan R, Blacklow SC. 2010. Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nat Struct Mol Biol 17:1312–1317.
- Aster JC, Pear WS, Blacklow SC. 2017. The varied roles of notch in cancer. Annu Rev Pathol 12:245–275.
- Bardot B, Mok LP, Thayer T, Ahimou F, Wesley C. 2005. The Notch amino terminus regulates protein levels and Delta-induced clustering of Drosophila Notch receptors. Exp Cell Res 304:202–223.
- Baron M, Norman DG, Harvey TS, Handford PA, Mayhew M, Tse AG, et al. 2008. The three-dimensional structure of the first EGF-like module of human factor IX: comparison with EGF and TGF-alpha. Protein Sci 1:81–90.
- Becam I, Fiuza UM, Arias AM, Milán M. 2010. A role of receptor Notch in ligand cis-inhibition in Drosophila. Curr Biol 20:554–560.
- Bray SJ. 1998. Notch signalling in Drosophila: three ways to use a pathway. Semin Cell Dev Biol. 9:591–597.
- Bray SJ. 2016. Notch signalling in context. Nat Rev Mol Cell Biol 17:722–735.
- Brosnan CF, John GR. 2009. Revisiting Notch in remyelination of multiple sclerosis lesions. J Clin Invest 119:10–13.
- Chapman G, Sparrow DB, Kremmer E, Dunwoodie SL. 2011. Notch inhibition by the ligand DELTA-LIKE 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum Mol Genet 20:905–916.
- Chastagner P, Rubinstein E, Brou C. 2017. Ligand-activated Notch undergoes DTX4-mediated ubiquitylation and bilateral endocytosis before ADAM10 processing. Sci Signal 10:eaag2989.
- Chiba S. 2006. Notch signaling in stem cell systems. Stem Cells 24:2437–2447.
- Chillakuri CR, Sheppard D, Ilagan MX, Holt LR, Abbott F, Liang S, et al. 2013. Structural analysis uncovers lipid-binding properties of Notch ligands. Cell Rep 5:861–867.
- Choi D, Park E, Jung E, Seong YJ, Yoo J, Lee E, et al. 2017. Laminar flow downregulates Notch activity to promote lymphatic sprouting. J Clin Invest 127:1225–1240.
- Choi SH, Wales TE, Nam Y, O'Donovan DJ, Sliz P, Engen JR, Blacklow SC. 2012. Conformational locking upon cooperative assembly of Notch transcription complexes. Structure 20:340–349.
- Choy L, Hagenbeek TJ, Solon M, French D, Finkle D, Shelton A, et al. 2017. Constitutive NOTCH3 signaling promotes the growth of basal breast cancers. Cancer Res 77:1439–1452.
- Conboy IM, Rando TA. 2005. Aging, stem cells and tissue regeneration: lessons from muscle. Cell Cycle 4:407–410.
- Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, et al. 2008. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol 15:849–857.
- Cordle J, Redfield C, Stacey M, van der Merwe PA, Willis AC, Champion BR, et al. 2008. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J Biol Chem 283:11785–11793.
- Cui XY, Hu QD, Tekaya M, Shimoda Y, Ang BT, Nie DY, et al. 2004. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J Biol Chem 279:25858–25865.
- Das S, Knust E. 2018. A dual role of the extracellular domain of Drosophila Crumbs for morphogenesis of the embryonic neuroectoderm. Biol Open 7:bio031435.
- de Celis JF, Barrio R, del Arco A, García-Bellido A. 1993. Genetic and molecular characterization of a Notch mutation in its Delta- and Serrate-binding domain in Drosophila. Proc Natl Acad Sci U.S.A 90:4037–4041.
- de Celis JF, Bray S. 1997. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124:3241–3251.
- Djiane A, Shimizu H, Wilkin M, Mazleyrat S, Jennings MD, Avis J, et al. 2011. Su(dx) E3 ubiquitin ligase-dependent and -independent functions of polychaetoid, the Drosophila ZO-1 homologue. J Cell Biol 192:189–200.
- Domínguez M, de Celis JF. 1998. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature 396:276–278.
- Downing AK, Knott V, Werner JM, Cardy CM, Campbell ID, Handford PA. 1996. Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell 85:597–605.
- Fleming RJ, Gu Y, Hukriede NA. 1997. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development 124:2973–2981.
- Fleming RJ, Hori K, Sen A, Filloramo GV, Langer JM, Obar RA, et al. 2013. An extracellular region of Serrate is essential for ligand-induced cis-inhibition of Notch signaling. Development 140:2039–2049.
- Foster GG. 1975. Negative complementation at the Notch locus of Drosophila melanogaster. Genetics 81:99–120.
- Fostier M, Evans DA, Artavanis-Tsakonas S, Baron M. 1998. Genetic characterization of the Drosophila melanogaster Suppressor of deltex gene: A regulator of Notch signaling. Genetics 150:1477–1485.
- Gómez-del Arco P, Kashiwagi M, Jackson AF, Naito T, Zhang J, Liu F, et al. 2010. Alternative promoter usage at the Notch1 locus supports ligand-independent signaling in T cell development and leukemogenesis. Immunity 33:685–698.
- Gomez-Lamarca MJ, Falo-Sanjuan J, Stojnic R, Abdul Rehman S, Muresan L, Jones ML, et al. 2018. Activation of the notch signaling pathway in vivo elicits changes in CSL nuclear dynamics. Developmental Cell 44:611–613.
- Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. 2009. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood 113:4381–4390.
- Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, et al. 2015. Mechanical allostery: evidence for a force requirement in the proteolytic activation of notch. Dev Cell 33:729–736.
- Gorman MJ, Girton JR. 1992. A genetic analysis of deltex and its interaction with the Notch locus in Drosophila melanogaster. Genetics 131:99–112.
- Greenwald I. 1998. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev 12:1751–1762.
- Hambleton S, Valeyev NV, Muranyi A, Knott V, Werner JM, McMichael AJ, et al. 2004. Structural and functional properties of the human notch-1 ligand binding region. Structure 12:2173–2183.
- Hamel S, Fantini J, Schweisguth F. 2010. Notch ligand activity is modulated by glycosphingolipid membrane composition in Drosophila melanogaster. J Cell Biol 188:581–594.
- Handford PA, Mayhew M, Baron M, Winship PR, Campbell ID, Brownlee GG. 1991. Key residues involved in calcium-binding motifs in EGF-like domains. Nature 351:164–167.
- He L, Si G, Huang J, Samuel ADT, Perrimon N. 2018. Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel. Nature 555:103–106.
- Heuss SF, Tarantino N, Fantini J, Ndiaye-Lobry D, Moretti J, Israël A, Logeat F. 2013. A glycosphingolipid binding domain controls trafficking and activity of the mammalian notch ligand delta-like 1. PLoS One 8:e74392
- Hori K, Sen A, Artavanis-Tsakonas S. 2013. Notch signaling at a glance. J Cell Sci 126:2135–2140.
- Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. 2011. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J Cell Biol 195:1005–1015.
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, et al. 2003. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell 115:163–175.
- Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssière C, et al. 1997. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet 350:1511–1515.
- Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. 1997. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development 124:2245–2254.
- Kakuda S, Haltiwanger RS. 2017. Deciphering the fringe-mediated notch code: identification of activating and inhibiting sites allowing discrimination between ligands. Dev Cell 40:193–201.
- Katic I, Vallier LG, Greenwald I. 2005. New positive regulators of lin-12 activity in Caenorhabditis elegans include the BRE-5/Brainiac glycosphingolipid biosynthesis enzyme. Genetics 171:1605–1615.
- Kelley MR, Kidd S, Deutsch WA, Young MW. 1987. Mutations altering the structure of epidermal growth factor-like coding sequences at the Drosophila Notch locus. Cell. 51:539–548.
- Kelly DF, Lake RJ, Middelkoop TC, Fan HY, Artavanis-Tsakonas S, Walz T. 2010. Molecular structure and dimeric organization of the Notch extracellular domain as revealed by electron microscopy. PLoS One 5:e10532
- Kidd S, Lieber T. 2002. Furin cleavage is not a requirement for Drosophila Notch function. Mech Dev 115:41–51.
- Klein T, Brennan K, Arias AM. 1997. An intrinsic dominant negative activity of serrate that is modulated during wing development in Drosophila. Dev Biol 189:123–134.
- Komatsu H, Chao MY, Larkins-Ford J, Corkins ME, Somers GA, Tucey T, et al. 2008. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol 6:e196
- Kopan R, Ilagan MX. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233.
- Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, et al. 2005. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol 170:983–992.
- Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. 2005. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development 132:2319–2332.
- Langridge PD, Struhl G. 2017. Epsin-dependent ligand endocytosis activates notch by force. Cell 171:1383–1396.
- Lampreia FP, Carmelo JG, Anjos-Afonso F. 2017. Notch signaling in the regulation of hematopoietic stem cell. Curr Stem Cell Rep 3:202–209.
- Lee TV, Pandey A, Jafar-Nejad H. 2017. Xylosylation of the Notch receptor preserves the balance between its activation by trans-Delta and inhibition by cis-ligands in Drosophila. PLoS Genet 13:e1006723
- Lehmann R, Dietrich U, Jiménez F, Campos-Ortega JA. 1981. Mutations of early neurogenesis in Drosophila. Wilehm Roux Arch Dev Biol 190:226–229.
- Liu L, Wada H, Matsubara N, Hozumi K, Itoh M. 2017. Identification of domains for efficient notch signaling activity in immobilized notch ligand proteins. J Cell Biochem 118:785–796.
- Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC. 2015. Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science 347:847–853.
- Luca VC, Kim BC, Ge C, Kakuda S, Wu D, Roein-Peikar M, et al. 2017. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science 355:1320–1324.
- Mašek J, Andersson ER. 2017. The developmental biology of genetic Notch disorders. Development 144:1743–1763.
- Mohtashami M, Shah DK, Nakase H, Kianizad K, Petrie HT, Zúñiga-Pflücker JC. 2010. Direct comparison of Dll1- and Dll4-mediated Notch activation levels shows differential lymphomyeloid lineage commitment outcomes. J Immunol 185:867–876.
- Micchelli CA, Rulifson EJ, Blair SS. 1997. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124:1485–1495.
- Morel V, Lecourtois M, Massiani O, Maier D, Preiss A, Schweisguth F. 2001. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol 11:789–792.
- Morgan TH. 1917. The theory of the gene. Am Naturalist 51:513–544.
- Mukherjee T, Kim WS, Mandal L, Banerjee U. 2011. Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science 332:1210–1213.
- Mund T, Graeb M, Mieszczanek J, Gammons M, Pelham HR, Bienz M. 2015. Disinhibition of the HECT E3 ubiquitin ligase WWP2 by polymerized Dishevelled. Open Biol 5:150185
- Mumm JS, Kopan R. 2000. Notch signaling: from the outside in. Dev Biol 228:151–165.
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. 2006. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124:973–983.
- Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME, Elowitz MB. 2018. Dynamic ligand discrimination in the notch signaling pathway. Cell 172:869–880.
- Nemetschke L, Knust E. 2016. Drosophila Crumbs prevents ectopic Notch activation in developing wings by inhibiting ligand-independent endocytosis. Development 143:4543–4553.
- Nguyen MB, Vuong LT, Choi KW. 2016. Ebi modulates wing growth by ubiquitin-dependent downregulation of Crumbs in Drosophila. Development 143:3506–3513.
- Ohata S, Aoki R, Kinoshita S, Yamaguchi M, Tsuruoka-Kinoshita S, Tanaka H, et al. 2011. Dual roles of Notch in regulation of apically restricted mitosis and apicobasal polarity of neuroepithelial cells. Neuron 69:215–230.
- Ohlstein B, Spradling A. 2007. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential Notch signaling. Science 315:988–992.
- Palmer WH, Jia D, Deng WM. 2014. Cis-interactions between Notch and its ligands block ligand-independent Notch activity. Elife 8 3:
- Panin VM, Papayannopoulos V, Wilson R, Irvine KD. 1997. Fringe modulates Notch-ligand interactions. Nature 387:908–912.
- Papayannopoulos V, Tomlinson A, Panin VM, Rauskolb C, Irvine KD. 1998. Dorsal-ventral signaling in the Drosophila eye. Science 281:2031–2034.
- Parks AL, Klueg KM, Stout JR, Muskavitch MA. 2000. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127:1373–1385.
- Parks AL, Stout JR, Shepard SB, Klueg KM, Dos Santos AA, Parody TR, et al. 2006. Structure-function analysis of delta trafficking, receptor binding and signaling in Drosophila. Genetics 174:1947–1961.
- Pei Z, Baker NE. 2008. Competition between Delta and the Abruptex domain of Notch. BMC Dev Biol 8:4.
- Poulson DF. 1937. Chromosomal deficiencies and the embryonic development of drosophila melanogaster. Proc Natl Acad Sci U.S.A 23:133–137.
- Preusze K, Tveriakhina L, Schuster-Gossler K, Gaspar C, Rosa AI, Henrique D, et al. 2015. Context-dependent functional divergence of the notch ligands DLL1 and DLL4 in vivo. PLoS Genet 11:e1005328
- Raffetseder U, Rauen T, Boor P, Ostendorf T, Hanssen L, Floege J, et al. 2011. Extracellular YB-1 blockade in experimental nephritis upregulates Notch-3 receptor expression and signaling. Nephron Exp Nephrol 118:e100–e108.
- Rauen T, Raffetseder U, Frye BC, Djudjaj S, Mühlenberg PJ, Eitner F, et al. 2009. YB-1 acts as a ligand for Notch-3 receptors and modulates receptor activation. J Biol Chem 284:26928–26940.
- Ramain P, Khechumian K, Seugnet L, Arbogast N, Ackermann C, Heitzler P. 2001. Novel Notch alleles reveal a Deltex-dependent pathway repressing neural fate. Curr Biol 11:1729–1738.
- Rao Z, Handford P, Mayhew M, Knott V, Brownlee GG, Stuart D. 1995. The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell 82:131–141.
- Raya A, Kawakami Y, Rodriguez-Esteban C, Ibanes M, Rasskin-Gutman D, Rodriguez LJ, et al. 2004. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature 427:121–128.
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. 1991. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67:687–699.
- Sakamoto K, Chao WS, Katsube K, Yamaguchi A. 2005. Distinct roles of EGF repeats for the Notch signaling system. Exp Cell Res 302:281–291.
- Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S. 2004. Drosophila Nedd4 regulates endocytosis of Notch and suppresses its ligand-independent activation. Curr Biol 14:2228–2236.
- Schneider M, Troost T, Grawe F, Martinez-Arias A, Klein T. 2013. Activation of Notch in lgd mutant cells requires the fusion of late endosomes with the lysosome. J Cell Sci 126:645–656.
- Schrijver I, Liu W, Brenn T, Furthmayr H, Francke U. 1999. Cysteine substitutions in epidermal growth factor-like domains of fibrillin-1: distinct effects on biochemical and clinical phenotypes. Am J Hum Genet 65:1007–1020.
- Schuster-Gossler K, Cordes R, Müller J, Geffers I, Delany-Heiken P, Taft M, et al. 2016. Context-dependent sensitivity to mutations disrupting the structural integrity of individual EGF repeats in the mouse notch ligand DLL1. Genetics 202:1119–1133.
- Seo D, Southard KM, Kim JW, Lee HJ, Farlow J, Lee JU, et al. 2016. A mechanogenetic toolkit for interrogating cell signaling in space and time. Cell. 165:1507–1518.
- Sharma A, Rangarajan A, Dighe RR. 2013. Antibodies against the extracellular domain of human Notch1 receptor reveal the critical role of epidermal-growth-factor-like repeats 25-26 in ligand binding and receptor activation. Biochem J 449:519–530.
- Shimizu H, Woodcock SA, Wilkin MB, Trubenová B, Monk NA, Baron M. 2014. Compensatory flux changes within an endocytic trafficking network maintain thermal robustness of Notch signaling. Cell 157:1160–1174.
- Shimizu H, Wilkin MB, Woodcock SA, Bonfini A, Hung Y, Mazaleyrat S, Baron M. 2017. The Drosophila ZO-1 protein Polychaetoid suppresses Deltex-regulated Notch activity to modulate germline stem cell niche formation. Open Biol 7: 160322.
- Shimizu K, Chiba S, Saito T, Kumano K, Hirai H. 2000. Physical interaction of Delta1, Jagged1, and Jagged2 with Notch1 and Notch3 receptors. Biochem Biophys Res Commun 276:385–389.
- Siebel C, Lendahl U. 2017. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev 97:1235–1294.
- Spadaro D, Le S, Laroche T, Mean I, Jond L, Yan J, Citi S. 2017. Tension-dependent stretching activates ZO-1 to control the junctional localization of its interactors. Curr Biol 27:3783–3795.
- Sörensen I, Adams RH, Gossler A. 2009. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood 113:5680–5688.
- Stephenson NL, Avis JM. 2012. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proc. Natl. Acad. Sci. U.S.A 109:E2757–E2765.
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. 2011. The mutational landscape of head and neck squamous cell carcinoma. Science 333:1157–1160.
- Suckling RJ, Korona B, Whiteman P, Chillakuri C, Holt L, Handford PA, Lea SM. 2017. Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands. Embo J 36:2204–2215.
- Taylor P, Takeuchi H, Sheppard D, Chillakuri C, Lea SM, Haltiwanger RS, Handford PA. 2014. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc Natl Acad Sci U.S.A 111:7290–7295.
- Thompson BJ, Mathieu J, Sung HH, Loeser E, Rørth P, Cohen SM. 2005. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell 9:711–720.
- Vaccari T, Bilder D. 2005. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell 9:687–698.
- Vaccari T, Rusten TE, Menut L, Nezis IP, Brech A, Stenmark H, Bilder D. 2009. Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J Cell Sci 122:2413–2423.
- Vooijs M, Schroeter EH, Pan Y, Blandford M, Kopan R. 2004. Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization. J Biol Chem 279:50864–50873.
- Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. 2011. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U.S.A 108:17761–17766.
- Wang X, Ha T. 2013. Defining single molecular forces required to activate integrin and notch signaling. Science 340:991–994.
- Weinmaster G. 1997. The ins and outs of notch signaling. Mol Cell Neurosci 9:91–102.
- Weisshuhn PC, Sheppard D, Taylor P, Whiteman P, Lea SM, Handford PA, Redfield C. 2016. Non-linear and flexible regions of the human notch1 extracellular domain revealed by high-resolution structural studies. Structure 24:555–566.
- Welshons WJ. 1971. Genetic basis for two types of recessive lethality at the notch locus of Drosophila. Genetics 68:259–268.
- Welshons WJ, Von Halle ES. 1962. Pseudoallelism at the Notch locus in drosophila. Genetics 47:743–759.
- Wesley CS. 1999. Notch and wingless regulate expression of cuticle patterning genes. Mol Cell Biol 19:5743–5758.
- Wesley CS, Saez L. 2000. Notch responds differently to Delta and Wingless in cultured Drosophila cells. J Biol Chem 275:9099–9101.
- Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. 1985. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell 43:567–581.
- Whiteman P, de Madrid BH, Taylor P, Li D, Heslop R, Viticheep N, et al. 2013. Molecular basis for Jagged-1/Serrate ligand recognition by the Notch receptor. J Biol Chem 288:7305–7312.
- Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, et al. 2004. Regulation of Notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol 14:2237–2244.
- Wilkin M, Tongngok P, Gensch N, Clemence S, Motoki M, Yamada K, et al. 2008. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev Cell 15:762–772.
- Wilson JJ, Kovall RA. 2006. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124:985–996.
- Wong CM, Wang Y, Lee JT, Huang Z, Wu D, Xu A, Lam KS. 2014. Adropin is a brain membrane-bound protein regulating physical activity via the NB-3/Notch signaling pathway in mice. J Biol Chem 289:25976–25986.
- Xu T, Artavanis-Tsakonas S. 1990. Deltex, a locus interacting with the neurogenic genes, Notch, Delta and Mastermind in Drosophila melanogaster. Genetics 126:665–677.
- Yamamoto S, Charng WL, Rana NA, Kakuda S, Jaiswal M, Bayat V, et al. 2012. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science 338:1229–1232.
- Yamamoto Y, Craggs LJ, Watanabe A, Booth T, Attems J, Low RW, et al. 2013. Brain microvascular accumulation and distribution of the NOTCH3 ectodomain and granular osmiophilic material in CADASIL. J Neuropathol Exp Neurol 72:416–431.
- Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, et al. 2001. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J Biol Chem 276:45031–45040.
- Zheng L, Conner SD. 2018. PI5P4Kγ functions in DTX1-mediated Notch signaling. Proc Natl Acad Sci U.S.A 115:E1983–E1990.
- Zheng L, Saunders CA, Sorensen EB, Waxmonsky NC, Conner SD. 2013. Notch signaling from the endosome requires a conserved dileucine motif. Mol Biol Cell 24:297–307.
