Abstract
The integral membrane protein Ptr2p transports di/tri-peptides into the yeast Saccharomyces cerevisiae. The sequence FYXXINXG (FYING motif) in the 5th transmembrane domain (TM5) is invariably conserved among the members of the PTR (Peptide TRansport) family ranging from yeast to human. To test the role of TM5 in Ptr2p function, Ala-scanning mutagenesis of the 22 residues comprising TM5 was completed. All mutated transporters, with the exception of the Y248A mutant, were expressed as determined by immunoblots. In peptide-dependent growth assays, ten mutants of the non-FYING residues grew as well as wild-type Ptr2p on all twelve different peptides tested. All of the FYING motif mutants, except the non-expressed Y248A, plus seven other mutants in TM5 exhibited differential growth on peptides including Leu-Leu and Met-Met-Met indicating that these mutations conferred substrate preference. In assays measuring direct uptake of the radioactive peptides 3H-Leu-Leu or 14C-Met-Met-Met, the F, I and G mutants of the FYING motif did not demonstrate accumulation of these peptides over a ten minute interval. The mutation N252A of the FYING motif, along with L240A, M250A, and L258A, exhibited differential substrate preference for Met-Met-Met over Leu-Leu. Other mutations (T239A, Q241A, N242A, M245A, and A260) resulted in preference for Leu-Leu over Met-Met-Met. These data demonstrate that TM5, in particular its conserved FYING motif, is involved in substrate preference of Ptr2p.
| Abbreviations | ||
| Ptr2p | = | the di/tri-peptide transport protein of Saccharomyces cerevisiae |
| TM | = | transmembrane domain |
| FYING | = | the conserved motif in TM5 of di/tri-peptide transport proteins in the PTR family |
| Oxa | = | oxalysine |
| Eth | = | ethionine |
Introduction
The ability of the yeast Saccharomyces cerevisiae to transport small peptides was first documented over 25 years ago (Becker & Naider [Citation1977]). In 1994, Ptr2p was identified as the protein responsible for peptide transport. This 601 amino acid, integral membrane protein mediates translocation of di/tri-peptides into the yeast cell in a proton-dependent manner (Perry et al. [Citation1994]). The identification and characterization of a mammalian peptide transporter (PEPT1) from rabbit small intestine was reported as well in 1994 (Fei et al. [Citation1994]). Subsequently, similar transporters were identified by sequence similarity in virtually all systems studied to date, ranging from archaea to human (Saier [Citation2000]).
The hallmarks of the PTR (Peptide TRansport) [also known as the POT (Proton-dependent Oligopeptide Transporter) family (TC 2.A.17)] (Busch & Saier [Citation2002], Saier [Citation2000]) are that they are integral membrane proteins of 12 membrane-spanning domains that function in the symport of protons and di/tri-peptides into the cell. The predicted topology of Ptr2p is presented in . This model is based on the experimentally determined topology of the di/tri-peptide transporter DtpT of Lactococcus lactis (Hagting et al. [Citation1997]) and that of hPEPT1 (Covitz et al. [Citation1998]), both of which indicate that the PTR transporters have 12 membrane-spanning domains and internal N- and C-termini. In addition to exhibiting broad substrate recognition of small peptides, many of these family members also recognize and transport peptidomimetic drugs such as beta-lactam antibiotics (Boll et al. [Citation1994], Ganapathy et al. [Citation1995]), dipeptide-like anti-neoplastic drugs (Saito et al. [Citation1996], Terada et al. [Citation1997]) and anti-viral prodrugs (Ganapathy et al. [Citation1998], Sugawara et al. [Citation2000]). It is the potential role of the peptide transporters, particularly the human intestinal PEPT1 and its renal counterpart PEPT2, as targets for drug delivery that has stimulated interest in the structure and function of these proteins (Gebauer et al. [Citation2003]), with recent emphasis on elucidating the residues involved in substrate translocation (Kulkarni et al. [Citation2003a], Kulkarni et al. [Citation2003b]).
Figure 1. (a) Cartoon of Ptr2p Topology. Filled circles represent amino acids 1–601 of native Ptr2p; open circles denote the placement of the C-terminal FLAG and His epitope tags. TM5 is indicated by the oval. (b) Enlarged representation of TM5 indicated by the oval in . The invariably conserved FYING residues are indicated in black. Highly, but not invariably, conserved residues are shaded grey. (c) Helical wheel representation of TM5. FYING residues are in black, and are all localized to the same half of the helix as indicated by the dotted line.
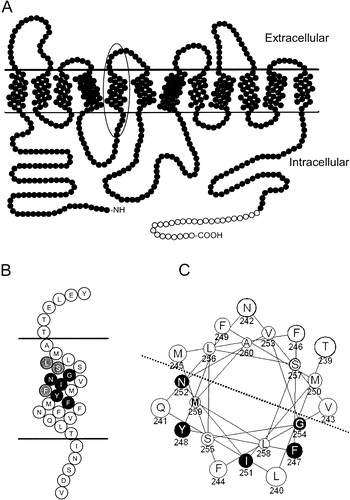
A direct comparison of the primary amino acid sequence for eukaryotic members of the PTR family reveals that within the fifth transmembrane domain (TM5) there exists an invariably conserved motif (), FYXXINXG, which we will refer to as the ‘FYING’ motif (Hauser et al. [Citation2001]). The universal nature of this signature sequence implies that these residues are important in the biology of the transporter. In the human peptide transporter hPEPT1, conversion of Y167 in the FYING motif to alanine, phenylalanine, serine, or histidine abolished the transport of the non-metabolized radiolabeled substrate glycyl-sarcosine (Yeung et al. [Citation1998]), even though the transporter was still detected in the membrane. In a recent study, Cys-scanning mutagenesis was applied to the entire TM5 of hPEPT1. The effect of the mutations on Gly-Sar transport was examined in both HEK293 cells and Xenopus oocytes. Based on the Substituted Cysteine Accessibility Method (SCAM) to identify solvent-accessible residues, it was postulated that TM5 lines the putative aqueous substrate translocation channel (Kulkarni et al. [Citation2003a]). Although this study provided important information regarding the involvement of TM5 in substrate translocation, it did not address the issue of how specific residues might be involved in substrate preference or recognition. As members of the PTR family have a broad range of substrate recognition (nearly 400 dipeptides and 8000 tri-peptides among the naturally-occurring amino acids) it is difficult to understand how peptide transporters can discriminate peptides from other substrates yet still retain a broad substrate recognition encompassing thousands of individual peptide molecules.
Figure 2. The FYING motif is invariably conserved in Ptr2p. The fifth transmembrane domain (TM5) of representative eukaryotic Ptr2ps for which peptide transport has been functionally verified were aligned using Clustal W. The FYING motif residues are highlighted. The superscript number before the first amino acid in each TM5 indicates the residue number of that particular protein. Names for genes encoding the transporters are listed in parenthesis.

To address this issue of substrate recognition, we have generated a series of Ala-scanned mutants encompassing the entire TM5 of Ptr2p in S. cerevisiae. Using S. cerevisiae it is possible to rapidly screen both wild-type Ptr2p and mutants with respect to peptide substrate preference. Strains of S. cerevisiae exist with a variety of auxotrophic requirements for many different amino acids, including leucine, lysine, histidine, methionine, and tryptophan. Amino acids can be supplied to these strains in the form of di- or tri-peptides containing any one of the auxotrophic amino acids thereby providing a simple growth assay to determine peptide uptake. Intact peptides containing the required amino acid must be transported into the cells, since there is no evidence that small (di-, tri-, tetra- or penta-) peptides are hydrolyzed in the yeast extracellular environment to yield free amino acids (Hauser et al. [Citation2000]). Once inside the cells, peptides are rapidly hydrolyzed to their component amino acids, thus satisfying the amino acid requirement of the strain and enabling its growth on minimal medium. Synthetic peptides containing toxic moieties (e.g., ethionine, oxalysine, p-fluorophenylalanine) can also be used for assays of peptide transport (Island et al. [Citation1987]). In this case, those cells expressing a peptide transporter with an affinity for a specific toxic peptide will accumulate the peptide and ultimately die. In addition to these trophic assays, direct measurement of radiolabeled di- or tri-peptide accumulation can be completed to assess preference for a particular peptide.
In this paper, we report that specific site-directed alanine mutations in TM5 of the S. cerevisiae peptide transporter Ptr2p result in differential alterations of transporter function, leading to changes in substrate preference. The FYING residues in TM5 appear to be particularly involved in substrate recognition, since mutagenesis of these residues resulted in the most striking changes in transporter activity.
Materials and methods
Strains, media and plasmids
The peptide-transport deficient strains BY4741-6009 and BY4742-16009 obtained from Invitrogen Corporation (Carlsbad, CA, USA), were used in all assays. The relevant genotype for strain BY4741-6009 is: MATa, ptr2 his leu met ura. For strain BY4742-16009, the genotype is: MATα, ptr2 his leu lys ura. The PTR2 gene was amplified by PCR from plasmid pJP9 (Perry et al. [Citation1994]) and cloned into pRS316 (Sikorski & Hieter [Citation1989]), a CEN-based yeast/bacterial shuttle vector containing a URA3 selectable marker and an F1 origin of replication. In this construct, PTR2 is under the control of its native promoter. FLAG and His tag sequences were fused in-frame to the 3′ end of the coding region. The tag sequences were PCR amplified from plasmid pNED (David et al. [Citation1997]) and added to the PTR2 sequence by in vivo ligation (Oldenburg et al. [Citation1997]) to yield plasmid pMS2. This was transformed by the method of Geitz (Gietz & Woods [Citation2002]) into the peptide-transport deficient strains BY4741-6009 and BY4742-16009. Transformants bearing pMS2 constructs were routinely maintained under uracil selection on medium containing 0.67% yeast nitrogen base (YNB) without amino acids, 2% glucose, 0.2% vitamin assay casamino acids, histidine (20 µg/ml), methionine (20 µg/ml), leucine (30 µg/ml), and lysine (30 µg/ml). To upregulate peptide transport activity, cells were grown under the ‘inducing conditions’ of nitrogen starvation (0.67% YNB without amino acids and ammonium sulphate plus 0.1% allantoin) in the presence of histidine, methionine, leucine, lysine, and the inducing amino acid tryptophan (20 µg/ml). All media components were obtained from Difco (Sparks, MD, USA) and amino acids from Sigma (St. Louis, MO, USA).
Site-directed mutagenesis
Single-stranded phagemid DNA of pMS2 was prepared by infecting E. coli strain CJ236 (ung−,dut−) carrying pMS2 with the helper phage M13KO7 (Vieira & Messing [Citation1987]). Oligonucleotide-directed mutagenesis of single-stranded phagemid DNA was completed as described by Kunkel et al. ([Citation1987]) such that each residue of TM5 was converted to alanine. Since alanine is normally present at position 260, this residue was converted to glycine. The product of the mutagenesis reaction mixture was transformed into E. coli strain DH5α (Invitrogen Corporation, Carlsbad, CA, USA), and transformants were selected on medium containing ampicillin. Plasmids were then isolated from transformants using the Wizard SV Miniprep kit (Promega, Madison, WI, USA). After sequence confirmation, constructs were transformed into yeast strain BY4741-6009 (ptr2-deletion strain), and transformants were selected by their growth on medium lacking uracil. Primers were purchased from Sigma/Genosys (The Woodlands, TX, USA) or IDT (Coralville, IA, USA). DNA sequencing was carried out in the Molecular Biology Resource Facility located on the campus of the University of Tennessee (Knoxville, TN, USA).
Growth assays
Peptides used in growth assays were obtained from Bachem (King of Prussia, PA, USA) and prepared as concentrated stocks in water. Cells expressing the various Ptr2p constructs were grown in liquid medium to mid-log phase under inducing conditions at 30°C, harvested, washed with water, and adjusted to a final concentration of 2×106 cells/ml. Cells were spotted (5 µl per spot) onto agar plates under inducing conditions, supplemented with peptide (20 and 160 µM) and auxotrophic amino acids, as required. Cells were scored for growth at 24 hour intervals over a three day time course. Spots exhibiting growth similar to wild-type were scored as ++, spots exhibiting reduced growth were scored as +, and spots which did not grow were scored as −. In all cases, the comparative growth among strains was similar whether determined at day one, two, or three. Growth assays were repeated three times with similar results. Data presented are from a representative experiment.
Membrane preparation and immunoblots
Cell expressing the various Ptr2p constructs were grown in liquid medium under inducing conditions overnight at 30°C. Cells were harvested at mid-log phage and washed with buffer (50 mM HEPES, pH 7.5, 5 mM EDTA) supplemented with protease inhibitors (1.5 µg/ml PMSF, 1 µg/ml leupeptin, 1 µg/ml pepstatin A). All steps were performed at 4°C. The cells were lysed by vortexing with glass beads. Following a low-speed spin (700×g, 5 min) to remove unbroken cells, cell wall debris and glass beads, the resulting supernatant was centrifuged at high speed (15,000×g, 30 min) to pellet membranes. The pellet was re-suspended in buffer (50 mM HEPES, pH 7.5, 20% glycerol) plus protease inhibitors and protein concentration determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA). The membranes were stored at –20°C until use. To assess the quality of the membrane preparation, proteins were fractionated by SDS-PAGE (10% acrylamide) and stained with Coomassie brilliant blue R-250 (0.2% in 50% methanol, 10% acetic acid). Gels were examined to verify that the lanes were equally loaded and that banding was sharp. For immunoblot analysis, membranes were solubilized in SDS sample buffer (10% glycerol, 5% 2-mercaptoethanol, 1% SDS, 0.03% bromophenol blue, 62.5 mM Tris, pH 6.8). Solubilized proteins (10 µg) were resolved by SDS-PAGE (10% acrylamide) using pre-stained Precision Plus protein standards (BioRad Laboratories, Hercules, CA, USA) and transferred to Immobilon™-P membrane (Millipore Corporation, Bedford, MA, USA). The blot was probed with Anti-FLAG M2 antibody (Eastman Kodak Company, New Haven, CT, USA), and bands were visualized with West Pico chemiluminescent detection system (Pierce Biotechnology, Inc., Rockford, IL, USA). The immunoblots were repeated twice, using two independently isolated sets of membranes, with similar results.
Peptide synthesis and uptake assays
L-leucyl-L-[3H]-leucine and L-methionyl-L-methionyl-L-[14C]-methionine were synthesized by standard solution phase techniques (Naider et al. [Citation1974]) and purified by reverse-phase HPLC to greater than 99% homogeneity. Yeast expressing the various pMS2 constructs were grown under inducing conditions to mid-log phase, harvested, and washed with 2% glucose. Cells were counted on a haemocytometer, adjusted to a final concentration of 2×108 cells/ml, divided into aliquots (60 µl), and kept on ice until used. Uptake assays were initiated by warming the aliquots to 30°C and mixing with an equal volume of 2X uptake medium. Uptake medium (2X) consisted of sodium citrate/potassium phosphate buffer (20 mM, pH 5.5), 2% glucose, and substrate. For di-leucine uptakes, the 2X substrate concentration was 320 µM L-leucyl-L-leucine supplemented with L-leucyl-L-[3H] leucine (2 µCi/ml). For tri-methionine uptakes, the substrate was L-methionyl-L-methionyl-L-[14C] methionine (0.2 µCi/ml) at a final concentration of 320 µM in the 2X uptake medium. Cells were incubated in uptake medium for 10 minutes. The assay was terminated by filtration of the cells over 0.45 µ HAWP membrane filters (Millipore, Bedford, MA, USA) followed by washing with ice cold water. Retained radioactivity was determined by liquid scintillation spectrometry and normalized to nmol/109cells. Uptake assays were repeated a minimum of three times. Each point was determined in triplicate and results reported as the mean of the replicate values plus or minus standard deviation. Statistical analysis (One Sample T-test) was completed using GraphPad Prism Version 4.0 (GraphPad Software Inc., San Diego, CA, USA).
Toxic peptide assays
The sensitivity of Ptr2p mutants to toxic peptides was assessed by placing sterile 6 mm filter disks (Becton, Dickinson and Company, Sparks, MD, USA) impregnated with toxic peptide onto lawns of cells in top agar. To prepare lawns, cells (BY4741-6009 or BY4724-16009) were grown in liquid culture under inducing conditions to mid-log phase. Cells were washed and resuspended in water to a final concentration of 5×106 cells/ml and combined with 3 ml molten noble agar (1.1%) tempered to 55°C. The agar-cell mixture was then poured onto plates (10 cm) of solid medium containing 0.67% YNB without amino acids and ammonium sulphate, 0.1% allantoin and tryptophan (20 µg/ml). For strain BY4741-6009, the solid medium was further supplemented with histidine, leucine, and methionine; for BY4742-16009 histidine, leucine, and lysine were added. The toxic peptides (alanyl-ethionine, lysyl-alanyl-ethionine and oxalysyl-glycine) were synthesized as previously described (Naider et al. [Citation1983]) and prepared as concentrated stocks (10 mM) in water, such that the desired molar amount of peptide (0.1 µmol) could be applied to the disk in a final volume of 10 µl. The toxic amino acids ethionine and oxalysine, prepared as a concentrated water stock, were used as controls. Peptide solution was applied to the disk, and then the disk was transferred onto the solidified top-agar lawn. Strain BY4742-16009 was used to test ethionine-containing peptides, and BY4741-6009 was used to test the oxalysine peptide. The plates were incubated at 30°C, and the diameter of any clear zone of inhibition surrounding the disk was measured. Experiments were repeated a minimum of three times and results reported as mean halo diameter±SD.
Results
Epitope tagging, mutagenesis and expression of Ptr2p
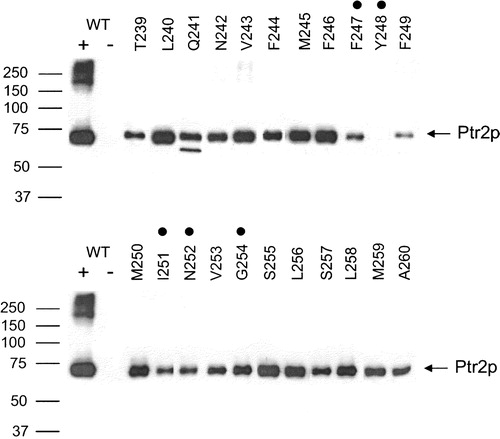
A FLAG and His-tagged construct of the peptide transporter Ptr2p () under the control of its native promoter was assembled and expressed from the plasmid pMS2. Addition of the epitopes to the C-terminus of Ptr2p did not affect its function; proteins expressing these tags were still fully competent with respect to peptide transport measured by both growth and uptake assays compared to wild-type Ptr2p (data not shown). This same construct was then used as the template for the Ala-scan mutagenesis of the residues T239–A260, which comprise the fifth transmembrane domain of Ptr2p. This domain contains the highly conserved FYING motif ( and ). To determine if the introduction of an alanine mutation eliminated or reduced expression of the transporter, membranes were isolated from yeast expressing the various Ala-scanned constructs. Equal amounts (10 µg) of solubilized protein were fractionated by SDS-PAGE. Epitope-tagged Ptr2p was detected as a single band of 71 kDa, in agreement with the predicted mass based on the primary amino acid sequence of the FLAG and His-tagged protein (). Some signal was also detected at higher molecular weight for wild-type Ptr2p, which suggested the presence of aggregated protein. No signal was detected in membranes from cells transformed with the empty vector. In all, twenty-two different mutants were examined. With the exception of Y248A, all of the mutant transporters were detected in the membrane preparations (). The expression levels of the mutant proteins varied from high (L240A and F246A) to low (F247A and F249A). Unlike the wild-type, aggregated material was not seen in any of the mutant proteins. For the mutant Q241A two discrete bands were detected. The first was at the expected size for Ptr2p, the second was somewhat smaller. Since the epitope tag is at the C-terminal end of the protein, this smaller species is an N-terminally cleaved form of the Q241A transporter.
Figure 3. Expression of wild-type and TM5 Ala-scanned mutant Ptr2p transporters in the membrane of Saccharomyces cerevisiae. Membranes were prepared from cells expressing TM5 Ala-scanned mutants (T239–A260). As controls membranes were also prepared from cells expressing wild-type (WT +) or no (WT −) Ptr2p. Membrane proteins (10 µg) were resolved by SDS-PAGE, blotted and immunodetected with anti-FLAG antibodies. Ptr2p was present in all lanes except the negative control (WT −) and the Y248A mutant. FYING mutants are indicated by dots over the corresponding residues.
TM5 mutants influence the recognition of dipeptide substrates
To assess the phenotypic consequences of Ala-scanning mutagenesis of TM5, peptide growth assays were performed. In these studies, an auxotrophic amino acid was supplied to the cells in the form of a di- or tri-peptide; cells that could transport the peptide containing the essential amino acid would grow, while cells that could not translocate this peptide would not grow. The strain used in the growth assay (BY4741-6009) had auxotrophic requirements for leucine, histidine and methionine. For each of the twenty-two different Ala-scanned mutants, twelve different peptides containing one of the auxotrophic amino acids were tested to determine if the mutation altered substrate recognition. Cells expressing the wild-type and mutant Ptr2p constructs were visually scored for growth as described in Experimental Procedures. An example of growth response on Leu-Leu as the sole source of leucine is shown (). Based on growth, the mutants were placed into one of three categories: wild-type growth (+ + ), reduced growth (+) or no growth (−). Cells expressing wild-type Ptr2p could utilize all of the tested peptides for growth; in contrast the negative control containing the empty vector did not grow on any of the substrates (). Ten of the mutants (T239A, L240A, Q241A, N242A, V243A, M245A, F246A, S255A, L256A, A260G) grew at wild-type levels on all substrates tested, which suggest that they function like native Ptr2p in their ability to recognize these peptides (data not shown). The remaining mutants exhibited some degree of substrate preference (). In all cases, the differences in growth among strains was similar whether scored at one, two, or three days post plating. All of these Ala-scanned mutants could grow on some peptide substrates, with the obvious exception of Y248A, which was not expressed in the membrane. The mutants F249A, M250A, V253A, S257A, L258A, and M259 grew on all twelve peptides, but grew at reduced levels on some of the peptides. Mutation of the highly-conserved residue F244 to Ala resulted in a lack of growth on Leu-Leu, Met-Leu, and Leu-Met. The F244A mutant grew at wild-type levels on Met-Ala and Ala-Met, but at reduced levels on Met-Met-Met and Met-Met and all histidine-containing peptides tested, except for Gly-His which did not support growth. In contrast, mutants of the FYING motif were more severely compromised in their ability to use peptides for growth. In particular, the G254A mutant was able to grow only on di- and tri-methionine at wild-type levels and grew poorly on Ala-Met; the I251A mutant did not grow at wild-type levels on any peptide tested, exhibiting only poor growth on di- and tri-methionine as well as on His-Leu and His-Ser. The F247A and N252A mutations of the FYING motif retained a broader range of substrate recognition, but were still reduced overall in growth.
Figure 4. Growth assay for wild-type and TM5 mutant Ptr2p transporters. Cells expressing (WT) or lacking (Vector) Ptr2p or expressing the various Ala-scanned mutants were assayed for growth on medium containing the dipeptide Leu-Leu as the sole source of leucine. Growth was scored by visual comparison of spot densities as wild-type growth (+ + ), reduced growth (+) or no growth (−) as indicated below each label. FYING residues are indicated in italics. Growth on other dipeptide substrates was determined in like manner and data are presented in .
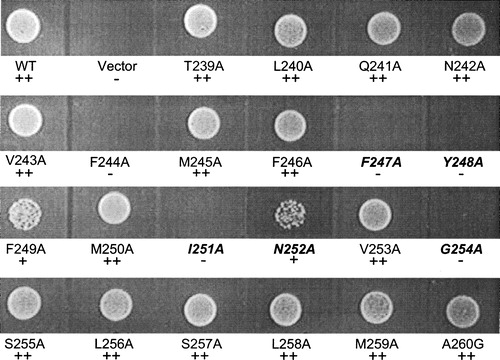
Table I. Ala-scanned mutants of TM5 which exhibit substrate preference. Yeast were plated onto media as described in experimental procedures. They were visually scored for growth by comparing spot densities (see ) to wild-type: ++, wild-type growth; +, reduced growth; −, no growth. Wild-type, positive control plasmid encoding wild-type PTR2. Vector, negative control plasmid without insert.
TM5 mutants change substrate recognition determined by transport assay
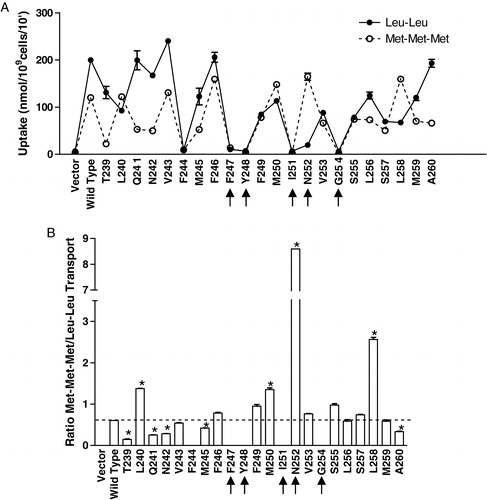
Growth assays measure the ability of the transporter to accumulate peptide over a long time course; even cells which express low levels of transport activity can potentially accumulate enough substrate to survive and exhibit some growth under these conditions. To determine how Ptr2p and the TM5 mutants interact with substrates in the short-term, transport assays to measure the uptake of two different radiolabeled peptides (Leu-Leu and Met-Met-Met) were completed. In the growth assays, Leu-Leu was a differentially recognized substrate, while Met-Met-Met could be utilized for growth by all expressed Ptr2p variants. The results of the uptake assay are presented in . For wild-type Ptr2p, the specific accumulations of Leu-Leu and Met-Met-Met were 200 nmol/109 cells/10 min and 120 nmol/109 cells/10 min, respectively. Since the immunoblot data indicate that the mutant transporters are each expressed at different levels in the membrane, direct comparison of peptide accumulation is not appropriate. Therefore, the relative accumulation of the two substrates, expressed as the ratio of Met-Met-Met to Leu-Leu transport, for the wild type and each mutant transporter were compared (). For the wild type transporter the ratio of Met-Met-Met to Leu-Leu transport was 0.6, reflecting the greater accumulation of Leu-Leu in comparison to Met-Met-Met. Mutants that differ significantly from this criterion value exhibit differential preference for the substrates relative to the preference of the wild-type. Alanine mutations at 8 positions (V243, F246, F249, V253, S255, L256, S257, and M259) did not differ significantly (p<0.01) from the wild-type with respect to substrate preference. Mutations at T239, Q241, N242, M245 and A260 resulted in increased preference for Leu-Leu over Met-Met-Met, relative to the wild-type preference. Tri-methionine was the preferred substrate for alanine mutants at L240, M250, N252 and L258; in contrast to wild-type, these mutants transported more Met-Met-Met than Leu-Leu. The enhanced preference for Met-Met-Met by the N252A mutant of the FYING motif was especially dramatic, with transport of Met-Met-Met over 8 times greater than that of Leu-Leu. Interestingly, the F, I, and G mutants of the invariably conserved FYING motif, as well as the highly conserved F244 residue () did not accumulate either peptide to levels significantly above that of the vector control (p<0.01).
Figure 5. Transport of radiolabeled Leu-Leu and Met-Met-Met by wild-type and TM5 mutant transporters. Cells expressing wild-type or Ala-scanned mutant (T239–A260) Ptr2p constructs were grown under inducing conditions as described in Experimental procedures. Each point was determined in triplicate and results reported as the mean of the replicate values plus or minus standard deviation. In some cases error bars are present but not visible. Cells lacking Ptr2p (Vector) were used as a negative control. (a) The 10 min. accumulation of 14C-Met-Met-Met (○) or 3H-Leu-Leu (•) was determined. (b) Ratio of Met-Met-Met/Leu-Leu accumulation for each mutant. Background (radioactivity associated with vector control) was subtracted from each mutant prior to calculating ratios. For mutants exhibiting zero transport of both substrates, no bar was plotted. Asterisks indicate that values are significantly different (p<0.01) from wild-type.
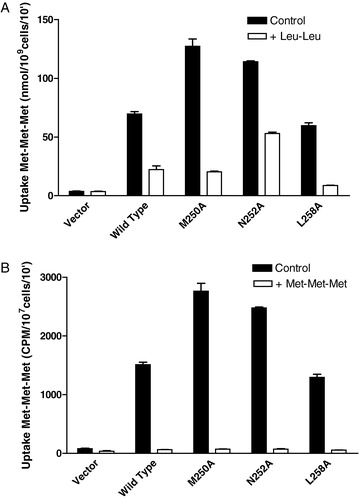
Since di-leucine and tri-methionine were differentially recognized by the mutant transporters M250A, N252A, and L258A relative to wild-type, the possibility existed that the substrates could be interacting with two distinct sites within the binding domain of the transporter. To determine if tri-methionine and di-leucine bind to the same site in the mutant transporters, a competition assay was completed. In this case, cells were incubated with radiolabeled tri-methionine in the presence or absence of a 10-fold molar excess of either cold di-leucine or cold tri-methionine. For competition with cold Leu-Leu, results are reported as nmols Met-Met-Met/109cells/10′. To take into consideration the dilution of specific activity in the competition assay for radioactive Met-Met-Met uptake by cold Met-Met-Met, the results for tri-methionine competition are reported as CPM/107cells/min. For the wild-type Ptr2p, excess di-leucine reduced the accumulation of tri-methionine by 68% () and excess tri-methionine reduced accumulated radioactivity by over 90%. Excess Leu-Leu reduced accumulation by the mutants M250A and L258A by over 80%; excess Met-Met-Met reduced accumulated radioactivity by over 90%. For the N252A mutant, the presence of a 10-fold molar excess of di-leucine reduced the transport of tri-methionine by 50% and Met-Met-Met reduced transport by over 90%. This result is consistent with the transport of Met-Met-Met and Leu-Leu observed for the N252A mutant (); compared to Met-Met-Met, Leu-Leu is a poor substrate for this transporter, and is therefore not as effective a competitor as it is for the wild-type, M250A and L258A transporters. However, since Leu-Leu is able to compete with Met-Met-Met, this suggests that the two substrates interact with the mutant and wild-type transporters in a similar manner, rather than interacting with two different substrate binding domains.
Figure 6. Effect of 10-fold excess unlabeled Leu-Leu or Met-Met-Met on 14C-Met-Met-Met transport in cells expressing wild-type or TM5 mutant transporters. Cells expressing the wild- type or mutant (F247A, M250A, L258A) Ptr2p constructs were grown under inducing conditions as described in Experimental procedures. Each point was determined in triplicate and results reported as the mean of the replicate values plus or minus standard deviation. In some cases error bars are present but not visible. Cells lacking Ptr2p (Vector) were used as a negative control. (a) The 10 min. accumulation of 14C-Met-Met-Met (160 µM) in the presence (open boxes) or absence (dark boxes) of 1.6 mM Leu-Leu. (b) The 10 min accumulation of 14C-Met-Met-Met (160 µM) in the presence (open boxes) or absence (dark boxes) of 1.6 mM Met-Met-Met.
N252A mutant was not sensitive to toxic peptides
To further explore the effect of mutations in TM5, the toxicity of di- and tri-peptides to various Ptr2p mutants was assessed. In the toxicity assay, a sterile filter disk containing a specific amount of a toxic compound is placed onto a lawn of cells. If the cells transport this compound, then a clear zone of inhibition (‘halo’) will form around the disk, the diameter of which is a function of the amount of toxic compound present. In this study we used the toxic peptides oxalysyl-glycine (Oxa-Gly), alanyl-ethionine (Ala-Eth), and lysyl-alanyl-ethionine (Lys-Ala-Eth) as well as the toxic amino acids oxalysine and ethionine to examine substrate preference for the mutants L240A, M250A, N252A, and L258A. These mutants were selected based on the previously determined differential transport of radiolabeled di-leucine and tri-methionine. All strains were sensitive to the toxic amino acid analogues ethionine and oxalysine (). For cells expressing the wild-type transporter and the mutants L240A, M250A, and L258A all three toxic peptides (Oxa-Gly, Ala-Eth, and Lys-Ala-Eth) produced halos of similar size when present at 0.1 µmol per disk. The greater toxicity of the Oxa-Gly di-peptide as compared to oxalysine alone was observed previously for Candida albicans and interpreted as the ability of the yeast cell to accumulate di-peptide to a greater extent than certain amino acids (Basrai et al. [Citation1992]). No halos were seen for the peptide-transport deficient control strain (vector) or for the mutant N252A, suggesting that this mutation results in a transporter which did not recognize or accumulate these toxic peptides.
Table II. Sensitivity to toxic dipeptides. Toxic peptide (0.1 µmols) or the corresponding toxic amino acid analogue (0.1 µmols) was applied to a sterile disk and placed on a top agar lawn of cells as described in experimental procedures. The diameter (mm) of the zone of inhibition was measured. Vector, negative control plasmid without insert. Wild-type, positive control plasmid encoding wild-type PTR2.
Discussion
Site-directed mutagenesis has been widely used to define the relationships which may exist between protein structure and function in membrane transport proteins (Abramson et al. [Citation2004], Kaback et al. [Citation2001]). For example, in hPEPT1 the conserved histidine residue H57 in TM 2, has been examined (Daniel & Kottra [Citation2004], Herrera-Ruiz & Knipp [Citation2003], Terada & Inui [Citation2004]) indicating that H57, located at the extracellular interface of TM2, is essential for transporter function and plays a role in proton-coupling (Chen et al. [Citation2000], Fei et al. [Citation1997], Uchiyama et al. [Citation2003]). Mutation to Ala, Pro, Ser, or His of the invariably conserved hPEPT1 tyrosine Y167 in TM5, a residue predicted by molecular modelling to be involved in substrate translocation, abolished Gly-Sar uptake into HEK293 cells (Bolger et al. [Citation1998], Yeung et al. [Citation1998]). In TM7, the mutant R282A exhibited little effect on Gly-Sar accumulation, suggesting that it played a minor role in transporter activity. However, recently it has been reported that that the mutation of R282 to glutamic acid in rabbit PEPT1 resulted in reduced transport of [3H]-D-Phe-L-Gln into Xenopus oocytes, in uncoupling of peptide and proton co-transport, and in the formation of a peptide-gated cation channel (Meredith [Citation2004]).
In the present study, we used Ala-scanning mutagenesis of the fifth transmembrane domain of Ptr2p to see if these residues, including the invariably conserved FYING motif, played a role in substrate preference. We found that specific mutations resulted in marked differences in substrate recognition, rather than completely eliminating transporter function. For example, the mutation N252A resulted in minimal transport of the dipeptide Leu-Leu. In contrast, this same mutation resulted in robust transport of the radiolabeled tri-peptide Met-Met-Met (). If only a single substrate had been assayed to determine transporter function, our interpretation of the results would have been quite different; if only di-leucine had been used to assess Ptr2p function, then we would have concluded that the mutation N252A, as well as L240A, M250A, or L258A severely compromised transporter function, when in fact these transporters are fully able to transport tri-methionine.
In most studies in which site-directed mutagenesis of specific amino acids in PTR family transporters has been employed, functionality of the transporter has been assessed using a single substrate. The most frequently used substrate is the non-metabolized peptide glycyl-sarcosine (Gly-Sar) (Ganapathy et al. [Citation1984]), but affinity for this compound will certainly not accurately reflect that for all of the 8400 potential different naturally-occurring di/tri-peptide substrates, especially in the context of mutant transporters. In one report, the conservative mutation of Y56F in TM2 of hPEPT1 caused only a slight decrease in the transport of [14C]-Gly-Sar into Xenopus oocytes; two electrode voltage clamp measurement of Imax indicated that Gly-Glu and Gly-Lys were transported at near wild-type levels, but transport of Gly-Leu was reduced by 5–7 fold (Chen et al. [Citation2000]). The large gap in our understanding of the effect of site-directed mutations on substrate specificity in peptide transporters prompted us to undertake the studies reported herein.
In this study, we looked at 12 different peptide substrates in growth assays. For many of the TM5 residues (10 of 22), Ala-scanning mutagenesis had no effect on substrate recognition, at least in this small sampling of peptides. However, for 12 of the 22 mutants, even within this small number of potential substrates, very distinct differences in substrate preferences were apparent. For example, the construct containing the N252A mutation had no obvious phenotypic difference from wild-type Ptr2p when grown on methionine-containing peptide substrates, however, it was unable to grow on any of the histidine-containing peptides with the exception of reduced growth on His-Leu. In contrast, the mutant F244A was able to utilize all of the histidine-peptides except Gly-His, but could not grow on Leu-Leu, Met-Leu, or Leu-Met. F249A could grow on the peptide His-Gly, but exhibited reduced growth on Gly-His, while the M250A mutant exhibited the opposite phenotype (i.e., growth on Gly-His and reduced growth on His-Gly).
Mutants of the FYING motif F247A, I251A, and G254A as well as the non-FYING, but phylogenetically highly conserved F244A mutant were expressed in the membrane, but did not accumulate appreciable amounts of either radiolabeled di-leucine or tri-methionine in the short term uptake assay. These transporters were, however, able to use some peptides, including tri-methionine for growth. This suggests that there are fundamental differences between the growth and uptake assays. Even though over the ten minute time course of the uptake assay very little radiolabeled tri-methionine was accumulated, over the course of the growth assay (48 + h) enough substrate must be accumulated to allow for near-normal growth. Tri-methionine is a rich source of the auxotrophic amino acid methionine. Even low levels of transport would result in the entry of sufficient methionine to support growth, since for every nmol of tri-methionine transported, 3 nmols of methionine are made available inside the cell.
The expression levels of the various mutants as determined by immunoblot analysis varied to some extent, depending on the mutation. However, reduced expression level did not appear to influence the function of cells expressing the mutant transporters in either growth or transport assays. For example, F249A was poorly expressed, however this mutant functioned well in both growth and transport assays (). In contrast, G254A was expressed at relatively high levels in the membrane, yet did not perform well in either growth or transport assays. This indicates that the amount of transporter present in the membrane is not a determining factor in the growth of cells expressing the mutant transporters. Another difference between the wild-type and mutant transporters observed upon immunoblot analysis is the presence of high molecular weight bands above the expected 71 kDa band, corresponding to monomeric Ptr2p, observed for the wild-type transporter (). This material, which was not observed in most of the mutants, may indicate formation of dimers and larger aggregates of the transporter. The absence of aggregates in the mutants may be due to their decreased expression level in comparison to the wild-type Ptr2p. Nevertheless, the inability to form such aggregates does not appear to affect transport function as determined by growth and transport assays as carried out in this study.
The Y248A transporter in S. cerevisiae was not detected in the membrane via immunoblot analysis suggesting that the mutation influenced proper protein folding or targeting to the membrane. Mutations of the corresponding residue in hPEPT1 (Y167) to cysteine (Kulkarni et al. [Citation2003a]), alanine, serine, phenylalanine or histidine (Yeung et al. [Citation1998]) did not result in loss of protein expression, but did result in loss of function as assayed by Gly-Sar uptake, therefore it appears that tyrosine is essential at this position for protein function. In S. cerevisiae Ptr2p the requirement for tyrosine at position 248 should be similarly explored by generating a panel of mutants encompassing the full range of amino acid functional groups.
In order to visualize the spatial relationships between the residues composing TM5, a helical wheel model was used (). In this model of S. cerevisiae Ptr2p, the FYING residues all fall on the same face of the helix. This model is agreement with that proposed by Kulkarni ([Citation2003a]) for TM5 of hPEPT1. In that model, the FYING residues were in a solvent-accessible region of the helix, determined by Cys-scanning mutagenesis coupled with accessibility to membrane-impermeant methanethiosulfonate derivatives (MTSEA and MTSET). In our study, mutagenesis of FYING residues F247, I251, and G254 to alanine resulted in dramatic decreases in the ability to transport radiolabeled Met-Met-Met. Transporters carrying these same mutations, or the mutation N252A, were also deficient in their ability to transport radiolabeled di-leucine. Coupled with the solvent accessibility data, these results provide compelling evidence that all residues of the FYING motif are involved in interactions with the substrate that affects peptide translocation.
The mutagenesis analysis undertaken in this study helps to define the region of the peptide binding site of the di/tri-peptide transporter of S. cerevisiae. Our substrate specificity analysis indicated that mutations spanning residues F244 to G254 had significant effect on substrate preference () and also dramatically influenced the uptake of either or both Leu-Leu or Met-Met-Met. Mutations of residues outside of this range had a much lower effect on both substrate preference and di/tri-peptide transport. Assuming that TM5 of Ptr2p is helical, 11 residues would span 3.3 turns or approximately 18 Å. In comparison, a tri-peptide in an extended structure would span approximately 11 Å. Thus the region defined in this study is certainly long enough to accommodate the substrates of the transporter. In addition, it is interesting that within this defined domain residues M245, F246, F249, M250, and V253 have virtually no effect on substrate preference (), and relatively little effect on Leu-Leu or Met-Met-Met transport. All of these residues lie on the face of the helix opposite from the FYING motif () and it is reasonable to surmise that their side chains are not directly involved in substrate recognition.
Since the PTR transporters are pharmacologically important targets for drug delivery, significant progress has been made in understanding the properties of peptides and peptidomimetics drugs which are essential for high affinity transport (Doring et al. [Citation1998c], Gebauer et al. [Citation2003], Knutter et al. [Citation2001], Knutter et al. [Citation2004], Payne et al. [Citation2000], Theis et al. [Citation2002a]). Our growth and toxic peptide assay system explores transporter-substrate interactions and provides a high-throughput means to rapidly assess the effect of specific site-directed mutations on substrate preference. While transgenic Pichia pastoris, a methylotrophic yeast, has been used to study the mammalian peptide transporters (Doring et al. [Citation1997], Doring et al. [Citation1998a]), including studies defining structural features of their substrates (Doring et al. [Citation1998c], Knutter et al. [Citation2004], Theis et al. [Citation2002a]) and identification of novel substrates and inhibitors (Doring et al. [Citation1998b], Foltz et al. [Citation2004], Knutter et al. [Citation2001], Theis et al. [Citation2002b]) the use of Pichia in growth assays is somewhat limited by the available auxotrophic markers (Lin Cereghino et al. [Citation2001]). Assays for the growth of S. cerevisiae on substrates for the peptide transporter Ptr2p, as well as for other peptide transporters expressed heterologously in this yeast, may provide a fast and inexpensive approach to rapidly screen the affect of mutations on peptide recognition, thus providing a useful model system for the study of substrate specificity in the PTR family.
This paper was first published online on prEview on 28 April 2005.
We would like to acknowledge the contributions of Serene Williams and Cagdas Son to the work presented in this manuscript.
References
- Abramson J, Iwata S, Kaback HR. Lactose permease as a paradigm for membrane transport proteins. Mol Membr Biol 2004; 21: 227–236
- Basrai MA, Zhang HL, Miller D, Naider F, Becker JM. Toxicity of oxalysine and oxalysine-containing peptides against Candida albicans: regulation of peptide transport by amino acids. J Gen Microbiol 1992; 138: 2353–2362
- Becker JM, Naider F. Peptide transport in yeast: Uptake of radioactive trimethionine in Saccharomyces cerevisiae. Arch Biochem Biophys 1977; 178: 245–255
- Bolger MB, Haworth IS, Yeung AK, Ann D, von Grafenstein H, Hamm-Alvarez S, Okamoto CT, Kim KJ, Basu SK, Wu S, Lee VH. Structure, function, and molecular modeling approaches to the study of the intestinal dipeptide transporter PepT1. J Pharmeceut Sci 1998; 87: 1286–1291
- Boll M, Markovich D, Weber WM, Korte H, Daniel H, Murer H. Expression cloning of a cDNA from rabbit small intestine related to proton-coupled transport of peptides, beta-lactam antibiotics and ACE-inhibitors. Pflugers Arch 1994; 429: 146–149
- Busch W, Saier MH, Jr. The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol 2002; 37: 287–337
- Chen XZ, Steel A, Hediger MA. Functional roles of histidine and tyrosine residues in the H(+)-peptide transporter PepT1. Biochem Biophys Res Comm 2000; 272: 726–730
- Covitz KM, Amidon GL, Sadee W. Membrane topology of the human dipeptide transporter, hPEPT1, determined by epitope insertions. Biochemistry 1998; 37: 15214–15221
- Daniel H, Kottra G. The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pflugers Arch 2004; 447: 610–618
- David NE, Gee M, Andersen B, Naider F, Thorner J, Stevens RC. Expression and purification of the Saccharomyces cerevisiae alpha-factor receptor (Ste2p), a 7-transmembrane-segment G protein-coupled receptor. J Biol Chem 1997; 272: 15553–15561
- Doring F, Theis S, Daniel H. Expression and functional characterization of the mammalian intestinal peptide transporter PepT1 in the methylotropic yeast Pichia pastoris. Biochem Biophys Res Comm 1997; 232: 656–662
- Doring F, Michel T, Rosel A, Nickolaus M, Daniel H. Expression of the mammalian renal peptide transporter PEPT2 in the yeast Pichia pastoris and applications of the yeast system for functional analysis. Mol Membr Biol 1998a; 15: 79–88
- Doring F, Walter J, Will J, Focking M, Boll M, Amasheh S, Clauss W, Daniel H. Delta-aminolevulinic acid transport by intestinal and renal peptide transporters and its physiological and clinical implications. J Clin Invest 1998b; 101: 2761–2767
- Doring F, Will J, Amasheh S, Clauss W, Ahlbrecht H, Daniel H. Minimal molecular determinants of substrates for recognition by the intestinal peptide transporter. J Biol Chem 1998c; 273: 23211–23218
- Fei YJ, Kanai Y, Nussberger S, Ganapathy V, Leibach FH, Romero MF, Singh SK, Boron WF, Hediger MA. Expression cloning of a mammalian proton-coupled oligopeptide transporter. Nature 1994; 368: 563–566
- Fei YJ, Liu W, Prasad PD, Kekuda R, Oblak TG, Ganapathy V, Leibach FH. Identification of the histidyl residue obligatory for the catalytic activity of the human H + /peptide cotransporters PEPT1 and PEPT2. Biochemistry 1997; 36: 452–460
- Foltz M, Meyer A, Theis S, Demuth HU, Daniel H. A rapid in vitro screening for delivery of peptide-derived peptidase inhibitors as potential drug candidates via epithelial peptide transporters. J Pharmacol Exp Therapeut 2004; 310: 695–702
- Ganapathy ME, Brandsch M, Prasad PD, Ganapathy V, Leibach FH. Differential recognition of beta-lactam antibiotics by intestinal and renal peptide transporters, PEPT 1 and PEPT 2. J Biol Chem 1995; 270: 25672–25677
- Ganapathy ME, Huang W, Wang H, Ganapathy V, Leibach FH. Valacyclovir: a substrate for the intestinal and renal peptide transporters PEPT1 and PEPT2. Biochem Biophys Res Comm 1998; 246: 470–475
- Ganapathy V, Burckhardt G, Leibach FH. Characteristics of glycylsarcosine transport in rabbit intestinal brush-border membrane vesicles. J Biol Chem 1984; 259: 8954–8959
- Gebauer S, Knutter I, Hartrodt B, Brandsch M, Neubert K, Thondorf I. Three-dimensional quantitative structure-activity relationship analyses of peptide substrates of the mammalian H(+)/peptide cotransporter PEPT1. J Medic Chem 2003; 46: 5725–5734
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 2002; 350: 87–96
- Hagting A, vd Velde J, Poolman B, Konings WN. Membrane topology of the di- and tripeptide transport protein of Lactococcus lactis. Biochemistry 1997; 36: 6777–6785
- Hauser M, Donhardt AM, Barnes D, Naider F, Becker JM. Enkephalins are transported by a novel eukaryotic peptide uptake system. J Biol Chem 2000; 275: 3037–3041
- Hauser M, Narita V, Donhardt AM, Naider F, Becker JM. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Mol Membr Biol 2001; 18: 105–112
- Herrera-Ruiz D, Knipp GT. Current perspectives on established and putative mammalian oligopeptide transporters. J Pharmaceut Sci 2003; 92: 691–714
- Island MD, Naider F, Becker JM. Regulation of dipeptide transport in Saccharomyces cerevisiae by micromolar amino acid concentrations. J Bacteriol 1987; 169: 2132–2136
- Kaback HR, Sahin-Toth M, Weinglass AB. The kamikaze approach to membrane transport. Nature Rev Mol Cell Biol 2001; 2: 610–620
- Knutter I, Theis S, Hartrodt B, Born I, Brandsch M, Daniel H, Neubert K. A novel inhibitor of the mammalian peptide transporter PEPT1. Biochemistry 2001; 40: 4454–4458
- Knutter I, Hartrodt B, Theis S, Foltz M, Rastetter M, Daniel H, Neubert K, Brandsch M. Analysis of the transport properties of side chain modified dipeptides at the mammalian peptide transporter PEPT1. Eur J Pharmaceut Sci 2004; 21: 61–67
- Kulkarni AA, Haworth IS, Lee VH. Transmembrane segment 5 of the dipeptide transporter hPepT1 forms a part of the substrate translocation pathway. Biochem Biophys Res Comm 2003a; 306: 177–185
- Kulkarni AA, Haworth IS, Uchiyama T, Lee VH. Analysis of transmembrane segment 7 of the dipeptide transporter hPepT1 by cysteine-scanning mutagenesis. J Biol Chem 2003b; 278: 51833–51840
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol 1987; 154: 367–382
- Lin Cereghino GP, Lin Cereghino J, Sunga AJ, Johnson MA, Lim M, Gleeson MA, Cregg JM. New selectable marker/auxotrophic host strain combinations for molecular genetic manipulation of Pichia pastoris. Gene 2001; 263: 159–169
- Meredith D. Site-directed mutation of arginine 282 to glutamate uncouples the movement of peptides and protons by the rabbit proton-peptide cotransporter PepT1. J Biol Chem 2004; 279: 15795–15798
- Naider F, Becker JM, Katzir-Katchalski E. Utilization of methionine-containing peptides and their derivatives by a methionine-requiring auxotroph of Saccharomyces cerevisiae. J Biol Chem 1974; 249: 9–20
- Naider F, Shenbagamurthi P, Steinfeld AS, Smith HA, Boney C, Becker JM. Synthesis and biological activity of tripeptidyl polyoxins as antifungal agents. Antimicrob Agents Chemother 1983; 24: 787–796
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 1997; 25: 451–452
- Payne JW, Grail BM, Gupta S, Ladbury JE, Marshall NJ, O'Brien R, Payne GM. Structural basis for recognition of dipeptides by peptide transporters. Arch Biochem Biophys 2000; 384: 9–23
- Perry JR, Basrai MA, Steiner HY, Naider F, Becker JM. Isolation and characterization of a Saccharomyces cerevisiae peptide transport gene. Mol Cell Biol 1994; 14: 104–115
- Saier MH, Jr. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev 2000; 64: 354–411
- Saito H, Terada T, Okuda M, Sasaki S, Inui K. Molecular cloning and tissue distribution of rat peptide transporter PEPT2. Biochim Biophys Acta 1996; 1280: 173–177
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989; 122: 19–27
- Sugawara M, Huang W, Fei YJ, Leibach FH, Ganapathy V, Ganapathy ME. Transport of valganciclovir, a ganciclovir prodrug, via peptide transporters PEPT1 and PEPT2. J Pharmaceut Sci 2000; 89: 781–789
- Terada T, Saito H, Mukai M, Inui K. Characterization of stably transfected kidney epithelial cell line expressing rat H + /peptide cotransporter PEPT1: localization of PEPT1 and transport of beta-lactam antibiotics. J Pharmacol Exp Therapeut 1997; 281: 1415–1421
- Terada T, Inui K. Peptide transporters: structure, function, regulation and application for drug delivery. Current Drug Metabol 2004; 5: 85–94
- Theis S, Hartrodt B, Kottra G, Neubert K, Daniel H. Defining minimal structural features in substrates of the H(+)/peptide cotransporter PEPT2 using novel amino acid and dipeptide derivatives. Mol Pharmacol 2002a; 61: 214–221
- Theis S, Knutter I, Hartrodt B, Brandsch M, Kottra G, Neubert K, Daniel H. Synthesis and characterization of high affinity inhibitors of the H + /peptide transporter PEPT2. J Biol Chem 2002b; 277: 7287–7292
- Uchiyama T, Kulkarni AA, Davies DL, Lee VH. Biophysical evidence for His57 as a proton-binding site in the mammalian intestinal transporter hPepT1. Pharmaceut Res 2003; 20: 1911–1916
- Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol 1987; 153: 3–11
- Yeung AK, Basu SK, Wu SK, Chu C, Okamoto CT, Hamm-Alvarez SF, von Grafenstein H, Shen WC, Kim KJ, Bolger MB, Haworth IS, Ann DK, Lee VH. Molecular identification of a role for tyrosine 167 in the function of the human intestinal proton-coupled dipeptide transporter (hPepT1). Biochem Biophys Res Comm 1998; 250: 103–107