Abstract
Despite being synthesized in the cytosol without a leader sequence, the soluble 253-residue mammalian protein CLIC4 (Chloride Intracellular Channel 4, or p64H1), a structural homologue of Ω-type glutathione-S-transferase, autoinserts into membranes to form an integral membrane protein with ion channel activity. A predicted transmembrane domain (TMD) near the N-terminus of CLIC4 could mediate membrane insertion, and contribute to oligomeric pores, with minimal reorganization of the soluble protein structure. We tested this idea by reconstituting recombinant CLIC4 in planar bilayers containing phosphatidyethanolamine, phosphatidylserine and cholesterol, recording ion channels with a maximum conductance of ∼15 pS in KCl under both oxidizing and reducing conditions. The channels discriminated poorly between anions and cations, incompatible with the current “CLIC” nomenclature, and their conductance was modified by the trans (external or luminal) redox potential, as previously observed for CLIC1. We then reconstituted a truncated version of the protein, limited to the first 61 residues containing the predicted TMD. This included a single trans cysteine residue in the putative pore-forming subunits, at the external entrance to the pore. The truncated protein formed non-selective channels with a reduced conductance, but they retained their trans-redox sensitivity, and could still be blocked or inactivated by trans (not cis) thiol-reative dithiobisnitrobenzoic acid. We suggest that oligomers containing the putative TMD are essential components of the CLIC4 pore. However, the pore is inherently non-selective, and any ionic selectivity in CLIC4 (and other membrane CLICs) may be attributable to other regions of the protein, including the channel vestibules.
Introduction
Chloride Intracellular Channel (CLIC) proteins are unique among putative eukaryotic ion channels in being able to assume both soluble and membrane forms. CLICs also bypass the conventional secretory pathway, and “autoinsert” directly into membranes Citation[1], Citation[2]. The proteins are related to p64, a putative anion channel purified from bovine kidney by drug affinity chromatography in pioneering work by Al-Awqati and his colleagues Citation[3] before being cloned Citation[4] and characterized in considerable detail (e.g. Citation[5]). However, it has yet to be established (by single-channel recording) that p64 alone is sufficient to form an ion channel, in contrast to the best-studied CLIC protein, CLIC1 Citation[6].
The first CLIC protein to be discovered, rat brain CLIC4 (p64H1, or p64 homologue 1), known in mouse as mitochondrial or mtCLIC Citation[7], was identified and cloned by homology to p64 Citation[8], Citation[9] as a potential candidate for an intracellular anion channel previously shown to be co-localized with rat brain ryanodine-sensitive calcium-release channels Citation[10]. CLIC4 isologues are now known to be very widespread, and the proteins are highly conserved in vertebrates ranging from fish to mammals Citation[11]. The structure of a soluble form of human CLIC4 (crystallized with a short random C-terminal extension) is similar Citation[12] to previously crystallized, soluble human CLIC1 Citation[13], as anticipated by modelling CLIC4 onto CLIC1 Citation[2]. Like CLIC1, soluble CLIC4 has an Ω-glutathione-S-transferase (GST) fold, but although Ω-GSTs and CLICs are structurally similar, and distantly related, they do not have overlapping functions. In particular, Ω-GSTs do not autoinsert into membranes and form ion channels Citation[14], and CLIC proteins appear to have little or no enzymatic activity.
In addition to brain and kidney, CLIC4 mRNA is also expressed in many other mammalian tissues including lung, liver, skeletal muscle, testis and skin Citation[7], Citation[9]. A putative “cytoplasmic domain” containing most of the protein formed complexes involving brain actin, dynamin I, tubulin and 14-3-3 proteins Citation[15], and cytoplasmic CLIC4 has been shown to co-localize with A-kinase anchoring proteins (AKAPs) in specific cellular microdomains, including centrosomes and the cortical actin cytoskeleton Citation[16]. The much less abundant Citation[15] membrane form of CLIC4 is an integral (not peripheral Citation[9]) membrane protein localized to several intracellular organelles, including the endoplasmic reticulum and outer nuclear membrane Citation[9], (inner) mitochondrial membranes Citation[7], and the membranes of dense core secretory vesicles Citation[17], caveolae and (possibly) the trans-Golgi network Citation[18]. It has also been localized to the plasma membrane Citation[18], Citation[19], especially near intercellular junctions Citation[16].
CLIC4 levels are dynamically regulated, and increase with tumour necrosis factor-α and p53 signalling Citation[7]. Interestingly, CLIC4 overexpression induces p53-mediated apoptosis associated with mitochondrial depolarisation, cytochrome C release and caspase activation Citation[20]. Although it is not yet clear whether this involves the soluble or membrane form of the protein, or both, possible implication of a putative mitochondrial ion channel in apoptosis could clearly be very significant. It is however notable that some cells, especially mammalian cells, express many different CLIC proteins, suggesting that some or all of their functions may be redundant. For example, transgenic mice lacking CLIC1 appear to be essentially normal apart from increased weight, splenomegaly and mild thrombocythaemia Citation[21].
In contrast to CLIC1, relatively little is known about CLIC4-associated ion channels. Although the incorporation of microsomal membrane vesicles containing recombinant CLIC4 into planar lipid bilayers gave rise to novel anion channel activity of 10–50 pS Citation[9], this could have been due to the activation of unidentified endogenous channels, especially since another CLIC, CLIC2, has been shown to be a channel modulator rather than an ion channel itself Citation[22]. CLIC4 (possibly with the short C-terminal extension required for crystallisation) gave rise to channel activity in “tip-dip” bilayers Citation[12], but the conductance of the channel was unclear (reported as both 31 pS and 57 pS). Novel ion channels specifically shown to contain FLAG-tagged CLIC4 have also been recorded by patch-clamping the plasma membrane of cells overexpressing the protein Citation[19]. In the presence of large cations (to limit endogenous cation currents), the conductance of individual CLIC4 channels appeared to be very low, of the order of 1 pS. Currents through CLIC1 are now known to be regulated by the external redox potential Citation[23], and if CLIC4 shows a similar mechanism, this may help to explain the very small single-channel currents observed by patch-clamping. Also, as discussed later, the use of large cations may have further reduced the CLIC4 currents.
We set out to investigate the single channel properties of CLIC4 under various redox conditions, because in contrast to a previous report Citation[12], the protein can form channels under both reducing and non-reducing conditions, like CLIC1 Citation[23]. We then tested a simple model for the transmembrane topology of CLIC4 that can be extrapolated to CLIC1 and other membrane CLICs, by reconstituting a truncated protein containing the N terminus of CLIC4 and its single putative transmembrane domain (TMD). We also determined whether CLIC4 is sensitive to the trans (extracellular or luminal) redox potential in the presence of a glutathione buffer, like CLIC1 Citation[23], and whether a critical cysteine residue on the trans side of membrane CLIC4 subunits, corresponding to the GSH-binding “G-site” of Ω-GSTs, mediates this unusual effect.
Materials and methods
Expression and purification of CLIC4
We cloned rat brain CLIC4 (p64H1, Citation[9]) into pHis8, a modified pET vector encoding an N-terminal octa-His tag and a thrombin cleavage site Citation[24], and inserted a stop codon into one clone by “QuikChange” PCR to truncate the expressed protein at CLIC4 position 61. The inserts were verified by DNA sequencing (MWG Biotech), and fusion proteins were expressed in E. coli BL21 (DE3) cells and recovered from cell lysates by Ni2 + -NTA affinity chromatography, with yields for the soluble thrombin-cleaved full-length and truncated proteins of 4.0±0.50 mg/l and 2.0±0.65 mg/l (means±SD, n=15 or 3), respectively (determined by the Lowry method using appropriate standards). The preparations were analysed by SDS-PAGE and gel-exclusion FPLC using Superdex 200 (by methods detailed in Citation[25]), and protein aliquots (which had at no stage been exposed to detergents) were stored for up to 3 months at −70°C in the presence of 5 mM DTT. The masses of proteins subjected to gel-exclusion FPLC were determined from a plot of log(Mr) vs. Kav (the corrected partition coefficient):where Ve, Vo and Vt represent the elution volume, the void volume and the packed bed volume, respectively.
Channel incorporation into planar lipid bilayers
Planar bilayers were prepared at room temperature (20°C) using lipids selected from: palmitoyl-oleoyl phosphatidylcholine (POPC), PO-phosphatidylethanolamine (POPE), PO-phosphatidylserine (POPS) and cholesterol (all from Avanti, AL, USA), as detailed in the Results section. Briefly, the lipids were suspended in n-decane (25 µg total lipid/µl), and films were cast across a 0.3 mm hole in a polystyrene partition separating two chambers. The chambers, designated cis and trans, contained 50 mM KCl with 10 mM Tris-HCl (pH 7.4) and 1 mM DTT, unless otherwise specified, and were connected by agar salt bridges to the headstage input or ground, respectively, of an Axopatch 200-B amplifier, minimising and offsetting liquid junction potentials as previously described Citation[26]. After the lipid film had thinned spontaneously to form a planar bilayer, monitored by measuring a relatively abrupt increase in membrane capacitance from <50 pF to >200 pF, the cis chamber was clamped at various holding potentials (HPs) relative to the trans chamber, and up to 25 ng/ml purified protein was stirred into the same chamber, followed by small aliquots of 5 M KCl to raise the KCl concentration to 500 mM. Transmembrane currents appeared within 10 min of adding the (full-length) protein, and were digitally recorded. Thereafter, the contents of the chambers were changed by perfusion (at least 10 volumes) as required.
Single-channel analysis
Single-channel currents (labelled following the standard electrophysiological convention, i.e., positive currents represent net cation flux from cis to trans), were low-pass filtered (8-pole, Bessel type response) at 50 Hz or 25 Hz and analysed with pClamp8 (Axon Instruments) and pStat (SPSS), using amplitude histograms as previously described Citation[23] to measure channel amplitudes and open probabilities (Po). Salt concentrations were converted to activities using standard tables, and (relative) anion permeabilities (P) were calculated under equilibrium conditions using the Nernst equation:where a is the activity coefficient of the relevant salt, Er is the reversal or equilibrium potential, and z, F, R and T have their usual significance. Relative anion vs. cation permeabilities (selectivities) were calculated from the following form of the Goldman-Hodgkin-Katz (GHK) voltage equation:
where n is the cis/trans salt activity ratio and k = RT/zF (26 mV).
Results
CLIC4 ion channels in fully-reducing conditions
CLIC4 recordings had an inconsistent appearance in experiments using POPC or equimolar POPE and POPS, but like CLIC1 Citation[23], the protein formed highly consistent channels in 80/83 bilayers containing POPE, POPS and cholesterol, 4:1:1 mol/mol, respectively, in the presence of 1 mM DTT, 5 mM GSH or 100 µM H2O2. In the latter case, the protein was also exposed to 100 µM H2O2 for 5–10 min. before incorporation. Channel activity appeared more rapidly at an acidic cis pH of 5.5. However, these recordings were very noisy, and low pH conditions were not pursued further. No channel-like events were seen during prolonged (up to 30 min.) observation of control bilayers in the absence of added protein (15 experiments).
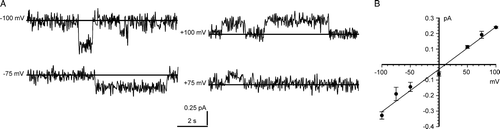
A shows examples of recordings obtained in 1 mM DTT, representing “fully-reducing” conditions. The slope conductance of the main open state in a cis:trans gradient of 500:50 mM KCl with 1 mM DTT in both chambers was 10.3±1.0 pS (mean±SD, n=15), calculated (by linear regression) from the linear region of the I/V plot (−100 mV to +70 mV) (B). We noted several substates, including the prominent ∼25% substate shown here. The reversal potential of the main open state was −12.2±3.3 mV (mean + SD, n=15) (B), corresponding to a mean Cl−/K+ selectivity of 0.54±0.09, i.e., a poorly selective or even mildly cation-selective channel. The Er for the ∼25% substate was similar, and we observed direct transitions to and from both states (inset in B). The main open state conductance was ohmic in symmetrical KCl solutions (e.g., inset I/Vs in ), but showed a complicated dependence on KCl activity (, main panel). Up to ∼350 mM, the relationship could be described as the sum of a hyperbolic and a linear component, but the maximum single-channel conductance was 13.8±0.60 pS (mean±SD, n=5), and it declined at higher KCl activities, consistent with self-block.
Figure 1. Bilayer reconstitution of CLIC4 in the presence of 1 mM DTT. Part A shows examples of recordings at selected holding potentials, with solid lines to indicate the closed levels, and an example of an all-points amplitude histogram from 30 sec of the +100 mV recording (the arrow indicates the maximum open level). Part B summarizes the I/V relationship of the main open level and a ∼25% substate, from 15 independent experiments with 500 mM KCl cis vs. 50 mM KCl trans. The bars indicate ±1 SD, and the line was fitted by eye (see text for linear regression analysis). The inset shows examples of direct transitions between the ∼25% substate, the main open level and the closed level, on an expanded time scale.
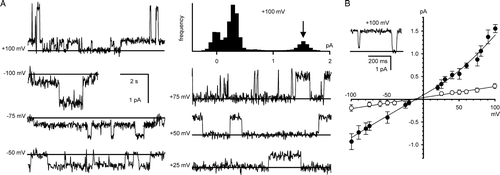
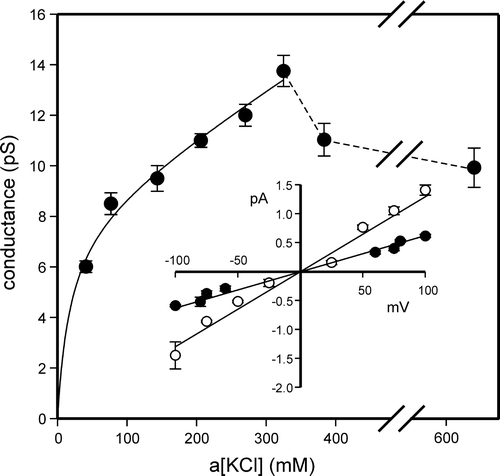
Figure 2. Conductance/activity relationship of CLIC4. The data in the main panel (shown as means±SD from 5 independent experiments) were fitted (by least squares) to the sum of a hyperbolic component with a gmax of 8.2 pS and a “Km” of 19.1 mM plus a non-saturating linear component of 0.017 pS per mM KCl. The relationship breaks down at KCl activities >325 mM (as shown by the dotted lines – note the break in the plot between 400–600 mM KCl). The inset I/V relationships provide examples of the I/V data used to calculate the conductance at low (40 mM, closed circles) and high (325 mM, open circles) activities (points are means±SD, n=5, lines fitted by linear regression).
We measured relative anion permeabilities at equilibrium under biionic conditions in the presence of 1 mM DTT by perfusing 50 mM KCl (corrected for activity) into the cis chamber, and different potassium salts with the same activity into the trans chamber. Relative anion permeabilities calculated from the equilibrium potential were the same for 5 test anions compared to Cl− (set to 1.0): SCN− (0.97±0.11); NO3− (1.1±0.50); I− (1.1±0.96); Br− (1.3±0.50); F− (1.4±0.95). The values in parentheses are the mean±SD for 4 independent comparisons in each case, and as expected they are not significantly different (p>0.5). Single-channel currents were also obtained in 500 mM:50 mM cis vs. trans TrisCl (pH 7.4) in the presence of 1 mM DTT (A). As shown in B, the conductance was reduced to 2.6±0.43 pS (mean±SD, n=7), but the reversal potential was +11±6.1 mV (mean±SD, n=7), giving a mean Cl−/Tris+ selectivity of 1.8±0.51 (not corrected for activities). The difference in relative anion vs. cation selectivity compared to KCl is highly significant (p<0.001).
CLIC4 ion channels in other redox conditions
H2O2-oxidized CLIC4 channels were reconstituted in the presence of 100 µM H2O2 (“fully-oxidizing” conditions). Ion channels (A) appeared more rapidly compared to the reduced protein, and had a slope conductance of 8.9±1.1 pS (mean±SD, n=5) in a cis:trans gradient of 500:50 mM KCl, measured (by linear regression) over the linear part of the I/V plot between −100 mV and +25 mV (B). This value was indistinguishable from the conductance of channels reconstituted in the presence of DTT (p>0.5). The oxidized channels displayed several substates, and the reversal potential of the main open state was +6.5±6.6 mV (mean±SD, n=5) under fully oxidizing conditions, corresponding to mildly anion-selective channels with a mean Cl−/K+ selectivity of 1.4±0.48.
Figure 4. Bilayer reconstitution of CLIC4 in the presence of 100 µM H2O2. Part A shows examples of recordings at selected holding potentials, and an example of an all-points amplitude histogram from 30 sec of the +100 mV recording (the arrow indicates the maximum open level). Part B summarizes the I/V relationship obtained from 5 independent experiments (means±1 SD, n=5). The line was fitted by eye (see text for linear regression analysis). All the data were obtained with 500 mM KCl cis vs. 50 mM KCl trans.
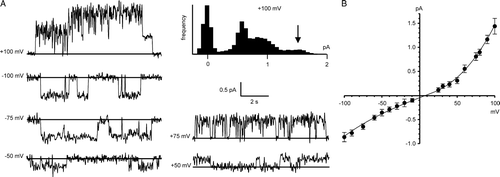
Channels were also obtained in a cis:trans gradient of 500:50 mM KCl from (non pre-oxidized) CLIC4 in 5 mM glutathione, corresponding to “physiological” redox conditions Citation[23], e.g., . The single-channel currents had higher amplitudes at positive holding potentials (i.e., they were outwardly-rectifying, if cis is equivalent to the cell cytosol), and the maximum slope conductance (with 5 mM GSH/0.5 mM GSSG cis and trans) was 14.8±1.6 pS (mean±SD, n=5, calculated by linear regression between −25 mV and +100 mV). The reversal potential of +8.8±4.0 mV (mean±SD, n=5) (B) corresponded to a mean Cl−/K+ selectivity of 1.5±0.27. As previously observed for CLIC1 Citation[23], the apparent single-channel slope conductance was modulated by the trans redox potential when this was manipulated in a GSH buffer. Sequential additions of GSSG to the cis chamber had no effect, but sequential additions of GSSG to the trans chamber (e.g., to give [GSH]/[GSSG] ratios of 2:1 or 1:1, as shown in A), markedly decreased the apparent single-channel currents. This could be reversed by reverting to a redox potential of −225 mV (5 mM GSH with 0.5 mM GSSG) in the trans chamber (also shown in A).
Figure 5. Bilayer reconstitution of CLIC4 in glutathione buffers. Part A shows recordings in 5 mM GSH (cis and trans), with GSSG added sequentially to the trans chamber to give the indicated GSH/GSSG ratios. Part B shows a combined I/V relationship from 5 independent experiments (means±SD) with 5 mM GSH/0.5 mm GSSG both cis and trans The line was fitted by eye (see text for results of linear regression analysis). The traces in part C (underfiltered, and on an expanded time scale), illustrate the appearance of substates (best-resolved where indicated by the arrowhead) immediately after replacing the cis glutathione buffer (5 mM GSH/0.5 mM GSSG) with 1 mM DTT by perfusion. All the data were obtained with 500 mM KCl cis vs. 50 mM KCl trans, and the recordings were carried out at a HP of +100 mV.
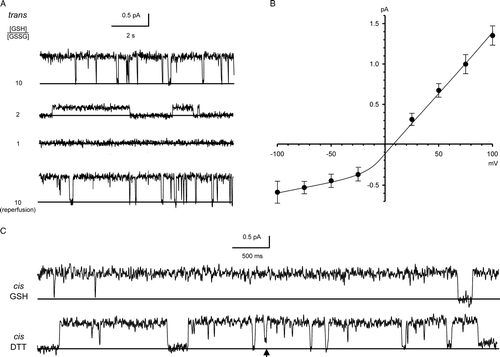
Like CLIC1 Citation[23], channel recordings in GSH buffers were less noisy, and contained fewer substates, than recordings under fully-reducing or fully-oxidizing conditions (i.e., in the presence of DTT or H2O2, respectively). The excess channel noise we observed in the absence of a glutathione buffer was largely attributable to short-lived or poorly-resolved substates, possibly suggesting that fully-oxidised or fully-reduced membrane CLICs failed to adopt a single, optimally-folded conformation. Consistent with this idea, replacing the cis glutathione buffer with 1 mM DTT appeared to induce substates following channel incorporation (e.g., C).
Single-channel properties of the putative TMD of CLIC4
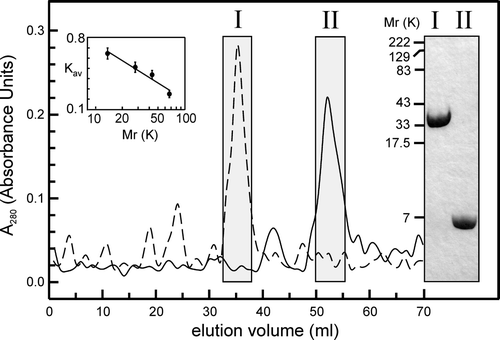
As shown in , the truncated protein, comprising the first 61 residues of CLIC4 (predicted Mr 6,800), including the N-terminus and the putative TMD Citation[2], Citation[11], showed appropriate mobilities on SDS-PAGE, and appropriate retention times during gel-exclusion FPLC (consistent with a monomeric solution, like the full-length protein, which is shown for comparison). We reconstituted truncated CLIC4 into bilayers of the same composition as before (POPE, POPS and cholesterol, 4:1:1 mol/mol, respectively) but it formed channels less readily, often taking 20–40 min. compared to less than 10 min for the full-length protein. However, once channels had appeared in the bilayer, “single-channel” recordings were highly consistent between experiments, typically showing infrequent low-amplitude currents of ∼0.5 pA at a HP or +100 mV. In some experiments we also observed occasional, large-amplitude currents of up to ∼1.5 pA at the same HP, especially in the presence of H2O2, including 3 independent experiments using only peak II fractions after FPLC of the truncated protein ().
Figure 6. FPLC and SDS-PAGE analysis of full-length and truncated CLIC4. The main panel shows typical gel-exclusion FPLC profiles for full-length (FL) and truncated (TR) proteins in the presence of 5 mM DTT (dashed and solid lines, respectively, shown on the same plot for comparison). Fractions from the boxed peaks (I and II, respectively) were collected and concentrated, and subjected to reducing 10% (w/v) SDS-PAGE and Coomassie-stained as shown. FL and TR proteins show Mr values of ∼30 K and ∼6–7 K, respectively. The FPLC analysis reveals similar values (Vt was 70 ml, Vo was 4.5 ml, and the Kav values for the FL and TR proteins are 0.73 and 0.47, respectively). The column was calibrated (inset plot) with: RNase A (13.7 K), pGEX vector GST (26 K), ovalbumin (43 K) and albumin (67 K).
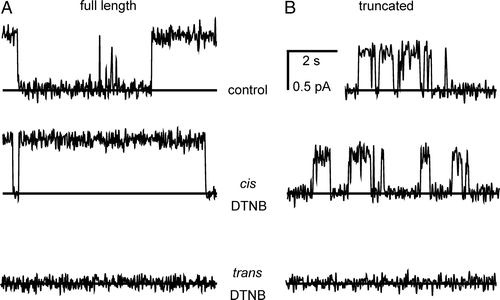
A compares the appearance of channels formed by the truncated protein under the different oxidizing and reducing conditions described earlier (all at +100 mV), and B summarizes their I/V relationships in a cis:trans gradient of 500:50 mM KCl. Currents through the truncated protein remained sensitive to the trans (but not the cis) redox potential, like the full-length protein (A). Overall, the conductances and anion vs. cation selectivities of the truncated protein were very similar in the presence of 1 mM DTT, or 5 mM GSH with 0.5 mM GSSG. From an analysis of several independent experiments, the single-channel slope conductances, calculated by linear regression between +25 mV and +100 mV, were 5.5±0.58 pS and 4.8±0.61 pS (both means±SD, n=5), respectively, for the two different conditions. The values are not significantly different (p>0.5). The relative anion vs. cation selectivities of the truncated proteins, determined from equilibrium potentials measured under the same conditions, were 1.1±0.27 and 1.0±0.45, respectively (both means±SD, n=5). Again, these values are indistinguishable (p>0.5), and in B all the points are fitted to a single line for both conditions.
Figure 7. Bilayer reconstitution of truncated CLIC4. Part A compares the appearance of channels at +100 mV in the presence of either 1 mM DTT, 100 µM H2O2 (with a star to indicate an unusual large-amplitude opening, see text), or the indicated GSH/GSSG ratios (with [GSH] set to 5 mM), all in a cis:trans gradient of 500:50 mM KCl. Part B summarizes the corresponding I/V relationships in the presence of 1 mM DTT (closed circles), 100 µM H2O2 (open circles) and a 10:1 GSH/GSSG buffer (open squares) (means±1 SD for 5 independent experiments in each case). The line was fitted by eye (see text for results of linear regression analysis).
![Figure 7. Bilayer reconstitution of truncated CLIC4. Part A compares the appearance of channels at +100 mV in the presence of either 1 mM DTT, 100 µM H2O2 (with a star to indicate an unusual large-amplitude opening, see text), or the indicated GSH/GSSG ratios (with [GSH] set to 5 mM), all in a cis:trans gradient of 500:50 mM KCl. Part B summarizes the corresponding I/V relationships in the presence of 1 mM DTT (closed circles), 100 µM H2O2 (open circles) and a 10:1 GSH/GSSG buffer (open squares) (means±1 SD for 5 independent experiments in each case). The line was fitted by eye (see text for results of linear regression analysis).](/cms/asset/5d4a412d-164d-40e7-84e8-cca5fa11ad38/imbc_a_192707_f0007_b.gif)
As noted earlier, pre-oxidized truncated proteins reconstituted in the presence of 100 µM H2O2 formed infrequent channels of both “low” and “high” conductance. The former had a slope conductance of 3.8±0.53 pS (mean±SD, n=5) measured over the same interval (+25 mV to +100 mV), and a relative anion vs. cation selectivity of 1.2±0.98 (mean±SD, n=5). Although these values are not significantly different from those found for the truncated protein in the presence of DTT or glutathione buffers (p>0.5), the experimental points (B, open circles) do not appear to align well with the corresponding data obtained using DTT or a glutathione buffer.
Orientation of membrane CLIC4
We previously investigated the bilayer orientation of CLIC1 containing an intact N-terminal His tag by attempting to block or inhibit the channel with 50 µM histidine-reactive Ni2 + from the cis and trans sides in turn, but in similar experiments with CLIC4 the metal ion left some channels unaffected, possibly because the N-terminal region before the putative TMD of CLIC4 is longer and more flexible. As an alternative, we deployed cysteine-specific reagents, also used previously, because the cysteine residues in CLIC4 are located asymmetrically with respect to the putative TMD, just as they are in CLIC1 Citation[2]. As shown later (), CLIC4 C35 lies in a cysteine-proline motif on the N-terminal side of the TMD, immediately before the postulated pore entrance. The three remaining cysteines in CLIC4 lie on the other side of the TMD, much further into the primary sequence. C35 is the only cysteine residue in the truncated from of CLIC4.
In contrast to similar experiments with CLIC1 Citation[23], 20 µM NEM had no effect from either side of the bilayer. However, 0.2 mM 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) blocked (or inactivated) CLIC4 channels from the trans side, but not the cis side (e.g., ). This applied equally to both full-length CLIC4 and the truncated protein (6/6 experiments in each case), and the effect was quantified by measuring the probability of a given channel being open (Po) over successive periods of 60 sec for each condition. The mean Po for full-length and truncated channels exposed to 5 mM GSH was 0.63±0.21 and 0.52±0.35, respectively (means±SD, n=6). After removing cis GSH by perfusion and adding 0.2 mM cis DTNB, the Po values were 0.65±0.26 and 0.53±0.34, respectively (means±SD, n=6). Following a similar procedure to add 0.2 mM trans DTNB, the mean Po values were reduced to 0.05±0.03 and 0.04±0.03, respectively (means±SD, n=6).
Figure 8. Inhibition of CLIC4 currents by DTNB. Parts A and B show examples of CLIC4 currents from full-length and truncated proteins, respectively, at +100 mV, obtained in the presence of 5 mM GSH (with 500 mM KCl cis vs. 50 mM KCl trans) before removing the GSH by perfusion with fresh GSH-free solutions. In each case, 0.2 mM DTNB had no effect from the cis side, but abolished channel activity when added from the trans side. See text for full statistical analysis.
Although the differences between the mean Po values for control and trans DTNB differed significantly, the differences in Po between control and cis DTNB, and between control and trans DTNB, were also analysed channel by channel by paired t-tests, because of the relatively large variation in control Po from channel to channel. This showed no significant difference between control and cis DTNB for both full-length and truncated CLIC4 (p>0.5), but significant differences between control and trans DTNB in each case (p<0.002 and p<0.02, respectively).
Discussion
Channel activity of CLIC proteins
Despite their structural similarities, Ω-GSTs and CLICs have very distinct functions. In particular, Ω-GSTs remain soluble and have no ion channel activity Citation[14]. However, they are unusual compared to other GSTs in having weak GSH-dependent thioltransferase activities Citation[28]. It has been speculated that this could involve thiol transfer between GSH bound at the G-site (containing the first cysteine residue in the primary sequence), and S-thiolated substrates (including proteins) in an adjacent ligand-binding site (the H-site). In contrast, CLICs appear to have little or no enzymatic activity, but CLIC1 can autoinsert into membranes and form novel ion channels Citation[6]. CLIC proteins contain a single predicted TMD near the N-terminus, which is notable by its absence in Ω-GSTs Citation[2]. In every mammalian CLIC protein, this putative TMD is preceded by an Ω-GST-like “G-site” cysteine residue, in a cysteine-proline motif.
The suggestion that an entire class of soluble eukaryotic proteins, including several mammalian proteins, can bypass the secretory pathway and autoinsert into membranes without any specific insertion machinery, remains highly controversial. In addition, there has been substantial disagreement concerning even the basic properties of the ion channels associated with CLIC1 (the best studied protein) Citation[6], Citation[23], Citation[29–35]. This includes disagreement over whether the protein can insert into membranes in both oxidized and reduced forms Citation[23], or whether it can only insert as an oxidized protein containing an intrachain disulphide bond Citation[34]. However, the importance of specific membrane lipids, and the sensitivity of reduced membrane CLIC1 to external (or luminal) oxidation, could help to explain many previous inconsistencies. We suggested that membrane CLIC1 oligomers are oxidised and reduced by thiol-disulphide exchange involving GSH, GSSG and external or luminal “G-site”-like subunit cysteines, equivalent to the G-site in Ω-type GSTs. This in turn could regulate channel gating and the overall ion flux through CLIC1 Citation[23]. Thus, channel function may depend on the external (or luminal) redox conditions, which are often poorly-controlled in experiments.
Like CLIC1, proper functional reconstitution of CLIC4 (distinct from simple protein autoinsertion, as shown previously for CLIC1 Citation[23]) required a specific lipid mixture (PE/PS/cholesterol, 4:1:1 mol/mol). Although other, untested, lipid mixtures may also be effective, the presence of cholesterol recalls the specific role of the sterol in cytolysin channel insertion or assembly Citation[36], Citation[37], and intracellular membranes contain a similar amount of the free sterol Citation[38]. Alternatively, given that cholesterol and the inverted-cone shaped phospholipid PE are both associated with “curvature stress” in planar membranes, they may promote channel assembly or activity by a physical role similar to the activation of membrane-associated protein kinase C Citation[39].
Channel formation by full-length CLIC4
CLIC4 contains 4 cysteine residues Citation[9] and we reconstituted the membrane protein under a wide range of oxidizing and reducing conditions, including DTT (with a standard redox potential of −330 mV, for “fully-reducing” conditions), GSH/GSSG (providing “physiological” redox potentials between −225 mV and −195 mV at a pH of 7.4), and 100 µM H2O2 (“fully-oxidising” conditions). The channels had a similar maximum conductance of ∼15 pS in a cis vs. trans gradient of 500 mM:50 mM KCl, but were noisier and displayed more substates in DTT and H2O2 compared to GSH. We speculated that the protein only folds optimally under physiological redox conditions. The single-channel conductance/activity relationship contained both hyperbolic and linear components at a[KCl] values up to ∼350 mM (in 1 mM DTT), with a paradoxical reduction in conductance above 350 mM, consistent with a multi-ion conduction mechanism (i.e., more than one ion at a time in the selectivity filter).
CLIC4 is poorly-selective between anions and cations, inconsistent with the widely-adopted “CLIC” nomenclature proposed by Citation[18] and Citation[40]. It may be necessary to re-evaluate CLIC proteins, especially CLIC4, as “chloride” channels, if they behave like non-selective channels. This could be due to a wide pore lacking specific ion-binding sites. However, a maximum conductance of ∼15 pS in KCl is inconsistent with a wide water-filled pore (as found for example in bacterial porins). Franciolini and Nonner Citation[41], Citation[42] noted similar paradoxical behaviour in neuronal “background” Cl− channels, and suggested that anions and cations crossed the membrane at least partly as counter ions. Based on similar ideas, if the putative pore lining of CLIC4 contains rings of arginine or lysine residues (as suggested in ), transient binding of permeant anions could provide an opportunity for counter ions (e.g., K+) to cross the membrane without encountering a prohibitive positive charge.
Figure 9. Organization of the putative CLIC TMD. Part A: selected regions of 3 CLICs aligned at the putative 18-residue TMD (underlined). The first cysteine residue in each sequence is numbered, and differences within the TMD are highlighted (rat and human CLIC4 are identical in this region). CLIC4 was truncated where indicated. Part B is a helical wheel projection of the TMD from N to C (D. Armstrong and R. Zidovetzki, http://rzlab.ucr.edu/scripts/wheel/wheel.cgi) with a “tetrameric pore” cartoon. Squares, circles and polygons indicate relatively hydrophobic or hydrophilic residues, or residues with positively charged side chains, respectively. The arrow and the arc indicate the direction of the hydrophobic moment, and suggested pore-lining residues, respectively. The alternative residues in CLIC1 (all 3) and CLIC5A (starred residue only) are also shown.
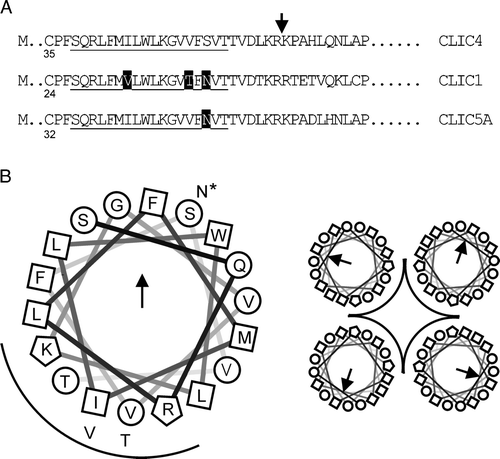
In a scheme like this, ion permeation requires the presence of anions, and they must be sufficiently small to penetrate the pore. Large, impermeant anions would prevent the passage of both anions and cations (regardless of the latter's size), whereas large, relatively impermeant cations would not prevent all ion permeation, but would make the channel more anion-selective. Consistent with this idea, we noted enhanced anion vs. cation selectivity in the presence of the relatively large cation Tris+ compared to K+. We also noted that CLIC4 was unable to discriminate between a range of different anions, again consistent with the idea that the pore itself is poorly-selective. However, CLIC1 is slightly but measurably more anion-selective than CLIC4 Citation[23], so if the putative pore-lining is not responsible for this difference (and the sequences are very similar, as shown in , suggesting in passing that detailed site-directed mutagenesis studies of putative TMD/pore-lining residues may not be very productive), could other parts of the protein be important for selectivity?
We also noted enhanced anion vs. cation selectivity in fully-oxidized CLIC4 (exposed to 100 µM H2O2) compared to the fully-reduced channel, although the channel remains essentially very poorly-selective. Oxidation of soluble CLIC1 induces a completely new protein conformation involving intrachain disulphide bond formation and non-covalent association into dimers Citation[34], but a similar mechanism has been ruled out for CLIC4 Citation[12], so we speculate that following membrane insertion the pore-lining regions of CLIC4 remain the same irrespective of oxidation or reduction. Instead, we suggest that the difference in selectivity observed here could be due to changes in the charged surface of the protein, away from the putative TMD, that affect the channel vestibule(s) or entrance(s). This suggestion is consistent with experiments involving truncated CLIC4.
Channel formation by truncated CLIC4
Several lines of evidence support the idea that membrane CLICs have a single TMD. Sequence alignments and hydrophobicity plots (e.g., Citation[2], Citation[11]) strongly predict a single TMD near the N-terminus, following the first cysteine residue in CLIC1 and CLIC4. This predicted TMD is very obviously absent in Ω-GSTs Citation[2], and it contains a tryptophan residue that could promote membrane autoinsertion. The presence of a single TMD is supported by the positioning of the N- and C-termini of CLIC1 on opposite sides of the membrane Citation[32], by protease digestion studies on membrane CLIC4 Citation[9], by truncation, mutagenesis and membrane localisation studies of the worm protein exc-4 Citation[27], Citation[43], and by the presence of potential (and functional) protein interaction sites throughout the “cytosolic” region of, especially, CLIC4 Citation[15]. Based on these findings, we suggest that CLIC4 (and CLIC1) channels are likely to be oligomers comprising at least 4 subunits.
We explored the possibility that a truncated version of CLIC4, comprising the N-terminus and the putative TMD, including a single remaining cysteine residue just before the putative TMD, could form functional ion channels in planar bilayers. Although channel formation proceeded less rapidly, the truncated protein formed ion channels with consistent appearances from experiment to experiment, with a reduced conductance compared to the full-length protein. This reduction is consistent with the idea that the missing parts of the protein form charged channel vestibules that “concentrate” permeant ions near the entrance to the pore Citation[44]. Channels formed from the truncated protein were non-selective, regardless of redox conditions, again consistent with the idea that the missing extramembranous regions of the protein, rather than specific ion binding sites in the pore, confer the (poor) ionic selectivity of full-length CLIC4. Overall, we suggest that under partially- or fully-reducing conditions, despite the absence of much of the protein, including all the potential “cytosolic” domains, the truncated subunits continue to associate (by non-covalent interactions) to form a functional pore.
The truncated protein also formed functional pores under fully-oxidizing conditions, although pore formation occurred much less frequently. Under these conditions, two conductances were recorded, one slightly less than the pores formed under reducing conditions, and another whose conductance was substantially higher. The oxidized proteins presumably insert into bilayers as pre-formed disulphide-linked dimers, and then associate with other similar dimers in the membrane to form functional channels. However, trans-oxidized CLIC4 is normally poorly-conducting (see below), so we speculate that these pores must be assembled from a relatively large number of pre-linked dimers in order to produce a patent pore, compared to fewer pairs of monomers for the reduced protein. Occasional even larger assemblies could explain the infrequent high conductances we observed.
Orientation and redox-regulation of membrane CLIC4
CLIC1 inserted into bilayers with a well-defined orientation, with its N-terminus facing the trans (“extracellular” or “luminal”) side of the membrane Citation[23]. The same topology was expected for CLIC4, especially given that all the protein interaction sites known to be located between the putative TMD and the C-terminus Citation[27] are cystosolic. This orientation places the first cysteine residue in membrane CLIC monomers in the trans chamber, immediately before the predicted TMD. Consistent with this orientation, CLIC4 was sensitive to the trans redox potential in glutathione buffers (as discussed below). However, in contrast to CLIC1, NEM had no effect on CLIC4, suggesting that in this case NEM may not have been able to react covalently with the relevant residue, or the reaction had no functional consequences. In contrast, DTNB inhibited the channel, and it only acted from the trans side. We speculate that the pore-associated cysteines in CLIC4 oligomers, equivalent to the “G-site” cysteine in Ω-GSTs that forms mixed disulphides with GSH Citation[1], can react with DTNB to form mixed disulphides that block the channel pore or interfere with channel opening.
The maximum slope conductance of full-length CLIC4 showed a substantial and reversible dependence on the trans redox potential in glutathione buffers. This is similar to the behaviour of CLIC1, which was analysed in detail Citation[23] and attributed to reversible disulphide bond formation involving pairs of subunits in the presence of GSH and GSSG. The “smooth” reduction in single-channel currents was consistent with rapid reduction and oxidation of the relevant thiol groups in the presence of the glutathione buffer system, at rates too fast to be resolved under the filtering conditions used. The requirement for relatively heavy filtering of CLIC channels in planar bilayers, and their relative long-lived open and closed times, precludes any meaningful channel gating analysis, because recordings would have to be very long in order to collect enough events. Truncated CLIC4 retained its sensitivity to the trans redox potential in glutathione buffers, strongly supporting the idea that the effect is mediated by C35, which is the only cysteine left in the truncated protein.
The sensitivity of the channels to extracellular (or luminal) redox conditions could have significant effects on the amplitude of CLIC4 currents recorded in vitro (e.g., Citation[9]) and in vivo (e.g., Citation[19]), making it difficult to compare recordings unless the trans redox potential is well controlled. In addition, our selectivity data and their interpretation suggest that large cations (e.g., Tris+) may have a dual effect – to reduce single-channel currents (especially with very large cations), and also make the channels more anion-selective. The anion vs. cation selectivities of both CLIC1 and CLIC4 may therefore be “artificially” improved in the presence of relatively large cations like N-methyl-D-glucamine+, often used in patch-clamp experiments to reduce endogenous cation currents.
Conclusion
Our results suggest that mammalian membrane CLIC4 forms poorly-selective, oligomeric ion channels modulated by luminal (or external) GSH-dependent transthiolation. Its redox-sensitivity may shed more light on the role of CLIC4 in apoptosis Citation[20], and at this stage it would be very helpful to establish the stoichiometry of the channel (the tetramer in is entirely speculative), and confirm the topology of its subunits. However, even if CLIC4 monomers contain just a single TMD, other parts of the protein may still contribute in unanticipated ways to the ion channel pore. In common with other transporters, a full understanding of ion permeation through membrane CLICs, and channel regulation by oxidation, awaits detailed structural analysis. As an added complication, it will be especially important to obtain membrane structures relevant to functional channels, with predictable electrophysiological properties, as opposed to the structures of proteins that insert into membranes without forming specific ion channels Citation[23].
This paper was first published online on prEview on 29 September 2006.
HS was supported by a University of Edinburgh College of Medicine and Veterinary Medicine Scholarship, and by the Overseas Research Students Award Scheme.
References
- Cromer BA, Morton CJ, Board PG, Parker MW. From glutathione transferase to pore in a CLIC. Eur Biophys J 2002; 31: 356–364
- Ashley RH. Challenging accepted ion channel biology: p64 and the CLIC family of putative intracellular anion channel proteins (Review). Mol Membr Biol 2003; 20: 1–11
- Landry DW, Akabas MH, Redhead C, Edelman A, Cragoe EJ, Al-Awqati Q. Purification and reconstitution of chloride channels from kidney and trachea. Science 1989; 244: 1469–1472
- Landry D, Sullivan S, Nicolaides M, Redhead C, Edelman A, Field M, Al-Awqati Q, Edwards J. Molecular cloning and characterization of p64, a chloride channel protein from kidney microsomes. J Biol Chem 1993; 268: 14948–14955
- Redhead C, Sullivan SK, Koseki C, Fujiwara K, Edwards JC. Subcellular distribution and targeting of the intracellular chloride channel p64. Mol Biol Cell 1997; 8: 691–704
- Warton K, Tonini R, Fairlie WD, Matthews JM, Valenzuela SM, Qiu MR, Wu WM, Pankhurst S, Bauskin AR, Harrop SJ, Campbell TJ, Curmi PM, Breit SN, Mazzanti M. Recombinant CLIC1 (NCC27) assembles in lipid bilayers via a pH-dependent two-state process to form chloride ion channels with identical characteristics to those observed in Chinese hamster ovary cells expressing CLIC1. J Biol Chem 2002; 277: 26003–26011
- Fernandez-Salas E, Sagar M, Cheng C, Yuspa SH, Weinberg WC. p53 and tumor necrosis factor alpha regulate the expression of a mitochondrial chloride channel protein. J Biol Chem 1999; 274: 36488–36497
- Howell S, Duncan RR, Ashley RH. Identification and characterisation of a homologue of p64 in rat tissues. FEBS Lett 1996; 390: 207–210
- Duncan RR, Westwood PK, Boyd A, Ashley RH. Rat brain p64H1, expression of a new member of the p64 chloride channel protein family in endoplasmic reticulum. J Biol Chem 1997; 272: 23880–23886
- Ashley RH. Activation and conductance properties of ryanodine-sensitive calcium channels from brain microsomal membranes incorporated into planar lipid bilayers. J Membr Biol 1989; 111: 179–189
- Shorning BY, Wilson DB, Meehan RR, Ashley RH. Molecular cloning and developmental expression of two Chloride Intracellular Channel (CLIC) genes in Xenopus laevis. Dev Genes Evol 2003; 213: 514–518
- Littler DR, Assaad NN, Harrop SJ, Brown LJ, Pankhurst GJ, Luciani P, Aguilar MI, Mazzanti M, Berryman MA, Breit SN, Curmi PM. Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4. FEBS J 2005; 272: 4996–5007
- Harrop SJ, DeMaere MZ, Fairlie WD, Reztsova T, Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Jankova L, Warton K, Bauskin AR, Wu WM, Pankhurst S, Campbell TJ, Breit SN, Curmi PM. Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-Å resolution. J Biol Chem 2001; 276: 44993–45000
- Dulhunty A, Gage P, Curtis S, Chelvanayagam G, Board P. The glutathione transferase structural family includes a nuclear chloride channel and a ryanodine receptor/calcium release channel modulator. J Biol Chem 2001; 276: 3319–3323
- Suginta W, Karoulias N, Aitken A, Ashley RH. Chloride intracellular channel protein CLIC4 (p64H1) binds directly to brain dynamin I in a complex containing actin, tubulin and 14-3-3 isoforms. Biochem J 2001; 359: 55–64
- Berryman MA, Goldenring JR. CLIC4 is enriched at cell-cell junctions and colocalizes with AKAP350 at the centrosome and midbody of cultured mammalian cells. Cell Motil Cytoskel 2003; 56: 159–172
- Chuang JZ, Milner TA, Zhu M, Sung CH. A 29 kDa intracellular chloride channel p64H1 is associated with large dense-core vesicles in rat hippocampal neurons. J Neurosci 1999; 19: 2919–2928
- Edwards JC. A novel p64-related Cl− channel: subcellular distribution and nephron segment-specific expression. Am J Physiol 1999; 276: F398–F408
- Proutski I, Karoulias N, Ashley RH. Overexpressed Chloride Intracellular Channel protein CLIC4 (p64H1) is an essential molecular component of novel plasma membrane anion channels. Biochem Biophy Res Comm 2002; 297: 317–322
- Fernandez-Salas E, Suh KS, Speransky VV, Bowers WL, Levy JM, Adams T, Pathak KR, Edwards LE, Hayes DD, Cheng C, Steven AC, Weinberg WC, Yuspa SH. mtCLIC/CLIC4, an organellular chloride channel protein, is increased by DNA damage and participates in the apoptotic response to p53. Mol Cell Biol 2002; 22: 3610–3620
- Qiu, M. Functional and molecular aspects of ion channels in macrophages. PhD Thesis, University of New South Wales, SydneyAustralia; 2003.
- Dulhunty AF, Pouliquin P, Coggan M, Gage PW, Board PG. A recently identified member of the glutathione transferase structural family modifies cardiac RyR2 substate activity, coupled gating and activation by Ca2 + and ATP. Biochem J 2005; 390: 333–343
- Singh H, Ashley RH. Redox regulation of CLIC1 by cysteine residues associated with the putative channel pore. Biophys J 2006; 90: 1628–1638
- Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochem 2000; 39: 890–902
- Findlay HE, McClafferty H, Ashley RH. Surface expression, single-channel analysis and membrane topology of recombinant Chlamydia trachomatis Major Outer Membrane Protein. BMC Microbiol 2005; 5: 5
- Hayman KA, Spurway TS, Ashley RH. Single anion channels reconstituted from cardiac mitoplasts. J Membr Biol 1993; 136: 181–190
- Berry KL, Bulow HE, Hall DH, Hobert O. A C. elegans CLIC-like protein required for intracellular tube formation and maintenance. Science 2003; 302: 2134–2137
- Board PG, Coggan M, Chelvanayagam G, Easteal S, Jermiin LS, Schulte GK, Danley DE, Hoth LR, Griffor MC, Kamath AV, Rosner MH, Chrunyk BA, Perregaux DE, Gabel CA, Geoghegan KF, Pandit J. Identification, characterization, and crystal structure of the omega class glutathione transferases. J Biol Chem 2000; 275: 24798–24806
- Valenzuela SM, Martin DK, Por SB, Robbins JM, Warton K, Bootcov MR, Schofield PR, Campbell TJ, Breit SN. Molecular cloning and expression of a chloride ion channel of cell nuclei. J Biol Chem 1997; 272: 12575–12582
- Tulk BM, Schlesinger PH, Kapadia SA, Edwards JC. CLIC-1 functions as a chloride channel when expressed and purified from bacteria. J Biol Chem 2000; 275: 26986–26993
- Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Warton K, Musgrove EA, Campbell TJ, Breit SN. The nuclear chloride ion channel NCC27 is involved in regulation of the cell cycle. J Physiol 2000; 529: 541–552
- Tonini R, Ferroni A, Valenzuela SM, Warton K, Campbell TJ, Breit SN, Mazzanti M. Functional characterization of the NCC27 nuclear protein in stable transfected CHO-K1 cells. FASEB J 2000; 14: 1171–1178
- Tulk BM, Kapadia S, Edwards JC. CLIC1 inserts from the aqueous phase into phospholipid membranes, where it functions as an anion channel. Am J Physiol 2002; 282: C1103–C1112
- Littler DR, Harrop SJ, Fairlie WD, Brown LJ, Pankhurst GJ, Pankhurst S, DeMaere MZ, Campbell TJ, Bauskin AR, Tonini R, Mazzanti M, Breit SN, Curmi PM. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J Biol Chem 2004; 279: 9298–9305
- Novarino G, Fabrizi C, Tonini R, Denti MA, Malchiodi-Albedi F, Lauro GM, Sacchetti B, Paradisi S, Ferroni A, Curmi PM, Breit SN, Mazzanti M. Involvement of the intracellular ion channel CLIC1 in microglia-mediated beta-amyloid-induced neurotoxicity. J Neurosci 2004; 24: 5322–5330
- Ramachandran R, Tweten RK, Johnson AE. Membrane-dependent conformational changes initiate cholesterol-dependent cytolysin oligomerization and intersubunit beta-strand alignment. Nat Struct Mol Biol 2004; 11: 697–705
- Tweten RK, Parker MW, Johnson AE. The cholesterol-dependent cytolysins. Curr Top Microbiol Immunol 2001; 257: 15–33
- van Meer G. Lipids of the Golgi membrane. Trends Cell Biol 1998; 8: 29–33
- Ho C, Slater SJ, Stagliano B, Stubbs CD. The C1 domain of protein kinase C as a lipid bilayer surface sensing module. Biochem 2001; 40: 10334–10341
- Heiss NS, Poustka A. Genomic structure of a novel chloride channel gene, CLIC2, in Xq28. Genomics 1997; 45: 224–228
- Franciolini F, Nonner W. Anion-cation interactions in the pore of neuronal background chloride channels. J Gen Physiol 1994; 104: 711–723
- Franciolini F, Nonner W. A multi-ion permeation mechanism in neuronal background chloride channels. J Gen Physiol 1994; 104: 725–746
- Berry K, Hobert O. Mapping functional domains of Chloride Intracellular Channel (CLIC) proteins in vivo. J Mol Biol 2006; 359: 1316–1333
- Brelidze TI, Niu X, Magleby KL. A ring of eight conserved negatively charged amino acids doubles the conductance of BK channels and prevents inward rectification. Proc Natl Acad Sci USA 2003; 100: 9017–9022