Abstract
Cranes are monomorphic birds and it is difficult to determinate their sexes. Polymerase chain reaction was conducted to amplify the female-specific segments on chromo-helicase-DNA binding 1 gene, using specially designed primers. The products were cloned into vector pMD-18T, transformed into Escherichia coli DH-5α and sequenced. The sequences have been submitted to National Center for Biotechnology Information (NCBI) (GenBank Accession Number: AY366489, AY366703, AY367004, AY367005, AY377921), and were aligned by Clustal W using DNAstar 2.0 Megalign. This highly conserved region in cranes is 176 bp, consisting of 87 bp intron b and 89 bp exon. This section is female-specific as shown by alignment in GenBank. The pair of primers in our experiment has been proved to be very useful to determine the sex in cranes.
Keywords:
Introduction
The sex identification of birds is of the utmost importance in effective mating and propagation. About 60% of the birds are monomorphic hence genders are hard to be distinguished only by their appearances and behaviours (Bermúdez-Humarán et al. Citation2002). Even in some sexually dimorphic birds, the two genders can only be differentiated on the morphology level only after sexual maturation.
Currently, many approaches have been applied to identify sexes (Khaledi et al. Citation2009), such as visual observation of sexual organ, cloacal examination, chromosome analysis of cells in vitro, the ratio assay between estrogen and androgen in faeces, biochemical examination, spermatogonium detection, etc. However, all of them have been considered to be inefficient, time-consuming, insensitive, harmful or expensive. As a result, it is imperative to develop an accurate, easy and less harmful way to determine sexes. Compared with other sex identification approaches, the advantages of DNA-based methods are early accessibility to samples (feather), reliability, convenience for popularisation and generalisation as the operation is simple, short-cycled and highly-universal. Finally, it is very effective because a large amount of samples can be tested simultaneously (Svensson et al. Citation2008).
Some experts have succeeded in diagnosing sexes of many bird species through molecular DNA probes linked to sex chromosomes. Chromo-helicase-DNA binding 1 (CHD), which regulates global transcriptional activation at the chromatin level, may be involved in the physiological determination of sexes (Ellegren Citation2000; Dawson et al. Citation2001; Lee et al. Citation2010). CHD gene was used to design probes for sex identification (Ramos et al. Citation2009; Lee et al. Citation2010). It encodes a protein involved in the regulation of global activity on the chromatin level in almost all birds. The avian CHD gene has two genomic copies, one of which (termed CHD-W) was located on the W chromosome in all-ratite species investigated (Handley et al. Citation2004) and the other is CHD-Z on the Z chromosome. The homology of different avian CHD-Z and CHD-W is about 90% (Fridolfsson and Ellegren Citation2000). CHD probes that were designed according to the conserved sequence in CHD gene have been successfully used to sex different bird species. Molecular sexing can be conducted in all-ratite birds through Southern blots with CHD probes (Chang et al. Citation2008).
Some laboratories have used specific primers to get the female-specific bands through PCR, and the segments of PCR products have been proved to be different from each other (Ramos et al. Citation2009). Meanwhile, the restriction maps between two sexes were also used to sex birds. Bermúdez-Humarán et al. (Citation2002) have designed a pair of primers to amplify CHD gene of Ara birds and have generated the restriction maps of CHD gene with DNA strider computational program to sex the birds. This experiment applied three pairs of primers designed by us to sex cranes. It has been proved that our own primers are very effective to determine the sexes of cranes.
Materials and methods
Feather follicle samples were collected from Balearica pavonina, Grus grus, Grus japonensis, Grus vipio, Grus leucogeranus, Antropoides virgo and Grus nigricollis in the Nanjing Zoo. Each sample was cut into tiny pieces and added to a 10 ml tube containing 4.5 ml lysis buffer. Equal volume of Tris phenol was added to the sample followed by centrifugation at 5000 rpm for 10 minutes at 4°C. The sample was extracted twice with phenol, followed by one volume of phenol:chloroform and one volume of chloroform: IAA (1:1). DNA precipitation was rinsed once with 1 ml 70% ethanol and dissolved in T10E1 for 16–20 h at 50°C and stored at −20°C. The absorbance (A) was measured by ultraviolet spectrophotometer, the ratio of A260/A280 was used to express the DNA purity, and the mass concentration was calculated according to A value. DNA was successfully collected from all follicle samples.
Genomic DNA was used as the template in PCR reactions in a volume of 10 µl containing 50 ng of genomic DNA, 0.2 mM dNTP, 1 µl 10 × buffer, 2.5 mM MgCl2, 1 U Taq DNA polymerase, 0.5 µM primer 1 (5′-AGA AAA AGA TGG TGT TAG AT-3′) and 0.5 µM primer 2 (5′-CAT AAC TCC TTA CCA CAT AT-3′). PCR reactions were carried out on PTC-100TM Programmable Thermal controller: 94°C for 5 minutes; 35 cycles of 94°C for 1 minutes, 54°C for 1 minutes and 72°C for 2 minutes; 72°C for 10 minutes. PCR products were separated by running on 8% polyacrylamide gels or 2% agarose gel and stained with Ethidium Bromide (EB).
The reported primers (P2: 5′-CTC TGC ATC GCT AAA TCC TTT-3′; P3: 5′-AGA TAT TCT GGA TCT GGA TCT GAT AGT GA-3′, Bermúdez-Humarán et al. Citation2002) were also used to amplify CHD gene. PCR reactions were also carried out on PTC-100TM: 94°C for 4 minutes; 30 cycles of 94°C for 1 minutes, 56°C for 1 minutes and 72°C for 3 minutes; 72°C for 7 minutes. Restriction enzymes DdeI, XhoI and HaeIII were used to cut PCR products. Enzyme digestions were performed in 20 µl reactions according to the suppliers’ specifications, visualised on 8% polyacrylamide gels and stained with AgNO3.
Finally, the reported primers (P4: 5′-CAC CCT GGA TTG GAC AAC CAT CTA TTT C-3′; P5: 5′-CAC TCT TTC CAG GAA ATC AA-3′, Li et al. Citation2001) were used to amplify CHD gene. PCR reactions were carried out on PTC-100TM: 94°C for 5 minutes; 35 cycles of 95°C for 80 seconds, 59°C for 90 seconds, and 72°C for 60 seconds; 72°C for 10 minutes. PCR products were separated by running on 8% polyacrylamide gels and stained with EB.
DNA mixed pool extracted from these 10 females with the 210 bp female-specific band served as positive control, and DNA pool from the 18 individuals without female-specific bands as negative control; then the 10 females were re-amplified with primer 1 and 2, respectively. PCR products from B. pavonina, G. grus, G. leucogeranus, G. vipio and G. japonensis were purified from the gels using a kit (Shanghai Shenggong Co. Ltd). The purified DNA was cloned into the vector pMD-18T and transformed into Escherichia coli DH-5α. Positive clones were sequenced in both directions using an ABI PRISMTM 377. The sequences were submitted to NCBI (GenBank Accession Number: AY366489, AY366703, AY367004, AY367005, AY377921).
The genetic identity and divergence of the sequences of CHD-W gene fragments from B. pavonina, G. grus, G. leucogeranus, G. vipio and G. japonensis were analyzed with DNAstar 2.0 Megalign. The common sequence was used as query sequence to search the database using BLAST in NCBI. Phylogenetic tree of CHD gene of cranes was also drawn with DNAstar 2.0, G. gallus serving as the outgroup.
Female cranes in this sex identification were mated with the male individuals, and their production indices and parameters (including fertilisation rate of eggs, hatching rate, average egg weight, and egg yield) were recorded for biological statistics.
Results and discussion
Results
Twenty eight cranes were sexed by PCR using primer 1 and 2 ( and ). Ten of them are female with the female-specific band. The results of sex identification are shown in .
Figure 1. PCR products amplified with the primers (1 and 2), separated on 8% polyacrylamide gels and stained with EB. *Lane 2, 14: G. leucogeranus; Lane 3, 5, 7, 9, 10, 15, 18, 19: G. japonensis; Lane 4, 17, 20: B. pavonina; Lane 12: G. vipio; Lane 13, 16: G. vipio; Lane 11: G. nigricollis; Lane M: DNA marker (908, 659, 521, 403, 281, 257, 226, 100, 90 bp).
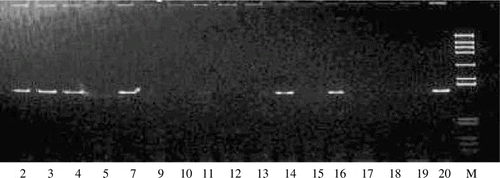
Figure 2. PCR products amplified with the primers (1 and 2), separated on 8% polyacrylamide gels and stained with EB. *Lane 1, 22, 27: G. grus; Lane 6, 21, 28: G. japonensis; Lane 8, 23, 29: G. vipio; Lane 24, 30: Balearica pavonina; Lane M: DNA marker (908 bp, 659 bp, 521 bp, 403 bp, 281 bp, 257 bp, 226 bp, 100 bp, 90 bp).
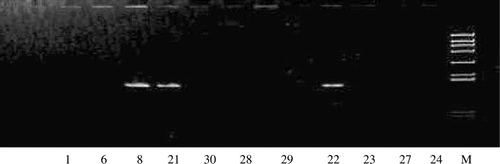
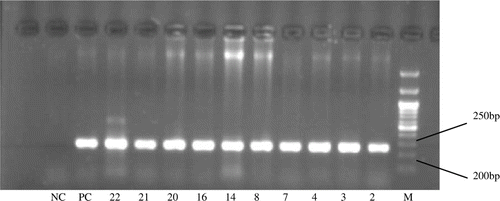
Figure 3. PCR products amplified with the primers (1 and 2), separated on 2% agarose gel and stained with EB. *DNA mixed pool from these 10 females (Sample 2, 3, 4, 7, 14, 16, 20, 8, 21 and 22) with the 210 bp female-specific band served as positive control (Lane PC), and DNA pool from 18 individuals without female-specific bands as negative control (Lane NC). Lane M: DNA marker. The 10 females included: two B. pavonina, one G. grus, three G. japonensis, three G. leucogeranus and one G. vipio.
The reported primers (P2 and P3) for sexing Ara birds could not successfully identify the sexes of cranes (); while (P4 and P5) did not consistently yield female-specific bands ( and ).
Figure 4. PCR products amplified with the reported primers (P2 and P3), separated on 8% polyacrylamide gels and stained with AgNO3. *The length of PCR products was 60 bp in all samples. Products were digested by DdeI, XhoI and HaeIII restriction enzymes, but no reported restriction maps appeared.
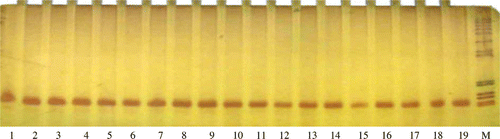
Figure 5. PCR products amplified with the reported primers (P4 and P5), separated on 8% polyacrylamide gels and stained with EB. *Lane M: DNA marker (908 bp, 659 bp, 521 bp, 403 bp, 281 bp, 257 bp, 226 bp, 100 bp, 90 bp).

Figure 6. PCR products amplified with the reported primers (P4 and P5), separated on 8% polyacrylamide gels and stained with EB. *Lane M: DNA marker (908 bp, 659 bp, 521 bp, 403 bp, 281 bp, 257 bp, 226 bp, 100 bp, 90 bp). Only four samples (4, 7, 14, 22) were found with the female-specific PCR bands (about 280 bp); Therefore, this pair of primers lacks consistency.

The five sequences were aligned with DNAstar 2.0 Megalign. The results are shown in . A total of 176 bp common sequence of cranes was used to search the database using BLAST in NCBI. Apparently, the region (89 bp) between 88th bp and 175th bp is an exon of CHD-W gene, and the region (87 bp) between 1st bp and 87th bp is intron b of CHD-W gene via our analysis. Phylogenetic tree of crane CHD gene was drawn in .
Figure 7. Alignment of the five sequences in this study using DNAstar 2.0 Megalign by Clustal W, showing substitutions, transversions and deletion of bases. *G. vipio, G→T on the site of the 11th bp. B. pavonina, C→T on the site of 31st bp and 83rd bp. G. grus, Deletion of C on the site of the 7th bp; A→T on the site of the 59th bp; T→A on the site of the 175th bp. G. japonensis, G→T on the site of the 11th bp; T→C on the site of the 32nd bp. G. leucogeranus, A→T on the site of the 84th bp; A→C on the sites of 86th bp.
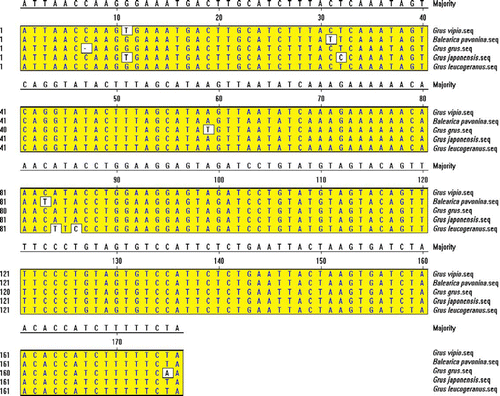
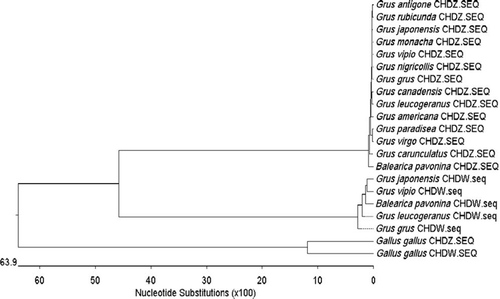
Figure 8. Phylogenetic tree of CHD gene. It is suggested that crane CHD-W and CHD-Z evolve toward two different trends. The percent identity of CHD-Z or CHD-W gene in different kinds of cranes is beyond 97%. CHD gene has a relatively distant phylogenic relationship with the chicken (G. gallus).
The results indicated that all the G. leucogeranus individuals are female, and all the A. virgo and G. nigricollis individuals are male (). These three kinds of identical sex cranes were exchanged with other zoos or introduced new individuals. The final 25 individuals were arranged according to different sex ratios. Their production traits were almost within normal ranges. All the mating females laid fertilised eggs which have been hatched out except A. virgo (maybe due to reproductive barriers).
Table 1. Animal reproduction records.
Discussion
Many ornithologists are eager to find a universal approach to sex their monomorphic birds (Bermúdez-Humarán et al. Citation2002). Various techniques are available, but the ones based on PCR are generally preferred. PCR is fast and only needs tiny DNA from various tissues, such as feathers, blood, etc. (Chang et al. Citation2008; Ramos et al. Citation2009). In this study, the first pair of primers apparently should be the best to identify crane sexes among the three. The male individuals lack this female-specific band because their introns b are different from those of the females (Dawson et al. Citation2001). We have also tried to sex other species such as Ciconia ciconia and Casuarius with it, but failed to acquire presentable results. It is suggested they are merely universal to sex cranes.
There are nine sites mutation in our PCR products (). The high mutation rate is caused by the rapid change of environmental factors in CYT (cytoplasmic) inducible multi-gene family, and it plays an important role in evolution (Duret et al. Citation2006). In addition, the majority of the rapid conversion of intron b of CHD-W tends to be G/C related conversion (5/8), and the increase of GC content is favourable for thermal tolerance of genes. The coding product of CHD is a DNA-binding protein (Lee et al. Citation2010), thus it should be thermal-stable.
reveals the evolutionary trend of crane CHD gene. It can be concluded that animal evolution could be studied by analyzing molecular phylogenetics of CHD genes on the avian sex chromosomes (Fridolfsson and Ellegren Citation2000). In birds, W chromosomes are passed from one generation to the next only in eggs, whereas Z chromosomes are transmitted just in sperm (Handley et al. Citation2004). High mutation of CHD-W gene is needed for the sake of the parallel evolution of both sexes (Ellegren Citation2000; Montell et al. Citation2001). Our research also indicated that CHD-W gene can be served as the marker of molecular phylogenetics due to their high mutation rate.
All in all, the first primers in our research are universal to sex cranes; and the mating and reproducing results afterwards were the same as predicted.
Acknowledgements
This study was supported by Anhui Natural Science Foundation (No KJ2009A039 and 10020303043) and Nanjing Zoo.
References
- Bermúdez-Humarán , LG , García-García , A , Leal-Garza , CH , Riojas-Valdes , VM , Jaramillo-Rangel , G and Montes-de-Oca-Luna , R . 2002 . Molecular sexing of monomorphic endangered Ara birds . Journal of Experimental Zoology , 292 : 677 – 680 .
- Chang , HW , Gu , DL , Su , SH , Chang , CC , Cheng , CA , Huang , HW , Yao , CT , Chou , TC , Chuang , LY and Cheng , CC . 2008 . High-throughput gender identification of Accipitridae eagles with real-time PCR using TaqMan probes . Theriogenology , 70 : 83 – 90 .
- Dawson , DA , Darby , S , Hunter , FM , Krupa , AP , Jones , IL and Burke , T . 2001 . A critique of avian CHD-based molecular sexing protocols illustrated by a Z-chromosome polymorphism detected in auklets . Molecular Ecology , 1 : 201 – 204 .
- Duret , L , Eyre-Walker , A and Galtier , N . 2006 . A new perspective on isochore evolution . Gene , 385 : 71 – 74 .
- Ellegren , H . 2000 . Evolution of the avian sex chromosomes and their role in sex determination . Trends in Ecology & Evolution , 15 : 188 – 192 .
- Fridolfsson , A and Ellegren , H . 2000 . Molecular evolution of the avian CHD1 genes on the Z and W sex chromosomes . Genetics , 155 : 1903 – 1912 .
- Handley , LJL , Ceplitis , H and Ellegren , H . 2004 . Evolutionary strata on the chicken Z chromosome: Implications for sex chromosome evolution . Genetics , 167 ( 1 ) : 367 – 376 .
- Khaledi , KJ , Panandam , JM , Maheran , AA and Siraj , SS . 2009 . Reliable and fast molecular sexing in Bos indicus cattle using amelogenin gene primers . Journal of Applied Animal Research , 35 : 83 – 85 .
- Lee , JC , Tsai , LC , Hwa , PY , Chan , CL , Huang , A , Chin , SC , Wang , LC , Lin , JT , Linacre , A and Hsieh , HM . 2010 . A novel strategy for avian species and gender identification using the CHD gene . Molecular and Cellular Probes , 24 : 27 – 31 .
- Li , M , Ding , CQ , Wei , FW , Meng , SJ , Xi , YM and Lu , BZ . 2001 . Sex-related gene and sex identification of Crested Ibis Nipponia nippon (Ciconiiformes: Threskiornithidae) . Chinese Science Bulletin , 46 ( 8 ) : 669 – 671 .
- Montell , H , Fridolfsson , AK and Ellegren , H . 2001 . Contrasting levels of nucleotide diversity on the avian Z and W sex chromosomes . Molecular Biology and Evolution , 18 ( 11 ) : 2010 – 2016 .
- Ramos , PS , Bastos , E , Mannan , RW and Guedes-Pinto , H . 2009 . Polymerase chain reaction-single strand conformation polymorphism applied to sex identification of Accipiter cooperii . Molecular and Cellular Probes , 23 : 115 – 118 .
- Svensson , EM , Götherström , A and Vretemark , M . 2008 . A DNA test for sex identification in cattle confirms osteometric results . Journal of Archaeological Science , 35 : 942 – 946 .