Abstract
The objective of this study was to develop an accurate and reliable method of sex identification in the yak. To achieve this goal, a duplex-nested polymerase chain reaction (PCR) was optimised using DNA from male and female yaks and primers specific for SRY and bHBB gene sequences. Males showed both 1 SRY band and 1 bHBB band, but only 1 bHBB band was present in the agarose gel electrophoresis of females. The result of blind test for confirming the accuracy of the nested PCR showed all sex identifications were in agreement with the actual sexes of the yaks (26/26) from which blood samples were obtained. Thus, this study showed that this nested PCR method can be applied to sex the yak.
Introduction
The yak (Bos grunniens) is very important to people who live on Qinghai-Tibet Plateau for meat, milk and draft power, where few other animals will survive. Milk production benefits from the birth of female offspring. For meat production systems, however, male offspring are generally advantageous because they tend to grow faster and have better feed efficiencies (Wiener et al. Citation2003). Therefore, identifying the embryos is very important for the embryo transfer industry (Zi et al. Citation2008, Citation2009).
Several approaches have been established for sex identification, such as karyotyping (King Citation1984), H-Y antigen detection (Anderson Citation1987), X-linked enzymatic determination (Williams Citation1986) and the polymerase chain reaction (PCR) (Schroeder et al. Citation1990; Fajfar-Whetstone et al. Citation1993; Mara et al. Citation2004; Fu et al. Citation2007) in many livestock species. Among these procedures, the amplification of SRY gene is the most sensitive, accurate, rapid and reliable technique, but such a procedure has not been established in the yak. Therefore, the objective of this study was to develop an accurate and reliable method of sex identification in the yak by simultaneous amplification of the SRY and bHBB genes.
Materials and methods
Blood samples and DNA extraction
Venous blood samples were obtained from 13 male and 13 female adult Maiwa yaks and collected in EDTA Vacutainer® tubes (Becton Dickinson, Plymouth, UK). The animals were grazed on natural grassland in Hongyuan County located in Qinghai-Tibet Plateau, Sichuan Province of China in 2010. Blood samples were transported to laboratory within 10 h under 4°C. Genomic DNA was extracted and purified using TIANamp Blood DNA Kit (Tiangen, Beijing, China). The integrity of the genomic DNA was checked by agarose gel (1%) electrophoresis and visualisation of the gel under UV light after staining with ethidium bromide. The purity of the genomic DNA was checked by means of spectrophotometric readings at OD260 and OD280. Concentration of each DNA solution was adjusted to 15, 10 and 5 ng/µL for PCR purpose.
Primer design
Primers were designed by using Primer Premier 5. The primer was designed according to the reported bHBB sequence (GenBank accession no. M63453) and SRY sequence (GenBank accession no. FJ373272.1) of the yak ().
Table 1. Primer sequences and PCR product size for sex identification in yak.
Optimisation of the duplex-nested PCR procedure
Amplification was performed on an Eppendorf PCR system AG 22331 (PE applied Biosystems, Hamburg, Germany). First amplification was carried out at 15–35 cycles as follows: denaturation at 94°C for 35 s, annealing at 59°C for 35 s and extension at 72°C for 35 s. The PCR was performed in 25 µL of reaction mixture, containing genomic DNA 1 µL, 1.5 µL each outer primer (0.6 µM) of SRY and bHBB, 12.5 µL 2×Taq DNA PCR Mastermix polymerase (Tiangen, Beijing, China). An aliguot (1 µL) of the first PCR product was submitted to 30 cycles in the nest amplification containing 1.5 µL each inner primer (0.6 µM) of SRY and bHBB using the similar other PCR conditions to the first amplification. Before the start of each of the two amplification cycles, the samples were heated to 94°C for 4 min to produce total DNA denaturation. At the end of the cycles, the extension step was prolonged for 8 min to complete the extension of all DNA strands. The PCR procedures were optimised with various concentrations of DNA template (15, 10 and 5 ng/µL) and outer primer (0.2, 0.4, 0.6 and 0.8 µM), and various cycles of first amplification. The blind test for confirming the accuracy of the optimised duplex-nested PCR procedure was carried out using three replicates for each sample of 26 yaks.
Results and discussion
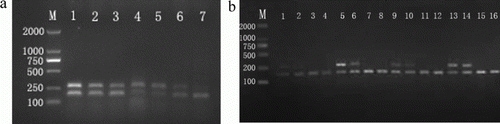
Although amplification of the SRY gene alone has been shown to be sufficient to identify gender in many animal species (Schroeder et al. Citation1990; Fajfar-Whetstone et al. Citation1993; Mara et al. Citation2004; Fu et al. Citation2007), to avoid false-negative result, the internal control gene (bHBB) was simultaneously amplified and the PCR assay was optimised to increase sensitivity and specificity in this study. The PCR using the SRY and bHBB primer sets with genomic DNA yielded the expected products. No non-specific amplification was observed (). Nested PCR of DNA from males displayed two bands (272 and 184 bp), whereas females one band only (184 bp). All samples yielded results consistent with their known sex down to 5 ng genomic DNA per reaction. However, the amount of DNA normally available during embryo sexing is still an order of magnitude lower, so modification of the procedure may be necessary. When the primer concentration was 0.4–0.6 µM, both the SRY and bHBB genes were amplified most clearly (a). With increasing cycle numbers after 20 cycles for the first PCR, no observable product increase was noticed for the nested PCR (b).
Figure 1. Identification of four pair primers. M: DL2000 DNA marker. Lanes 1–4 contain male yak DNA and outer primer (404 bp) or inner primer (272 bp) of SRY gene. Lanes 5–8 contain female yak DNA and outer primer (373 bp) or inner primer (184 bp) of bHBB gene.
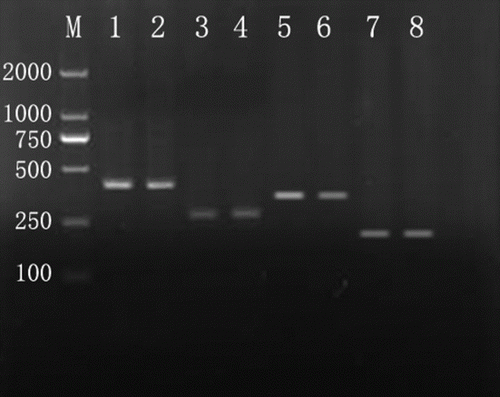
Figure 2. Optimisation of PCR protocol for sex identification in the yak by simultaneous amplification of the SRY (272 bp) and bHBB (184 bp) genes. An aliguot (1 µL) of the first PCR product was submitted to 30 cycles in the nest amplification containing 1.5 µL each inner primer (0.6 µM) of SRY and bHBB. (a) The nested PCR products amplified with various concentrations of male yak DNA and outer primer in the first PCR. 1–3: DNA 15, 10, and 5 ng/µL; 4–7: outer primer 0.2, 0.4, 0.6 and 0.8 µM; M: DL2000 marker. (b) The nested PCR products amplified with various cycles of the first PCR. 1–4: 15 cycles; 5–8: 20 cycles; 9–12: 25 cycles; 13–16: 30 cycles; M: DL2000 DNA marker. Lanes 1, 2, 5, 6, 9, 10, 13, 14 contain male yak DNA; Lanes 3, 4, 7, 8, 11, 12, 15, 16 contain female yak DNA.
The results of blind test for confirming the accuracy of the duplex-nested PCR showed all sex identifications were in agreement with the actual sexes of the yaks (26/26) from which blood samples were obtained. It indicates that the method is 100% reproducible and reliable. This is the first work to identify yak sex by the nested PCR to our knowledge.
In conclusion, the presented nested PCR protocol is reliable, accurate, reproducible and efficient for yak sexing.
Acknowledgements
This research was funded by Science and Technology Bureau of Sichuan Province (No. 07JY029-128), PR China; and Southwest University for Nationalities (2011XWD-S071007).
References
- Anderson , GB . 1987 . Identification of embryonic sex by detection of H-Y antigen . Theriogenology , 27 ( 1 ) : 81 – 97 .
- Fajfar-Whetstone , CJ , Lane Rayburn , IIIA , Schook , LB and Wheeler , MB . 1993 . Sex determination of porcine preimplantation embryos via Y-chromosome specific DNA sequences . Animal Biotechnology , 4 ( 2 ) : 183 – 193 .
- Fu , Q , Zhang , M , Qin , WS , Lu , YQ , Zheng , HY , Meng , B , Lu , SS and Lu , KH . 2007 . Cloning the swamp buffalo SRY gene for embryo sexing with multiplex-nested PCR . Theriogenology , 68 ( 9 ) : 1211 – 1218 .
- King , WA . 1984 . Sexing embryos by cytological method . Theriogenology , 21 ( 1 ) : 7 – 17 .
- Mara , L , Pilichi , S , Sanna , A , Accardo , C , Chessa , B , Chessa , F , Dattena , M , Bomboi , G and Cappai , P . 2004 . Sexing of in vitro-produced ovine embryos by duplex PCR . Molecular Reproduction and Development , 69 ( 1 ) : 35 – 42 .
- Schroeder , A , Miller , JR , Thomsen , PD , Roschlau , K , Avery , B , Poulsen , PH , Schmidt , M and Schwerin , M . 1990 . Sex determination of bovine embryos using the polymerase chain reaction . Animal Biotechnology , 1 ( 2 ) : 121 – 123 .
- Wiener , G , Han , JL and Long , RJ . 2003 . The yak , 2nd ed , Bangkok : The Regional Office for Asia and the Pacific of the Food and Agriculture Organization of the United Nations .
- Williams , TJ . 1986 . A technique for sexing mouse embryos by a visual colorimetric assay of the X-linked enzyme, glucose 6-phosphate dehydrogenase . Theriogenology , 25 ( 5 ) : 733 – 739 .
- Zi , XD , Lu , H , Yin , RH and Chen , SW . 2008 . Development of embryos after in vitro fertilization of bovine oocytes with sperm from either yaks (Bos grunniens) or cattle (Bos taurus) . Animal Reproduction Science , 108 ( 1 ) : 208 – 215 .
- Zi , XD , Yin , RH , Chen , SW , Liang , GN , Zhang , DW and Guo , CH . 2009 . Developmental competence of embryos derived from reciprocal in vitro fertilization between yak (Bos grunniens) and cattle (Bos taurus) . Journal of Reproduction and Development , 55 ( 5 ) : 480 – 483 .