Abstract
In this study, a simple PCR-based tetra-primers amplification refractory mutation system (ARMS)-PCR technique has been developed to screen one of the synonymous Single Nucleotide Polymorphism (SNPs) 1551A > G found in buffalo TLR8 gene. The technique was employed to screen 152 genomic DNA samples represented by various riverine and swamp buffaloes. Analysis of data revealed presence of all the three AA, A/G and GG genotypes in Chilika buffalo – whereas only genotype GG was found in all other riverine and swamp buffaloes. The results indicate allele ‘A’ to be Chilika specific and that the technique of tetra-primers ARMS-PCR can be used as a cost-effective, alternate method to differentiate the genotypes.
Introduction
Toll-like receptors (TLRs) belong to a family of transmembrane (TM) proteins playing a critical role in the activation of innate as well as adaptive immune response by differentially recognizing pathogen associated molecular patterns (PAMPs). Mammalian TLRs are composed of an ectodomain consisting mainly of tandem luecine rich repeat motifs, a single TM, and a cytoplasmic region Toll/interleukin-1 receptor domain and a C-terminal tail (Takeda et al. Citation2003; West et al. Citation2006). To date 13 TLRs have been identified in mice and 11 in human (Kawai and Akira Citation2008), whereas, in cattle all the 10 reported TLRs have been mapped and among them TLR8 is reported to be present on chromosome X.
Buffaloes play an important role in agricultural economy of south-east Asia, India in particular, contributing in terms of milk and meat production and also as a draft animal. Buffalo TLR8 Open Reading Frame (ORF) comprises single exon of 3102 nucleotides long, coding for 1033 amino acids, structurally similar to many other livestock TLR8 gene including bovine (GenBank accession no. GQ499854). Due to the central role of TLRs in the innate immune response, genetic variation in this gene family is predicted to alter susceptibility to infections in a PAMP-specific manner based on the ligand recognition of the individual TLR. Variation in the immune response genes of livestock species may also be useful in guiding genetic selection for disease resistance and designing a genotyping protocol for the differentiation of polymorphic nucleotides is required for further investigation of an animal population for which phenotypic data are generated. The workers in the past have used simple techniques like PCR-restriction fragment length polymorphism (RFLP), single strand conformation polymorphism (SSCP), tetra-primers ARMS (amplification refractory mutation system)-PCR, etc. for the genotyping of specific polymorphic nucleotide loci. Among these the tetra-primers ARMS-PCR described by Ye et al. (Citation2001) could be a useful tool for genotyping, since the possibility of getting a restriction site for an enzyme could be rare for genotyping by RFLP and SSCP may not be repeatable some times. In this study, we report an efficient and a cost-effective allele specific tetra-primers ARMS-PCR method for the detection of polymorphism in TLR8 gene of water buffaloes.
Materials and methods
Blood samples were collected from the native breeding tracts of different riverine buffalo breeds (Murrah, Mehsana, Toda, Chilika, Niliravi) and swamp types of Northeast Indian states of Assam, Manipur and Mizoram bordering China and Myanmar. The genomic DNA was isolated using the standard SDS-proteinaseK method (Sambrook and Russell Citation2001) and quality and quantity of DNA were evaluated by agarose gel electrophoresis as well as on Nanodrop ND-1000 spectrophotometer.
For the detection of polymorphism in buffalo TLR8 gene, primers were designed from reported sequence of cattle TLR8 gene (EF076719) to screen the entire coding region. Amplification was carried out in eight overlapping fragments using 20–48 genomic DNA samples of different riverine and swamp buffaloes and amplified products were sent for sequencing to Vimta Labs, Hyderabad, India. The sequence data generated were screened for the presence of polymorphic nucleotide sites using MegAlign programme of Lasergene software (DNASTAR Inc., Madison, WI, USA).
For the genotyping of one of the synonymous nucleotide variations 1551A > G, a tetra-primers ARMS-PCR was developed using four sets of primers () as described by Ye et al. (Citation2001). Primers were designed from the buffalo TLR8 sequence (accession no. GQ499854) using web-based software accessible from the website http://cedar.genetics.soton.ac.uk/public_html/primer1.html. The PCR was performed on 152 riverine and swamp buffalo genomic DNA samples in a 20 µl reaction volume containing about 50 ng of template DNA, 0.5 µl of 10 pmol of each inner primer, 0.1 µl of 10 pmol of each outer primer, 200 µM dNTPs, 2 µL 10× PCR buffer and 1 U of Taq DNA polymerase (Bangalore Genei, India). Amplification was carried out using the PCR conditions 95°C for 2.5 min followed by 32 cycles of 94°C for 30 sec, annealing at 60°C for 30 sec and 72°C for 1 min followed by final extension at 72°C for 5 min. Samples generated were checked on ethidium bromide stained 2% agarose gel and genotypes were recorded.
Table 1. Sequences of tetra-ARMS primers designed for genotyping of TLR8 gene SNP 1551A > G.
Results and discussion
Since the discovery of polymorphism at nucleotide levels, the SNPs in the immune response genes are gaining importance now to find out their association with disease susceptibility. We utilized the simple tetra-primers ARMS-PCR technique described by Ye et al. (Citation2001) for the genotyping of one of the polymorphic loci in buffalo TLR8 gene. In this method, a mismatch is deliberately introduced at the 3′ end of each of the two allele specific primers to increase the specificity of the reaction. The primers can be designed to amplify the fragments of differing sizes corresponding to each allele to enable easy resolution of the allele-specific bands on an agarose gel.
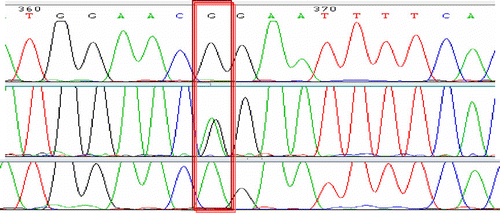
Sequencing of TLR8 coding region of riverine buffaloes represented by different breeds/populations amplified in overlapping fragments helped in identification of a synonymous mutation 1551A > G. This variation was observed only in Chilika buffaloes with one animal having ‘A’ and three having ‘G’ as in other animals. For genotyping by tetra-primers ARMS-PCR, primers were designed to amplify fragments that differ in size sufficiently (190, 233, and 368 bp) to be easily resolved by agarose gel electrophoresis. PCR of 152 riverine and swamp buffaloes showed presence of genotype AA only in one Chilika animal and two Chilika animals being heterozygous AG. Remaining riverine, including 26 of Chilika and also 42 swamp buffaloes, had GG genotype (). After genotyping by tetra-primers ARMS-PCR, Chilika animals representing the three genotypes were also confirmed by sequencing of samples representing different genotypes ().
Figure 1. Tetra-ARMS-PCR to differentiate three genotypes of 1551A > G SNP in Chilika buffalo (A) and fixation of GG genotype in other riverine and swamp buffaloes (B).
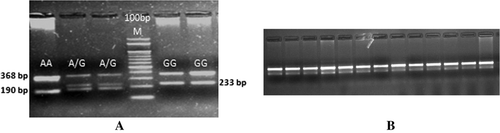
Figure 2. Confirmation of three different genotypes of 1551A > G SNP by direct sequencing of PCR products.
The Chilika buffaloes have unique behaviour of feeding on vegetation in the salty waters of Chilika lake of Orissa state and are reported to have the ability to withstand the marshy environment very well, as they are raised in the open under hot sun and heavy rains throughout the year with no medication (Patro et al. Citation2003). Since the results have indicated 1551A > G variation to be Chilika specific, it can be termed as population private SNP, with minor allele frequency observed being >1%, which makes this variation as true SNP (Brookes Citation1999).
Though the allele and genotype frequencies of this mutation do not show any significant differences among the buffalo breeds screened, nevertheless the technique was found to be a simpler one and also cost-effective that can be easily adopted. Since it is not possible to utilize the costlier techniques like sequencing for the genotyping of SNPs, the simple and cheaper methods of SSCP, PCR-RFLP, etc. have been utilized by many workers to find the allelic variation in TLR genes for different purposes (Zhang et al. Citation2009; Baris et al. Citation2010; Dubey et al. Citation2010). Since many workers have found correlation of polymorphism in TLR8 gene with the susceptibility to various diseases in different species (Davila et al. Citation2008; Mikula et al. Citation2010), this variation needs to be confirmed for such association if any, which further needs the generation of phenotypic data on disease resistance and susceptibility.
Conclusion
The technique of tetra-primers ARMS-PCR described here is simple and cost effective for the genotyping of polymorphic nucleotides. The SNP reported is a novel one identified in buffalo TLR8 gene, which needs to be confirmed for its association with phenotypic character if any.
Acknowledgements
The authors wish to thank National Agricultural Innovation Project of ICAR for financial support to carry out this work under project C2153. Technical assistance received from Mr. Naresh Kumar is also thankfully acknowledged.
References
- Baris , I , Etlik , O , Koksal , V and Arican-Baris , ST . 2010 . Rapid diagnosis of spinal muscular atrophy using tetra-primers-ARMS-PCR assay: simultaneous detection of SMN1 and SMN2 deletion . Molecular and Cellular Probes , 24 : 138 – 141 .
- Brookes , AJ . 1999 . The essence of SNPs . Gene , 234 : 177 – 186 .
- Davila S , Hibberd ML , Dass HR , Wong HEE , Sahiratmadja E . 2008 . Genetic association and expression studies indicate a role of toll-like receptor 8 in pulmonary tuberculosis . PLoS Genetics 4 , e1000218 . doi: 10.1371/journal.pgen.1000218 .
- Dubey , PK , Selvakumar , M , Kathiravan , P , Yadav , N , Mishra , BP and Kataria , RS . 2010 . Detection of polymorphism in exon 2 of Toll-like Receptor 4 gene of Indian buffaloes using PCR–SSCP technique . Journal of Applied Animal Research , 37 : 265 – 268 .
- Kawai , T and Akira , S . 2008 . Toll-like receptor and RIG-I-like receptor signaling . Annals of the New York Academy of Sciences , 1143 : 1 – 20 .
- Mikula , I , Mangesh , B , Silvia , P and Mikula , I . 2010 . Characterization of ovine TLR7 and TLR8 protein coding regions, detection of mutation and Maedi Visna virus infection . Veterinary Immunology and Immunopathology , 138 : 51 – 59 .
- Patro , BN , Mishra , PK and Rao , PK . 2003 . Chilika buffaloes in Orissa: a unique germplasm . Animal Genetics Resources Information , 33 : 73 – 79 .
- Sambrook , J and Russel , DW . 2001 . Molecular cloning; a laboratory manual , 3rd ed , New York : Cold Spring Harbour Laboratory Press .
- Takeda , K , Kaisho , T and Akira , S . 2003 . Toll-like receptors . Annual Review of Immunology , 21 : 335 – 376 .
- West , AP , Koblansky , AA and Ghosh , S . 2006 . Recognition and signaling by Toll-like receptors . Annual Review of Cell and Developmental Biology , 22 : 409 – 437 .
- Ye , S , Dhillon , S , Ke , X , Collins , AR and Day , INM . 2001 . An efficient procedure for genotyping single nucleotide polymorphisms . Nucleic Acids Research , 29 : E-88
- Zhang , LP , Gan , QF , Ma , TH , Li , HD , Wang , XP , Li , JY , Gao , X , Chan , JB , Ren , HY and Xu , SZ . 2009 . Toll-like receptor 2 gene polymorphism and its relationship with SCS in dairy cattle . Animal Biotechnology , 20 : 87 – 95 .