Abstract
Nedd4 family ubiquitin ligases regulate trafficking and degradation of numerous target substrates in different cellular compartments, including at the plasma membrane, in endosomes, in the secretory pathway and in the nucleus. WWP1 is a Nedd4 family protein closely related to mouse Itch and Drosophila Su(dx), both of which have been shown to regulate the Notch receptor. To investigate the possibility that WWP1 is also associated with Notch signalling we coexpressed human Notch1 and WWP1 in mouse myoblast cells. We found that WWP1 could localize to both the nucleus and cytoplasm in a context dependent manner. Coexpression of human Notch1 (hN1) depleted WWP1 from the nucleus to colocalise with hN1 in early endosomes, dependent on the presence of the C-terminal HECT domain. Furthermore we found that full-length expressed WWP1 could interact in vitro with the cytoplasmic domain of human Notch1. The Notch receptor has multiple roles in development, mediating a short-range signal that controls cell fate and pattern formation. The canonical Notch signal involves proteolytic release of the soluble Notch intracellular domain and the activation by the latter of the transcription factor Suppressor of Hairless/CBF-1 in the nucleus. This pathway does not however account for all of the activity of Notch. The ability of Notch to regulate the nuclear localization of WWP1 suggests a possible alternative mechanism by which Notch may communicate a signal to the nucleus. Drosophila Notch similarly regulated the nuclear localization of the Drosophila Nedd4 family protein, Suppressor of deltex, implying conservation of this mechanism during evolution.
Introduction
The Nedd4 family of E3 ubiquitin ligases are characterized by a similar domain structure with an N-terminal, membrane interacting C2 domain, two to four WW domains that regulate target specificity, and a C-terminal HECT domain that confers ubiquitin ligase activity (Ingham et al. [Citation2004]). Collectively Nedd4 proteins have been shown to have diverse functions in different subcellular compartments, including targeting substrates for proteosomal degradation, stimulating endocytosis from the cell surface, and diverting proteins from the secretory to the endosomal pathway (Ingham et al. [Citation2004]). WWP1 is a human Nedd4 family protein (Pirozzi et al. [Citation1997], Flasza et al. [Citation2002]), which has been shown to display both nuclear and cytoplasmic distribution (Malbert-Colas et al. [Citation2003]) and has been linked to the regulation of the transcription factors, KLF2 and NF-E2 (Mosser et al. [Citation1998]; Conkright et al. [Citation2001], Zhang et al. [Citation2004]). Cytoplasmically localized WWP1 has also been shown to ubiquitinate and downregulate Transforming growth factor-β receptor signalling after being recruited to the receptor through binding to Smad7 (Komuro et al. [Citation2004]).
WWP1 belongs to the subset of Nedd4 family proteins that contain two pairs of WW domains (Flasza et al. [Citation2002]). This subset includes mouse Itch and Drosophila Su(dx) both of which have been shown to down-regulate the developmentally important Notch receptor signalling pathway (Qiu et al. [Citation2000], McGill & McGlade [Citation2003], Cornell et al. [Citation1999], Wilkin et al. [Citation2004]). Itch promotes Notch ubiquitination and degradation in mammalian cell culture, while Su(dx) regulates sorting of Notch from the early to late endosome, resulting in its down-regulation. Here we show that WWP1 is also capable of interacting with Notch in vitro. Furthermore in C2C12 cells we found that WWP1 localized to the nucleus in a context dependent manner and that its localization changed when coexpressed with full-length human Notch1. In the latter case, WWP1 colocalized with Notch in early endosomes. The HECT domain of WWP1 was found to be both necessary and sufficient both for the localization of WWP1 in the nucleus and for its regulation by Notch. Similar results were found when we investigated the localization of Drosophila Su(dx) and Notch in S2 cells. These data suggest a novel alternative means by which Notch could communicate with the nucleus that has been conserved during evolution.
Materials and methods
Expression constructs
Full-length WWP1, and deletion constructs lacking the C2 domain, intact HECT domain, or sequences N-terminal to the HECT domain (WWP1ΔC2, WWP1ΔHECT and WWP1HECT respectively), were cloned in frame with the C-terminal V5 epitope tag of the pcDNA3.1-/V5-His vector (Invitrogen). Amino acids from the WWP1 open reading frame included in each construct are depicted in H. Expression constructs for full-length human Notch1 (hN1) in pcDNAI/Amp (Invitrogen) and soluble cytoplasmic domain of human Notch1 (NICD), in pcDNA3 (Invitrogen) were a gift from Spyros Artavanis-Tsakonas (Harvard Medical School, Massachusetts General Hospital). The expression vector for human integrin-αV, in pcDNA3, was a gift from Martin Humphries (University of Manchester). To express full-length Drosophila Su(dx), the V5 epitope tag was fused in frame with the C-terminus of the Su(dx) open reading frame (ORF) and the complete ORF was cloned into pRamH3 which contains the copper inducible metallothionein enhancer (Otto et al. [Citation1987]). To express the Su(dx) HECT domain, sequences N-terminal to amino acid 560 (Cornell et al. [Citation1999]) were replaced with a HA epitope tag and the fusion construct cloned into pRamH3. For GST fusion protein pull-down experiments, full-length WWP1 was cloned into pGex4T-1 vector (Amersham Biosciences) to create GST-WWP1 fusion protein.
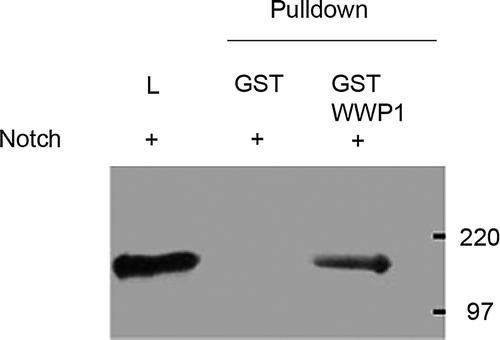
Figure 1. An in vitro pull-down assay using GST-fusion protein showed that GST-WWP1, but not GST alone, was able to pull-down NICD from cell lysates of Hela cells overexpressing NICD. The left lane shows staining for NICD present in total cell lysate before pull-down (L). The two adjacent lanes show the in vitro pull-down of Notch with GST alone and GST-WWP1 respectively. NICD was detected only with the GST-WWP1 pull-down showing a specific interaction between NICD and WWP1.
Cell culture and transfections
Hela cells and undifferentiated murine skeletal muscle derived C2C12 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% heat inactivated foetal calf serum (FCS), penicillin-streptomycine and 1mM L-glutamine. Cultures were incubated at 37oC in a humidified atmosphere consisting of 5% CO2, 95% air. Undifferentiated cells were grown on glass coverslips and transfected by standard calcium phosphate precipitation method. Immunostaining was performed 17 h after transfection. To examine WWP1 localization in differentiated C2C12 cells, undifferentiated cells were transfected, and 17 h afterwards were induced to differentiate by transfer to medium containing 1% FCS. Cells were then immunostained after 48 h in differentiating medium. S2 cells were cultured in Shields and Sang M3 medium, supplemented with 10% FCS. A stable S2 cell line expressing full-length Drosophila Notch under inducible control of the metallothionein enhancer was a gift from Spyros Artavanis-Tsakonas and was selectively maintained by addition of 4.9 µg/ml of methotrexate. S2 cells were transfected using a standard calcium phosphate precipitation method and expression induced by adding CuSO4 to the medium to a concentration of 500 µM.
GST fusion proteins and pull-down assays
GST-WWP1 fusion proteins were expressed in Escherichia coli BL-21 (DE3) and purified on glutathione-sepharose beads as described by the manufacturer (Amersham Biosciences). Equal amounts of isolated GST alone or GST fusion proteins were incubated with lysate from transfected Hela cells in the presence of protease inhibitors (Roche). Hela cell lysate was obtained by incubating the cells in Lysis buffer (50 mM HEPES, 5 mM EDTA, 50 mM NaCl, 1% Triton X-100, protease inhibitors) for 30 min on ice and cleared from cell debris by centrifugation at 14000 g for 10 min. After 1 h incubation, the column was washed three times in Modified lysis buffer (50 mM HEPES, 5 mM EDTA, 50 mM NaCl, 0.1% Triton X-100, protease inhibitors) and bound proteins were eluted in 2X SDS (20% glycerol, 20% SDS, 100 mM Tris-HCl, pH 6.8, 0.2% bromophenol blue and 10% mercaptoethanol) protein loading buffer and resolved by SDS-PAGE and immunoblots were probed with anti-Notch antibodies (C-20, 1:500; Santa Cruz).
Antibodies and immunostaining
Mouse monoclonal antibodies to human Notch1 (anti-hN1 or G18, 1:10), Drosophila Notch (9C6) and rabbit polyclonal anti-Drosophila Notch (anti-ICN, 1:100), were a gift from Spyros Artavanis-Tsakonas. Rabbit polyclonal anti-integrin-αV (1:50) was a gift from Martin Humphries. Anti-V5 mouse monoclonal antibodies were from Invitrogen (1:250) and rabbit polyclonal anti-HA was from Clontech (1:200). For the vesicular compartment characterization, we used rabbit polyclonal anti-M6PR (late endosome), anti-PDI (ER), anti-GM130 (Golgi apparatus), anti-Lpg120 (lysosome) and anti-Hrs (Drosophila early endosome) antibodies as specific markers, which were kindly provided by Neil Bulleid, Martin Lowe, Philip Woodman and Hugo Bellen. The early endosome marker, rabbit polyclonal anti-EEA1 antibodies, was purchased from Abcam. Secondary antisera were FITC conjugated anti-mouse and Texas Red conjugated anti-rabbit (Jackson Immunoresearch Laboratories Inc., 1:200), used accordingly. Cells were grown on coverslips, fixed with cold methanol (−20oC) for 10 min, blocked in 5% normal goat or donkey serum in PBS for one hour, incubated with primary antisera for 60 min at room temperature before being rinsed in PBS and incubated with secondary antisera for 45 min. Slides were washed in PBS and mounted in Vectashield (Vector Lab) containing 100 ng/ml DAPI. Fluorescence images were captured using a Zeiss Axioskop microscope equipped with a Hamamatsu CA742-95 digital camera and Improvision Openlab deconvolution software. Serial Z sections were obtained at 0.5 µm intervals and deconvolution of sections was performed using two nearest neighbours.
Pulse chase labelling of endocytosed Notch in S2 cells
A suspension of live S2 cells was centrifuged at 500 g for 5 min and the pellet was briefly cooled to 4°C and resuspended in cold complete medium containing anti-Notch extracellular domain antibodies (Developmental Studies Hybridoma Bank, C458.2H; 1:100). The cells were incubated at 4°C for 2 h with constant mixing, in order to label proteins at the plasma membrane in conditions where endocytosis is inhibited. The cells were then washed twice in cold complete medium and incubated at room temperature with constant mixing for 1 h to allow the labeled protein to be internalized. The cells were immobilized on a coverslip, fixed and permeabilized for immunostaining.
Results
Human Notch1 and WWP1 interact in vitro
The related ubiquitin ligases, Su(dx) and Itch, have been shown previously to interact with Notch proteins in vitro. We therefore investigated if there was an interaction between human Notch1 and WWP1 proteins by using an in vitro pull-down assay. Hela cells were transfected with a vector encoding the cytoplasmic domain of human Notch1 (NICD). Cell lysates were mixed separately with a glutathione-sepharose resin bound to either GST alone or a GST-WWP1 fusion construct and pull-downs tested for the presence of Notch1 on a western blot. GST-WWP1 fusion protein but not GST alone was found to interact with Notch1 ().
Context dependent WWP1 localization in the nucleus
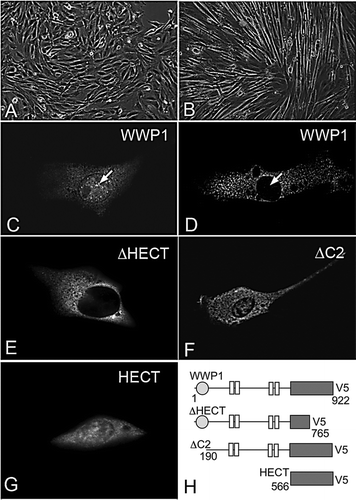
We assayed WWP1 localization by expressing a V5 epitope-tagged full-length WWP1 construct (WWP1) in the mouse C2C12 myoblast cell line. When we expressed WWP1 in non-differentiated C2C12 cells, WWP1 was distributed in both the nuclear and cytoplasmic compartments (A, C). However when C2C12 cells were allowed to differentiate by serum depletion, WWP1 staining became exclusively cytoplasmic, indicating that its localization is context dependent (B,D). To examine the domain dependence of WWP1 nuclear localization we examined the localization of three deletion constructs: WWP1ΔC2 (lacking C2 domain), WWP1ΔHECT (lacking intact HECT domain) and WWP1HECT (HECT domain only) (E-H). All constructs apart from WWP1ΔHECT displayed both cytoplasmic and nuclear localization, similar to full-length WWP1, while WWP1ΔHECT was localised exclusively to the cytoplasm. The HECT domain is therefore both necessary and sufficient for the nuclear localisation of WWP1.
Figure 2. Localization of WWP1 in C2C12 cells. (A) Undifferentiated C2C12 cells, and (B) morphologically distinct, differentiated C2C12 cells, 48 h after transfer to low serum containing medium. (C) When expressed in undifferentiated C2C12 cells, WWP1 is localized in the cytoplasm and in the nucleus (arrow). (D) In differentiated C2C12 cells WWP1 was only present in the cytoplasm and excluded from the nucleus (arrow). (E) When WWP1ΔHECT is expressed in undifferentiated cells it localizes to the cytoplasm. (F) WWP1ΔC2 and (G) WWP1HECT are both localized within the cytoplasm and nucleus, similar to full-length WWP1, when expressed in undifferentiated C2C12 cells. (H) Schematic diagram summarizing the WWP1 constructs used. Numbers refer to amino acids from the full-length protein sequence. All WWP1 constructs contain a C-terminal V5 epitope tagged used for immunostaining in C–G.
Notch regulates WWP1 nuclear localization
Coexpression of human Notch1 with WWP1 in undifferentiated C2C12 cells resulted in two different outcomes. In rare cases there was no colocalization of WWP1 and Notch1, and WWP1 was present in both the nucleus and the cytoplasm (A–C). In these cells Notch was predominantly observed in a perinuclear compartment. When found in the latter distribution, Notch was found to colocalize with a marker for the endoplasmic reticulum (data not shown). In the majority of transfected cells however, Notch1 and WWP1 displayed virtually complete colocalization in punctate vesicular-like structures in the cytoplasm, and in these cells WWP1 was no longer localized in the nucleus (D–F). To determine whether the observed relocalization of WWP1 was a specific effect of Notch1, or resulted from a non-specific consequence of increased expression of a membrane protein, we coexpressed WWP1 with integrin-αV. The latter had no effect on WWP1 localization and displayed very little colocalization with WWP1 (G–I). Next we investigated whether WWP1 relocalization from the nucleus resulted from a downstream response to an increase in Notch signalling. We coexpressed WWP1 with a soluble cytoplasmic domain of Notch1 (NICD). The latter localizes to the nucleus and has previously been shown to be constitutively active for Notch signalling by activation of the transcription factor Suppressor of Hairless/CBF-1 (Reviewed in Baron et al. [Citation2002], Wilkin & Baron [Citation2005]). In C2C12 cells NICD localized to the nucleus, but in all coexpressing cells WWP1 localization was both nuclear and cytoplasmic and indistinguishable from when WWP1 was expressed alone (J–L). This confirmed that the relocalization of WWP1 from the nucleus could not be a downstream consequence of transcriptional activation in response to increased Notch signalling. Also high levels of NICD in the nucleus did not sequester increased levels of WWP1 in the nucleus. This, together with the fact that in a proportion of the cotransfected cells WWP1 and full-length Notch1 did not show any colocalization, further argues that their interaction is context dependent.
Figure 3. Notch regulates WWP1 localization in the nucleus. Full-length WWP1 was coexpressed with full-length human Notch1 (hN1) (A–F), integrin-αV (Int-αV) (G–I), or soluble Notch1 intracellular domain (NICD) (J–L). When WWP1 was coexpressed with Notch, two classes of distribution were observed. A small proportion of cells showed no colocalization (A–C) with WWP1 being present in both cytoplasm and nucleus. The majority of cotransfected cells showed a strict colocalization of Notch1 and WWP1 with the latter excluded from the nucleus (D–F). When WWP1 was coexpressed with integrin-αV (G–I), or NICD (J–L), WWP1 was localized as in singly transfected cells. Expressed WWP1ΔC2 (M–O) or WWP1HECT (P–R) also became depleted from the nucleus in the majority of cells coexpressing Notch1, and both colocalized with the latter. (S–U) When WWP1ΔHECT was coexpressed with Notch, no colocalization with the latter was observed and the distribution of WWP1ΔHECT was exclusively cytoplasmic, as in singly transfected cells. WWP1 and its deletion constructs were visualized by staining for the V5 epitope (shown in green). hN1, NICD and integrin-αV immunostaining is shown in red. DAPI stained nuclei are also shown in some images (blue). Third column displays merged images.
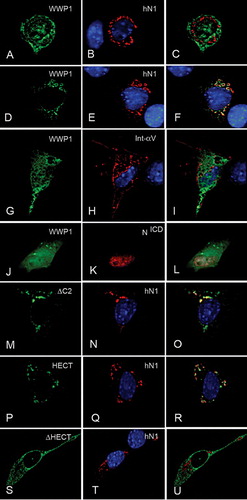
To determine the region of WWP1 necessary for its localization to be affected by Notch1, we coexpressed the different WWP1 deletion constructs with Notch1. When WWP1ΔC2 and WWP1HECT were coexpressed with Notch1 we observed the same two classes of distributions as we observed for full-length WWP1 (M–R). Colocalization of either WWP1 deletion construct with Notch1 and nuclear depletion was observed in the majority of cotransfected cells. When we coexpressed WWP1ΔHECT and Notch1 however, we only observed cells where Notch1 and WWP1 were not colocalized. In these cells WWP1ΔHECT localized, as before, only to the cytoplasm (S–U). Therefore the presence of the HECT domain appears to be both necessary and sufficient for the colocalization of WWP1 with Notch1 in C2C12 cells.
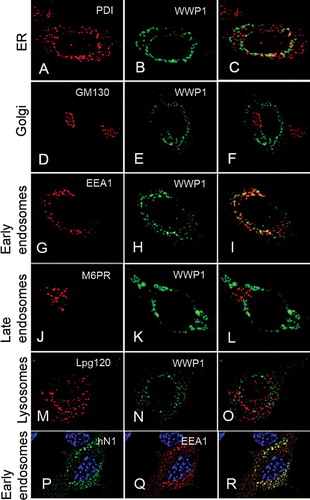
To identify the subcellular compartment in which WWP1 and Notch interacted, we used a panel of compartmental markers and found the Notch and WWP1 containing vesicles to be early endosomes (). In contrast the vesicular cytoplasmic localisation of Notch1 did not appear to be affected by the coexpression of WWP1. Notch1 was found predominantly in early endosomes even when WWP1 was not cotransfected (data not shown), although we cannot rule out the possibility that a more subtle shift of Notch between subdomains of the early endosome may occur in response to WWP1.
Figure 4. Human Notch1 and WWP1 colocalize in the early endosome. (A–O) C2C12 cells were cotransfected with human Notch1 and WWP1. 17 hours post-transfection, the cells were stained with anti-V5 (green) antibodies to detect WWP1 protein (B, E, H, K, N) and costained with a panel of different antibodies that label cellular compartments (red) including: (A) endoplasmic reticulum (anti-PDI), (D) Golgi (anti-GM130), (G) early endosomes (anti-EEA1), (J) late endosome (anti-M6PR), and (M) lysosome (anti-Lpg120). (C, F, I, L, O) Merged images of each costaining. When WWP1 is depleted from the nucleus, its cellular localization displays significant colocalization only with the early endosomes in cotransfected cells. (P, Q, R) In C2C12 cells coexpressing human Notch1 (hN1) and WWP1, hN1 (P, green) is colocalized with EEA1 (Q, red). (R) Merged image.
Drosophila Su(dx) localization is similarly regulated by Notch
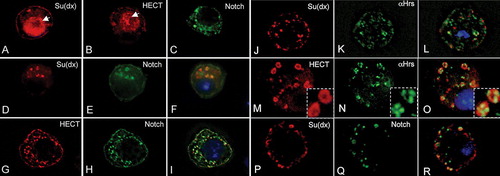
We have recently shown in the Drosophila wing imaginal disc that Su(dx) regulates the sorting of Notch within the early endosome, diverting the receptor into the late endosomal down-regulatory pathway (Wilkin & Baron [Citation2005]). We were interested to determine whether, in addition, Drosophila Notch also had a consequence on Su(dx) localization, similar to that observed for human Notch1 on WWP1. We therefore examined the localization of Su(dx) in S2 cells in the presence and absence of Notch. The results show that, as with WWP1, both full-length Su(dx) and its isolated HECT domain localize to the nucleus and cytoplasm in the absence of Notch expression, but only localize cytoplasmically when Drosophila Notch is coexpressed (A–I). As with human Notch1 and WWP1, Drosophila Notch and Su(dx) localized in early endosomes (J–O). The endocytic origin of colocalised Notch and Su(dx) was confirmed by surface labelling Notch in non-permeabilized cells using an antibody to its extracellular domain and following its endocytic uptake in a pulse chase labelling experiment (P–R).
Figure 5. Drosophila Notch redirects the nuclear localization of Su(dx) to the early endosome. (A) Full-length Su(dx) and B) Su(dx)HECT both display nuclear (arrows) and cytoplasmic staining when expressed in S2 cells. (C) Drosophila Notch localization when expressed in S2 cells is localized at the cell surface and in intracellular vesicles. (D–F) When coexpressed, Su(dx) (red) and Notch (green) co-localize in cytoplasmic vesicular compartments and Su(dx) is excluded from the nucleus. (G–I) When the HECT domain (red) was coexpressed with Notch (green), both proteins colocalized and this was correlated with depletion of Su(dx)HECT from the nucleus. The nucleus in the merged images F and I is shown stained with DAPI (blue). Su(dx) and Su(dx)HECT are stained using a C-terminal V5 epitope and N-terminal HA-epitope tags, respectively. (J–O) S2 cells were cotransfected with Notch and full-length Su(dx) (J–L) and the HECT domain of Su(dx) (M–O), and costained for Su(dx) and the early endosome marker Hrs. Both full-length Su(dx) and the HECT domain construct were associated with Hrs positive vesicles. An enlarged view of the vesicular compartments showing the localization of HECT and Hrs together is shown in insets (M-O). (P–R) Surface labeled Notch, stained with anti NECD (Q) becomes internalized into an endocytic compartment where it colocalizes with Su(dx) (P and merged image, R).
Discussion
The Notch receptor plays an essential role in metazoan development, regulating the timing and outcome of many cell fate decisions (Baron et al. [Citation2002], Wilkin & Baron [Citation2005]). Notch is also required in the adult for the maintenance of certain stem cell populations and the regulation of their cell fate. Misregulation of Notch is associated with tumour formation and a number of developmental disorders (Harper et al. [Citation2003]). A canonical signalling pathway for Notch has been elucidated which involves ligand-dependent cleavages of the Notch receptor that release the soluble cytoplasmic domain for translocation to the nucleus, where it binds to and activates the transcription factor Su(H)/CBF-1. A number of studies in Drosophila and mammalian cells have concluded that not all of the biological functions of Notch can be accounted for by this Su(H)/CBF-1 dependent pathway (Ordentlich et al. [Citation1998], Lawrence et al. [Citation2001], Ramain et al. [Citation2001], Hu et al. [Citation2003]). Here we suggest one possible alternative mechanism by which Notch can communicate with the nucleus.
We found that, like Itch and Su(dx) (Qiu et al. [Citation2000], Wilkin et al. [Citation2004]), WWP1 was able to interact with Notch by using an in vitro pull-down assay. In C2C12 cells, WWP1 showed a nuclear and cytoplasmic localization. Coexpression of full-length Notch however depleted WWP1 from the nucleus in most cotransfected cells, and this was correlated with the colocalization of Notch and WWP1 in early endosomes. Similar results were obtained using the isolated HECT domain. In contrast in rare cells where Notch and WWP1 failed to colocalize we found that Notch distribution in a perinuclear distribution similar to that of the ER. This suggests that the context dependence of the interaction reflects the different progression of Notch through the secretory and endocytic pathways in different cells. The expression of the nuclearly localized and constitutively active Notch intracellular domain did not promote a stronger accumulation of WWP1 in the nucleus, which supports the conclusion that the subcellular localisation of Notch determines its ability to interact with WWP1. The latter experiment also ruled out the possibility that WWP1 relocalization was a downstream response to increased Notch signalling through the canonical pathway. It is surprising that constructs lacking the WW domain region showed colocalization with Notch, since another Nedd4 family protein, Itch, was found, in vitro, to associate with Notch through its WW domains (Qiu et al. [Citation2000]), although the latter interaction has not been proven to be a direct one. Further work will be necessary to determine the nature of the interaction between the WWP1 HECT domain and Notch. It will also be interesting to determine whether the localizations of other HECT domain proteins are also modulated by Notch.
Since WWP1 has been shown to negatively regulate transcription factor activity (Conkright et al. [Citation2001], Zhang et al. [Citation2004]), then it follows that modulation of the nuclear localization of WWP1 may have a downstream consequence on gene regulation. Thus the interaction between WWP1 and Notch, by altering the levels of WWP1 in the nucleus, potentially constitutes an alternative mechanism by which Notch may signal. Furthermore the ability of WWP1 to regulate Smad7 localization and degradation (Komuro et al. [Citation2004]) suggests the possibility should be investigated as to whether there may be cross talk between Notch and TGF-β signalling pathways at the protein level. WWP1 has been shown to recruit Smad7 from the nucleus with Smad7 acting to target WWP1 to the TGF-β receptor. It will be interesting to determine whether these events are influenced by Notch, with WWP1 acting as an adaptor protein between components of the two pathways. Interestingly cross talk between Notch and Wingless signalling pathways has recently been proposed via the ability of Notch to promote armadillo/β-catenin degradation by an unknown mechanism that does not involve canonical Notch signalling (Hayward et al. [Citation2005]). Thus the possibility that endocytic trafficking of Notch can be linked to down-regulation of other developmental signals through recruitment of different pathway components deserves further investigation. Interestingly we obtained similar results when we examined the cellular localization of Drosophila Su(dx) and its response to the presence of Notch. This indicates that the ability of Notch to regulate Nedd4 family proteins has been conserved between Drosophila and mammals. Further work will now be necessary to determine if this potential alternative Notch signalling pathway is utilised in vivo, to identify its downstream consequences, and to understand its regulation.
We thank Spyros Artavanis Tsakonas, Hugo Bellen, Neil Bulleid, Martin Humphries, Martin Lowe and Philip Woodman for expression constructs, antibodies and cell lines, and Ann Canfield and Ged Brady for helpful advice. We acknowledge financial support from the BBSRC, MRC and Wellcome Trust. MF and SC were in part supported by a University of Manchester Overseas Research Studentship.
References
- Baron M, Aslam H, Flasza M, Fostier M, Higgs JE, Mazaleyrat SL, Wilkin MB. Multiple levels of Notch signal regulation (review). Mol Membr Biol 2002; 19: 27–38
- Chen X, Wen S, Fukuda MN, Gavva NR, Hsu D, Akama TO, Yang-Feng T, Shen CK. Human Itch is a coregulator of the hematopoietic transcription factor NF-E2. Genomics 2001; 73: 238–241
- Conkright MD, Wani MA, Lingrel JB. Lung Kruppel-like factor contains an autoinhibitory domain that regulates its transcriptional activation by binding WWP1, an E3 ubiquitin ligase. J Biol Chem 2001; 276: 29299–29306
- Cornell M, Evans DAP, Mann R, Fostier M, Flasza M, Monthatong M, Artavanis-Tsakonas S, Baron M. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 1999; 152: 567–576
- Flasza M, Gorman P, Roylance R, Canfield AE, Baron M. Alternative splicing determines the domain structure of WWP1, a Nedd4 family protein. Biochem Biophys Res Commun 2002; 290: 431–437
- Harper JA, Yuan JS, Tan JB, Visan I, Guidos CJ. Notch signaling in development and disease. Clin Genet 2003; 64: 461–472
- Hayward P, Brennan K, Sanders P, Balayo T, DasGupta R, Perrimon N, Martinez Arias A. Notch modulates Wnt signalling by associating with Armadillo/beta-catenin and regulating its transcriptional activity. Development 2005; 132: 1819–1830
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, Takeda Y, Chia W, Sankar N, Ng YK, Ling EA, Maciag T, Small D, Trifonova R, Kopan R, Okano H, Nakafuku M, Chiba S, Hirai H, Aster JC, Schachner M, Pallen CJ, Watanabe K, Xiao ZC. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell 2003; 115: 163–175
- Ingham RJ, Gish G, Pawson T. Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 2004; 23: 1972–1984
- Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K., Miyazawa K. Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1). Oncogene 2004; 23: 6914–6923
- Lawrence N, Langdon T, Brennan K, Arias AM. Notch signaling targets the Wingless responsiveness of a Ubx visceral mesoderm enhancer in Drosophila. Curr Biol 2001; 11: 375–385
- Malbert-Colas L, Fay M, Cluzeaud F, Blot-Chabaud M, Farman N, Dhermy D, Lecomte MC. Differential expression and localisation of WWP1, a Nedd4-like protein, in epithelia. Pflugers Arch 2003; 447: 35–43
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem 2003; 278: 23196–23203
- Mosser EA, Kasanov JD, Forsberg EC, Kay BK, Ney PA, Bresnick EH. Physical and functional interactions between the transactivation domain of the hematopoietic transcription factor NF-E2 and WW domains. Biochemistry 1998; 37: 13686–13695
- Ordentlich P, Lin A, Shen CP, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol 1998; 18: 2230–2239
- Otto E, Allen JM, Young JE, Palmiter RD, Maroni GA. DNA segment controlling metal-regulated expression of the Drosophila melanogaster metallothionein gene Mtn. Mol Cell Biol 1987; 7: 1710–1715
- Pirozzi G, McConnell SJ, Uveges AJ, Carter JM, Sparks AB, Kay BK, Fowlkes DM. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem 1997; 272: 14611–14616
- Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, Fang D, Hunter T, Liu YC. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem 2000; 275: 35734–35737
- Ramain P, Khechumian K, Seugnet L, Arbogast N, Ackermann C, Heitzler P. Novel Notch alleles reveal a Deltex-dependent pathway repressing neural fate. Curr Biol 2001; 11: 1729–1738
- Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, Myat A, Evans DA, Cornell M, Baron M. Regulation of Notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol 2004; 14: 2237–2244
- Wilkin MB, Baron M. Endocytic regulation of Notch activation and down-regulation (Review). Mol Membr Biol 2005; 22: 280–289
- Zhang X, Srinivasan SV, Lingrel JB. WWP1-dependent ubiquitination and degradation of the lung Kruppel-like factor, KLF2. Biochem Biophys Res Commun 2004; 316: 139–148