Abstract
To explore polymorphisms at exon V–VII, encompassing complete TM4 and part of TM3 and TM5 of buffalo SLC11A1 gene, 150 animals belonging to three breeds of buffalo (Bubalus bubalis) viz., Murrah, Surti and Mehsana were genotyped by PCR-RFLP using five restriction enzymes (REs) (Alu I, Taq I, Rsa I, Sac I and Bse GI). PCR–RFLP revealed monomorphic restriction pattern among the three buffalo breeds. Although, sequence comparison revealed no variations at nucleotide level among the buffalo breeds under study, however, two synonymous substitutions at amino acid position 145 (A→C transversion) and 151 (T→C transition) of TM3 were observed compared to cattle sequence (NM-174652). No changes in TM4 and synonymous substitutions at TM3 reflect the conserve nature of this region specially TM4 in buffalo. Future studies may be directed to explore polymorphisms throughout the entire buffalo SLC11A1 gene and to ascertain their suitability as potential genetic marker for resistance against various diseases.
Keywords:
Introduction
The solute carrier family 11A member 1 (SLC11A1) gene (formerly NRAMP1), coding for an iron/divalent cation transporter (Blackwell et al. Citation2003), has been reported to confer resistance or susceptibility to a number of antigenically different intracellular pathogens namely Salmonella typhimurium, Leishmania donovani and Mycobacterium bovis in mice (Vidal et al. Citation1993). A G169D point mutation of SLC11A1 gene located within the fourth transmembrane domain (TM4) is invariably associated with resistance and susceptible phenotypes, respectively. Although, the point mutation (G169D) has never been documented in cattle or other mammalian species, however, several polymorphisms within coding and non coding regions of SLC11A1 gene have been associated with resistance/susceptibility to infectious agents and to autoimmune disorders in human (Blackwell et al. Citation2003; Awomoyi Citation2007), Brucella abortus in water buffalo (Capparelli et al. Citation2007; Ganguly et al. Citation2008), Rhodococcus equi pneumonia in horses (Halbert et al. Citation2006), leishmaniasis in dog (Sanchez-Robert et al. Citation2005) and Salmonella choleraesuis in pig (Tuggle et al. Citation2005).
Considering the immense importance of this gene towards innate resistance against various diseases and the economic importance of buffalo in Indian context, the present study was undertaken with the objective of identifying and comparing polymorphisms in and around TM4 of SLC11A1 gene in different buffalo breeds using PCR–RFLP as well as nucleotide sequencing and to characterise the polymorphic region.
Materials and methods
In the present study a total of 150 animals belonging to three different breeds of riverine buffalo (Bubalus bubalis) viz. Murrah (n=60), Mehsana (n=50) and Surti (n=40) were included. Genomic DNA was isolated from the venous blood using standard phenol chloroform extraction method (Sambrook et al. Citation1989). A fragment of 954 bp, encompassing the complete TM4, was amplified using PCR (). PCR was carried out with a final reaction volume of 50 µl containing 100 ng of template DNA, 1× PCR assay buffer, 2.0 mM of Mg2 +, 200 µM dNTPs, 10 pM of each primer and 1.0U of Taq DNA polymerase. To rule out PCR carryover, a negative control without template (genomic DNA) was always included. After amplification, PCR products and DNA molecular weight ladder were electrophoresed on agarose gels (1%) containing ethidium bromide, in order to confirm the anticipated fragment sizes.
Table 1. Descriptions of the primers, amplified region, product length and RE used to explore polymorphism of Slc11A1 gene in buffalo.
Animals were genotyped by PCR–RFLP using five REs (Alu I, Taq I, Rsa I, Sac I and Bse GI). The restriction digestion was carried out in 25 µl final volume following manufacturer instructions overnight in a water bath at 37°C except Taq I (65°C) and Bse G I (55°C). Briefly, 10U RE was used for every 15 µl PCR product. Restriction patterns were analysed by agarose gel (2%) electrophoresis in 1× Tris-acetate-EDTA (TAE) buffer for 2–3 hours at 5 V/cm and visualised under UV-transilluminator. Corresponding amplicons of representative random samples of each breed were sequenced. Partial exons of submitted sequences were compared with the corresponding region of available cDNA and mRNA sequences of other species. Sequence divergence and phylogenetic tree was constructed.
Results and discussion
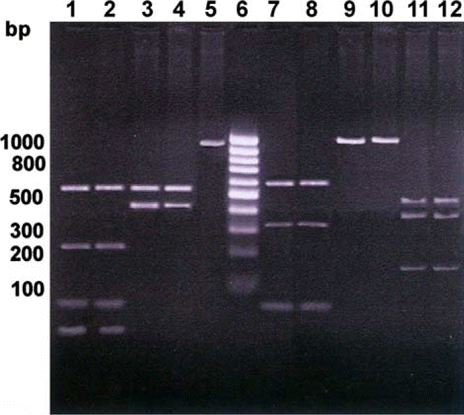
The region was chosen in the present investigation because of the fact that TM4, within exon V–VII, harbour one important mutation (G169D) in murine Nramp 1 protein, which makes individual susceptible towards a number of intracellular pathogens. PCR-generated amplicons were also included part of preceding and succeeding TM domain (). In this study no polymorphism was detected within animals across the buffalo breeds. Single PCR–RFLP pattern was resulted in all the animals with respect to each RE utilised (, ). Fragments <30 bp (in Alu I digestion) could not be resolved properly in agarose gel due to its small size.
Figure 1. PCR–RFLP patterns of buffalo SLC11A1 gene exon V–VII. Lane 1–2: Alu I digestion pattern (541, 255, 79 and 51 bp); Lane 3–4: Taq I digestion pattern (542 and 414 bp); Lane 5: Uncut (954 bp); Lane 6: 100 bp DNA ladder; Lane 7–8: Rsa I digestion pattern (559, 334 and 61 bp); Lane 9–10: Sac I digestion pattern (954 bp) and Lane 11–12: Bse GI digestion pattern (422, 385 and 147 bp).
Several studies have been initiated to explore new DNA polymorphisms of Slc11A1 gene and to ascertain their association with the host resistance/susceptibility against different intracellular pathogens in human (Blackwell et al. Citation2003; Awomoyi Citation2007), cattle (Kumar et al. Citation2005; Martinez et al. Citation2008), buffalo (Capparelli et al. Citation2007; Ganguly et al. Citation2008), horse (Halbert et al. Citation2006) and pig (Tuggle et al. Citation2005). However, lack of polymorphism at TM4 of Slc11A1 gene among Murrah, Surti and Mehsana breeds once again revealed that buffalo is mostly monomorphic in comparison to other species especially to cattle as observed in Kappa casein (Pipalia et al. Citation2001), IGFBP-3 (Kumar et al. Citation2006), DGAT1 and ABCG2 (Tantia et al. Citation2006) and other important gene polymorphism studies. In contrast to the above finding, we observed Alu I and Taq I polymorphisms in intron 6 and intron 5 of SLC11A1 gene and their association with brucellosis resistance in Indian zebu (Bos indicus) and crossbred (Bos indicus X Bos taurus) cattle was studied (Kumar et al. Citation2011).
Sequencing results from all the three buffalo breeds in the present study confirmed the amplicon length of 954 bp. Comparison of nucleotide sequences (AY707989, AY860618 and AY860620) demonstrated no differences among buffalo breeds. These sequences are the first report in Indian Buffaloes. Comparative study of Murrah/Mehsana/Surti SLC11A1 coding sequences with corresponding region of available buffalo (U27105) and cattle (NM-174652) cDNA sequences at nucleotide and amino acid level was carried out (). Two synonymous substitutions at TM3 were observed in all buffalo sequences, one at amino acid position 145 (A→C transversion) and another at amino acid position 151 (T→C transition), respective to available cattle sequence (NM-174652) (). No changes in TM4 and synonymous substitutions at TM3 reflect the conserve nature of this region specially TM4. Synonymous mutations do not alter coding sequences, and therefore they are not expected to change the physical properties, expression and function of the protein concern. However, recently, Nackley et al. (Citation2006) and Capon et al. (Citation2004) have provided evidence that synonymous single-nucleotide polymorphisms (SNPs) can effect protein expression and thereby function by modulating mRNA stability. Moreover, a recent finding claimed that a ‘silent’ polymorphism do change substrate specificity (Kimchi-Sarfaty et al. Citation2007). Therefore, it is difficult at present to rule out any possible role of synonymous substitution at buffalo TM3 and mRNA stability of SLC11A1 gene compare to cattle.
Table 2. Nucleotide changes (with or without amino acid changes) in Buffalo (Bubalus bubalis and Bubalus arnee bubalis) compared to B. taurus sequence (NM_174652).
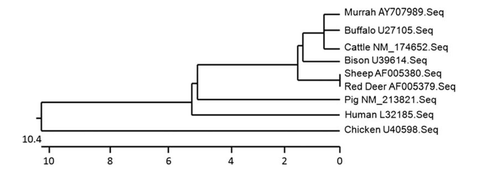
Sequence analysis of all the three buffalo breeds revealed the presence of a Gly at exon VI, which is the same amino acid of the murine NRAMP1-resistant phenotype at amino acid position 169 of TM4. As expected, cattle and buffalo were found to form single monogenic group as compared to other species (). The species next to these two species were sheep, followed by red deer, bison, pig, human and chicken. Partial exon V, complete exon VI and partial exon VII resulted a 64 amino acid sequence. It showed 100% similarity with available buffalo (U27105) and cattle (NM-174652) sequences. The amino acid sequence similarity of sheep, red deer, bison, pig human and chicken with Murrah sequence was found to be approximately 98.4%, 98.4%, 96.9%, 95.3%, 90.6% and 93.8%, respectively.
In conclusion, although analysis of TM4 of SLC11A1 gene by PCR–RFLP and automated DNA sequencing revealed monomorphic pattern and no variation in coding region among the three riverine buffalo breeds, however, two synonymous substitutions at amino acid position 145 (A→C transversion) and 151 (T→C transition) of TM3 were observed in buffaloes as compared to B. taurus (NM_174652).
Acknowledgements
We are grateful to the Director of the Indian Veterinary Research Institute, Izatnagar for providing laboratory facilities. Financial Assistance provided to IG in the form of Senior Research Fellowship (ICAR, New Delhi) is duly acknowledged.
References
- Awomoyi , AA . 2007 . The human solute carrier family 11 member 1 protein (SLC11A1): linking infections, autoimmunity and cancer . FEMS Immunology and Medical Microbiology , 49 : 324 – 329 .
- Blackwell , JM , Searle , S , Mohamed , H and White , JK . 2003 . Divalent cation transport and susceptibility to infectious and autoimmune disease: continuation of the Ity/Lsh/Bcg/Nramp1/Slc11a1 gene story . Immunology Letters , 85 : 197 – 203 .
- Capon , F , Allen , MH , Ameen , M , Burden , AD , Tillman , D , Barkerand , JN and Trembath , RC . 2004 . A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups . Human Molecular Genetics , 13 : 2361 – 2368 .
- Capparelli , R , Alfano , F , Amoroso , MG , Borriello , G , Fenizia , D , Bianco , A , Roperto , S , Roperto , F and Iannelli , D . 2007 . Protective effect of the Nramp1 BB genotype against Brucella abortus in water buffalo (Bubalus bubalis) . Infection and Immunity , 75 : 988 – 996 .
- Coussens , PM , Coussens , MJ , Tooker , BC and Nobis , BC . 2004 . Structure of the bovine natural resistance associated macrophage protein (NRAMP 1) gene and identification of a novel polymorphism . DNA Seq , 15 : 15 – 25 .
- Ganguly , I , Sharma , A , Singh , R , Deb , SM , Singh , DK and Mitra , A . 2008 . Association of microsatellite (GT)n polymorphism at 3′UTR of NRAMP1 with the macrophage function following challenge with Brucella LPS in buffalo (Bubalus bubalis) . Veterinary Microbiology , 129 : 188 – 196 .
- Halbert , ND , Cohen , ND , Slovis , NM , Faircloth , J and Martens , RJ . 2006 . Variations in equid SLC11A1 (NRAMP1) genes and associations with Rhodococcus equi pneumonia in horses . Journal of Veterinary Internal Medicine , 20 : 974 – 979 .
- Kimchi-Sarfaty , C , Oh , JM , Kim , IW , Sauna , ZE , Calcagno , AM , Ambudkar , SV and Gottesman , MM . 2007 . A “silent” polymorphism in the MDR1 gene changes substrate specificity . Science , 315 : 525 – 528 .
- Kumar , N , Ganguly , I , Singh , R , Deb , SM , Kumar , S and Sharma , A . 2011 . DNA Polymorphism in SLC11A1 Gene and its Association with Brucellosis Resistance in Indian zebu (Bos indicus) and Crossbred (Bos indicus X Bos taurus) Cattle . Asian Australasian Journal of Animal Sciences , 24 ( 7 ) : 898 – 904 .
- Kumar , N , Mitra , A , Ganguly , I , Singh , R , Deb , SM , Srivastava , SK and Sharma , A . 2005 . Lack of association of brucellosis resistance with (GT)13 microsatellite allele at 3′UTR of NRAMP1 gene in Indian zebu (Bos indicus) and crossbred (Bos indicus X Bos taurus) cattle . Veterinary Microbiology , 111 : 139 – 143 .
- Kumar , P , Choudharya , V , Kumar , KG , Bhattacharya , TK , Bhushan , B , Sharma , A and Mishra , A . 2006 . Nucleotide sequencing and DNA polymorphism studies on IGFBP-3 gene in sheep and its comparison with cattle and buffalo . Small Ruminant Research , 64 : 285 – 292 .
- Martinez , R , Toro , R , Montoya , F , Burbano , M , Tobón , J , Gallego , J , Dunner , S and Cañón , J . 2008 . Bovine SLC11A1 3' UTR SSCP genotype evaluated by a macrophage in vitro killing assay employing a Brucella abortus strain . Journal of Animal Breeding and Genetics , 125 : 271 – 279 .
- Nackley , AG , Shabalina , SA , Tchivileva , IE , Satterfield , K , Korchynskyi , O , Makarov , SS , Maixner , W and Diatchenko , L . 2006 . Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure . Science , 314 : 1930 – 1933 .
- Pipalia , DL , Ladani , DD , Brahmkshtri , BP , Rank , DN , Joshi , CG , Vataliya , PH and Solanki , JV . 2001 . Kappa-casein genotyping of Indian buffalo breed using PCR-RFLP . Buffalo Journal , 2 : 195 – 202 .
- Sambrook J , Fritsch EF , Maniatis T . 1989 . Molecular cloning: a laboratory manual . Vol 3 . Cold Spring Harbour Library Press , New York , 6.3 – 6.4 .
- Sanchez-Robert , E , Altet , L , Sanchez , A and Francino , O . 2005 . Polymorphism of Slc11a1 (Nramp1) gene and canine leishmaniasis in a case–control study . The Journal of Heredity , 96 : 755 – 758 .
- Tantia , MS , Vijh , RK , Mishra , BP , Mishra , B , Kumar , STB and Sodhi , M . 2006 . DGAT1 and ABCG2 polymorphism in Indian cattle (Bos indicus) and buffalo (Bubalus bubalis) breeds . BMC Veterinary Research , 2 : 32
- Tuggle CK , Marklund L , Stabel TJ , Mellencamp MA , Stumbaugh A . 2005 . Genetic markers for screening animals for improved disease resistance (nramp) . United States Patent, 6844159B2. http://ddr.nal.usda.gov/handle/10113/6983
- Vidal , SM , Malo , D , Vogan , K , Skamene , E and Gros , P . 1993 . Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg . Cell , 73 : 469 – 485 .