Abstract
The present study was carried out to characterise the serum lysozyme gene in Muzaffarnagri sheep. Total RNA was extracted from the white blood cells and serum lysozyme cDNA was synthesised by Reverse transcription-polymerase chain reaction, then cloned using pGEMT-cloning vector and sequenced. The sequencing of cDNA revealed that the length of serum lysozyme gene was 447 bp with 46% Guanine and Cytosine (GC) content. The comparative sequence analysis of serum lysozyme gene of cattle and buffalo with sheep revealed differences at 24 and 21 places, respectively. The molecular weight of serum lysozyme of sheep is 16.48 kDa. The sheep serum lysozyme cDNA sequence was 94.6% similar to cattle gene sequence, whereas it was 95.3% similar to buffalo gene sequence. Similarly, the derived amino acid sequence of sheep cDNA showed 90.6% similarity with cattle and 92.6% similarity with buffalo. Phylogenetic analysis revealed that sheep and buffalo sequences formed a cluster and evolutionary closer than the cattle sequences.
Keywords:
Introduction
Lysozymes are 1,4,β-N-acetyl muramidase that enzymatically degreades a glycosidic linkage between C-1 of N-acetyl muramic acid and C-4 of N-acetylglucosamine in the bacterial peptidoglycan component of cell wall and acts as an important antimicrobial component in serum and body fluids. Because of its ubiquitous presence lysozyme is considered to be a constituent of primitive unspecific defence mechanism associated with the monocyte-macrophage system (Osserman and Lawlor Citation1966). The role of lysozyme as an antibacterial agent appears to be mediated through its direct bacteriolytic action as well as via stimulatory effect on macrophage phagocytic function. Several studies have provided evidences that lysozyme can be used as index of macrophage functional status (Di Luzio Citation1979). The infection protective function of serum/milk protein-like lysozyme has been established beyond doubt (Jolles and Jolles Citation1984). Seyfert et al. (Citation1996) have suggested that lysozyme gene can be used as candidate gene for improvement of mastitis resistance. This enzyme usually functions in association with lactoferrin or immunogenic A. Lysozyme is effective against Escherichia coli in concert with IgA. It causes lysis of some species of Salmonella in association with ascorbate and peroxide. In addition, it can limit the migration of neutrophils into damaged tissue and functions as anti-inflammatory agent. Hence, considering lysozyme gene as a potential marker for general immune response as well as mastitis resistance, this gene needs to be characterised. Despite its tremendous potential, very little work has been done so far, except scattered information of its recombinant product in cattle and goat (Maga et al. Citation2006) and polymorphism study in cattle (Weikard et al. Citation1996; Pareek et al. Citation2003; Prusinowska et al. Citation2003) and buffalo (Sahoo Citation2007). So far, no report is available on genetic characterisation of serum lysozyme gene in any breed of indigenous sheep. Hence, present investigation was undertaken to clone and characterise the cDNA of serum lysozyme gene in Muzaffarnagri breed of sheep.
Materials and methods
Blood samples were collected from five animals of Muzaffarnagri sheep for synthesis of cDNA of serum lysozyme gene. Total RNA was isolated from the white blood cells following standard protocol (Sambrook and Russell Citation2001). Reverse transcription-polymerase chain reaction (RT–PCR) using oligo dT primer was carried out for the synthesis of first strand of cDNA. For amplification of full length cDNA specific primers were designed on the basis of gene sequences of cattle (Accession No. U25810) available publically, at NCBI. The primers used for amplification were 5′-ATGAAGGCTCTCATTATTCTG-3′ as forward and 5′-TTACACTCCACAACCCTGAA-3′ as reverse primer. PCR reaction is performed in a total volume of 25 µl with 100 ng of template cDNA, 15 pmoles of each primer, 1 mM of MgCl2, 200 µM of each dNTP, 1X PCR reaction buffer and 1.25 U of taq DNA polymerase. Template cDNA was initially denatured for 5 min at 95°C, then 34 cycles of denaturation for 30 s at 95°C, annealing for 60 s at 45°C and extension for 60 s at 72°C followed by final extension for 10 min at 72°C. The amplified product was checked in 1.2% agarose gel along with a standard DNA molecular weight marker (MBI Fermentas).
The amplified products were run in 0.8% agarose gel having long combed well. The product was visualised under transilluminator and the gel having the DNA fragment of interest was cut and DNA was eluted from the gel using Gel Extraction Kit (Qiagen, Germany) following the standard protocol. Thymine and adenine (TA) cloning strategy using pGEMT easy vector system (Qiagen, Germany) was used for cloning of serum lysozyme gene. Purified PCR products were ligated into pGEMT Easy vector system for transformation into E. coli strain DH5∝. The ligation reaction was set at 4°C overnight in 0.2 ml-PCR tube, adding 2X rapid ligation buffer having 1 U T4 DNA ligase, pGEMT easy vestor (50 ng/ml) and purified PCR product. Competent cells were prepared using TransformAid bacterial Transformation Kit (Fermentas) following standard protocol. For transformation, 5 µl of ligation mixture was added into new micro centrifuge tubes and chilled on ice for 2 min. Fifty microlitres of the prepared competent cells were added into each tube containing ligation mixture, mixed and incubated on ice for 5 min. The transformed cells were spreaded on pre-warmed LB agar plate containing ampicillin (100 µg/ml), X-Gal (20 mg/ml) and IPTG (20% w/v). Appropriate positive and negative controls were processed simultaneously. Plates were incubated overnight at 37°C and later stored at 4°C. Recombinant clones were identified by the screening of blue/white colonies. Positive clones were confirmed by colony and plasmid PCR using same pairs of primer.
Recombinant colonies were selected from the master plate and sequencing was performed by the Sanger's dideoxy chain termination sequencing method in an Automatic ABI Prism DNA sequencer. These sequences were compared with available sequences of NCBI Gene Bank (www.ncbi.nlm.nih.gov/BLAST) using Laser gene Software (DNASTAR) and submitted to gene NCBI (Accession No. GQ860955).
Results and discussion
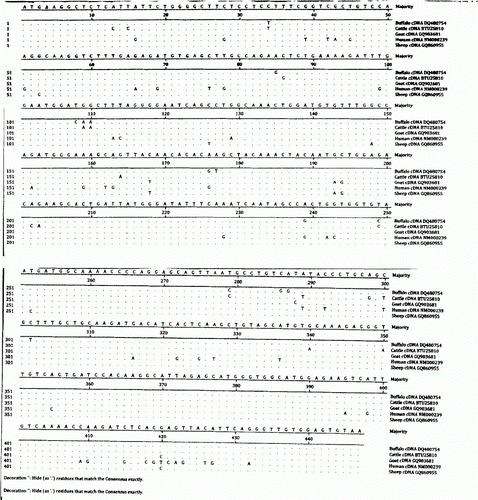
Sequencing of recombinant clones revealed that the size of serum lysozyme gene is 447 bp. The open reading frame (ORF) was of 447 bp bearing usual start codon ATG and stop codon TAA with GC content of 46%. Henke et al. (Citation1996) and Sahoo (Citation2007) also reported that serum lysozyme in cattle and Buffalo is also of 447 bp. The sequence of full length cDNA of sheep serum lysozyme gene was compared with the available sequences of other ruminant species using laser gene software. Alignment study revealed that cDNA length of serum lysozyme gene in all the ruminants and human is of 447 bp (). However, the comparative sequence analysis of serum lysozyme gene of cattle (Bos tarus) and sheep (Ovis aries) revealed difference at 24 places and buffalo (Bubalus bubalis) and sheep (O. aries) at 21 places (). Similarly, the sequence alignment of serum lysozyme gene of sheep and human revealed differences at 50 places ().
Table 1. Comparison of cDNA nucleotide differences among sheep, cattle and buffalo.
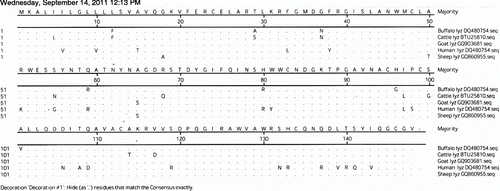
The ORF of cDNA was translated to get the amino acid sequence. The cDNA of lysozyme gene in sheep encodes a precursor polypeptide of 148 amino acids. The derived amino acid sequences of the lysozyme gene of different species were compared () and the differences are presented in . The molecular weight of serum lysozyme of sheep is 16.48 kDa. Priyadarshini and Kansal (Citation2002) also reported the molecular weight of lysozyme in buffalo as 16 kDa. However, the molecular weight of bovine lysozyme was reported as 18 kDa, which is higher than that of sheep (Eitenmiller et al. Citation1975). Mature peptide of serum lysozyme contains 11 strongly acidic residues (D and E) and 14 strongly basic residues (K and R), so as to make the iso-electric point of lysozyme protein in sheep in the basic side that is 8.123.
Table 2. Comparison of the amino acid differences among buffalo, cattle and sheep.
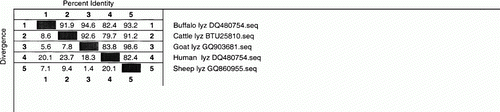
The nucleotide sequence of the cDNA of serum lysozyme gene of sheep was subjected to BLAST analysis (www.ncbi.hlm.nih.gov/BLAST) to retrieve similar sequences of mammalian origin. The nucleotide sequences were subjected to multiple alignments and phylogenetic analysis was performed to know the homology and divergence of sheep with the economically important ruminant species that is cattle and buffalo. The sheep serum lysozyme cDNA sequence was 94.6, 95.3, 98.9 and 87.9% similar to cattle, Buffalo, goat and human gene sequences, respectively (). Similarly, the derived amino acid sequence of sheep cDNA showed 91.2, 93.2, 98.6 and 82.4% similarity with cattle, buffalo, goat and human sequences, respectively (). This indicates that sheep serum lysozyme is more similar to buffalo lysozyme than the cattle lysozyme.
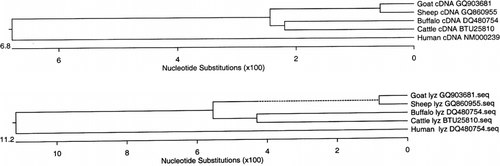
Phylogenetic tree was constructed based on the nucleotide and derived amino acid sequences of cDNA of the lysozyme gene of different species. It was observed that sheep and goat sequences formed one cluster and cattle and buffalo formed another cluster, whereas human sequence diverged earlier than both the clusters (a). Phylogenetic tree construction based on derived amino acid sequences of the lysozyme gene also showed similar evolutionary relationship among different species (b).
References
- Di Luzio , NR . 1979 . “ Lysozyme activity: an index of macrophage functional status ” . In Lysozymes in biology and pathology , Edited by: Dingle , JT , Jacques , PJ and Shaw , JH. Vol. 6 , 447 – 462 . Amsterdam. New York , Oxford : North Holland Publishing Company .
- Eitenmiller , RR , Friend , BA and Sahani , KM . 1975 . Relationship between composition and stability of bovine milk lysozyme . Journal of Dairy Science , 59 : 834 – 839 .
- Henke , M , Hobom , G , Senft , B and Seyfert , HM . 1996 . Structural deviations in a bovine low expression lysozyme-encoding gene active in tissues other than stomach . Gene , 178 ( 1-2 ) : 131 – 137 .
- Jolles , P and Jolles , J . 1984 . What's new in lysozyme research? Always a model system, today as yesterday . Molecular Cell Biochemistry , 63 : 165 – 189 .
- Maga , EA , Shoemaker , CF , Rowe , JD , Bon Durant , RH , Anderson , GB and Murray , JD . 2006 . Production and processing of milk from transgenic goats expressing human lysozyme in the mammary gland . Journal of Dairy Science , 89 : 518 – 524 .
- Osserman , EF and Lawlor , DP . 1966 . Serum and urinary lysozyme in monocyte leukemia . Journal of Experimental Medicine , 124 : 921 – 951 .
- Pareek , CS , Seyfert , HM , Walawski , K , Pareek , RS and Schwerin , M . 2003 . The 5-promoter and coding region of the macrophage expressed lysozyme encoding gene do not reveal variants associated with high serum lytic activity in Polish Black-and-White cattle . Journal of Animal Breeding and Genetics , 120 : 132 – 136 .
- Priyadarshini , S and Kansal , VK . 2002 . Purification, characterization, antibacterial activity and N-terminal sequencing of buffalo-milk lysozyme . Journal of Dairy Research , 69 : 419 – 431 .
- Prusinowska , I , Pareek , CS and Walawski , K . 2003 . Relationship between immunorelevant lysozyme gene polymorphism and differentiation of serum lysozyme activity in cattle . Bulletin of Veterinary Institute Pulawy , 47 : 523 – 531 .
- Sahoo NR. 2007 Molecular characterization of serum lysozyme gene in riverine buffalo [Ph.D thesis] IVRI Deemed University.
- Sambrook , J and Russell , DW . 2001 . Molecular cloning, a laboratory manual , 3rd ed , New York , NY : Cold Spring Harbor Laboratory Press .
- Seyfert , HM , Henke , M , Interhal , H , Klussmann , U , Koczan , D , Natour , S , Pusch , W , Senft , B , Stenhoff , UF Tuckoricz , A . 1996 . Defining candidate genes for mastitis resistance in cattle: the role of lactoferrin and lysozyme . Journal of Animal Breeding and Genetics. , 113 : 169 – 176 .
- Weikard , R , Henke , M , Kuhn , C , Barendse , W and Seyfert , HM . 1996 . A polymorphic microsatellite within the immunorelevant bovine lysozyme-encoding gene . Animal Genetics , 27 : 125