Abstract
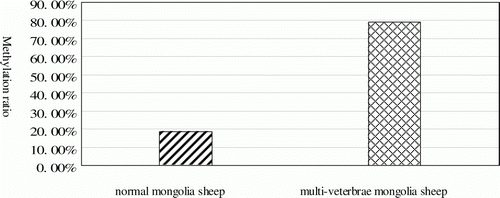
The multi-vertebrae phenotype of the Mongolian sheep is not explained by Mendel's law. To test the hypothesis that the multi-vertebrae phenotype is related to HOXC-8 mutation or genomic imprinting, the sequences of exon-1 and exon-2 of the HOXC-8 gene of normal and multi-vertebrae Mongolian sheep were analysed. Bisulphite sequencing Polymerase Chain Reaction (PCR) was employed to compare the methylation status of cytosine-guanine (CpG) sites in HOXC-8 exon-1 region from normal and multi-vertebrae Mongolian sheep. No mutations were observed in HOXC-8 exon-1 or exon-2 of normal and multi-vertebrae Mongolian sheep. However, the level of DNA methylation in exon-1 of the HOXC-8 gene was much lower in the 13-vertebrae sheep than in the 14-vertebrae sheep. In the CpG island (293 bp), there were six, three and six methylated CpGs in the normal sheep samples and 23, 20 and 21 in the multi-vertebrae sheep samples, respectively. The ratio of methylated to non-methylated CpG dinucleotides in normal and multi-vertebrae sheep were 18.50±0.064 and 79.03±0.056% (P<0.01, by a t-test), respectively.
Introduction
The trait of multi-vertebrae of Mongolian sheep (normal Mongolian sheep have 13 thoracic vertebrae, but the multi-vertebrae Mongolian sheep has 14 thoracic vertebrae) warrants investigation because of the sheep's excellent meat performance. However, there has been no systematic genetic study of Mongolia sheep. The multi-vertebral characteristic of Mongolian sheep is potentially linked to parental imprinting (Zhang et al. Citation2000). DNA methylation, a stable epigenetic change that regulates chromatin structure and gene expression, is involved in processes such as X chromosome inactivation, imprinting, gametogenesis and silencing of repetitive DNA elements (Gopalakrishnan et al. Citation2008). Cytosine–guanine (CpG) methylation has been proposed as a mechanism of gene imprinting (McLachlan et al. Citation2001). CpG islands are often several hundred to 1000 bp in length and are frequently located in the 5′-promoter and exon-1. A previous study reported that extra vertebrae developed in mice in whom the HOXC-8 gene had been knocked out, indicating an involvement of HOXC-8 in the control of the development of thoracic vertebrae (Griffiths et al. Citation1996). To test the hypothesis that the multi-vertebrae phenotype was related to a HOXC-8 mutation or genomic imprinting, HOXC-8 exon-1 and exon-2 sequences of normal and multi-vertebrae Mongolian sheep were analysed. Bisulphite sequencing PCR (BSP) was employed to compare the methylation status of the CpG island in exon-1 of the HOXC-8 gene in normal and multi-vertebrae Mongolia sheep.
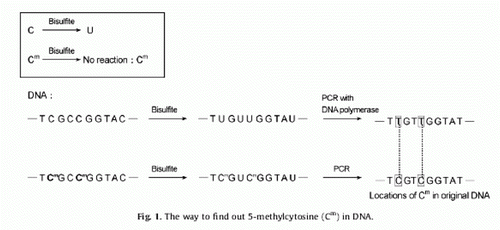
Bisulphite sequencing PCR (Frommer et al. Citation1992) is a powerful tool that has been used to uncover the methylation profile of each CpG within a region of interest and offers a continuous readout of the entire, detailed, base-by-base methylation map of a genomic DNA sequence (Han et al. Citation2006). The principle of BSP is shown in . We analysed the methylation status and number of CpGs in exon-1 of the HOXC-8 gene using BSP to explore the relationship between the multi-vertebrae trait of Mongolia sheep and HOXC-8 gene methylation.
Figure 1. Schematic representation of the bisulphite PCR technique. Cytosines (C) but not methylated cytosines (Cm) are deaminated to U by bisulphite, under appropriate conditions (Hayatsu Citation2008). When clones derived from PCR-amplified bisulphite-treated DNA were sequenced, the only cytosines present in the sequence represent those that were methylated in the original sample: all other cytosines were converted to thymine.
Material and methods
Genomic DNA extraction and bisulphite treatment
Genomic DNA was purified and extracted from the blood of normal and multi-vertebrae Mongolian sheep using a Wizard Genomic DNA purified kit (Promega) and purified.
The bisulphite treatment procedure was based on the method of Olek et al. (Citation1996), with some modifications. Following boiling for 15 min, the DNA sample was denatured with 0.3N NaOH at 50°C for 10 min, and then mixed with low melting point agarose to form beads. The beads then were treated with freshly prepared 5 M bisulphite solution (2.5 M sodium metabisulphite, Merck; 125 mM hydroquinone, Sigma; pH 5) at 50°C for 3.5 h, in the dark. Following desulphonation in 0.2N NaOH, the beads were washed with TE and H2O and stored at –20°C until use.
Bisulphite sequencing PCR
To design post-bisulphite treatment primers for the bisulphite-treated DNA, the sequence of exon-1 of the HOXC-8 gene from Mongolian sheep was electronically bisulphite-converted (converting all cytosines to thymines, but leaving CpG sites as ambiguously cytosine guanine (CG) or thymines guanine (TG)).
Bisulphite sequencing PCR amplification of the CpG-rich islands was performed in a reaction mixture containing 4 µl bisulphite-modified DNA beads, 4 µl dNTPs (2.5 mM each), 5 µl 10× PCR buffer, 3 µl MgCl2 solution, 0.5 µl Taq polymerase (5 U/µl), 1 µl forward and reverse primer (10 µM), respectively, and ddH20 to a total volume of 50 µl. The PCR used the following cycle conditions: 94°C denaturation for 4 min; 35 cycles of 94°C for 20 s, 55°C for 20 s and 72°C for 20 s; and a final extension of 5 min at 72°C. The primer sequences were 5′-AATTTTTTGTTTTTTAAGTATAAAGG-3′ (sense primer) and 5′-TACTAAACCCCATAAAAAAACTATCTA-3′ (antisense primer).
The PCR products were cloned into a pMD18T-simple vector (Takara) and transformed into Escherichia coli. White colonies were picked out and identified by PCR using vector primers. Ten positive clones were sequenced by Invitrogen Corp.
Statistical analysis
All statistical analyses were conducted using SPSS Version 13.0. For comparison between two groups, a t-test was used and a P-value < 0.05 was considered statistically significant.
Results
Identification of CpG Islands
Exon-1 and exon-2 of the HOXC-8 gene were analysed for the presence of a CpG Island using MethPrimer (http://www.urogene.org//methprimer). The following parameters were used: (1) Island size >100 bp; (2) GC percent >50.0%; (3) P(CpG)/(expected) P(CpG) >0.6.
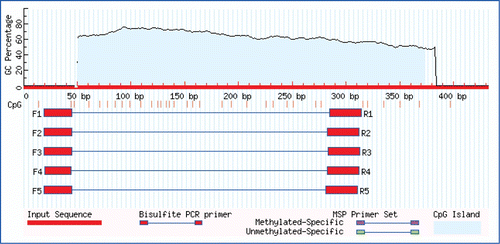
A CpG island (326 bp) was found in exon-1 (); however, no CpG island was found in exon-2. Therefore, only the methylation profile of exon-1 of the HOXC-8 gene was analysed.
Bisulphite sequencing of HOXC-8 exon-1 in Mongolian sheep
To monitor methylation in detail, a 293 bp region within the multi-vertebrae and normal Mongolian sheep Hoxc-8 exon-1 sequence was chosen as the target for methylation analysis (). There were 27 CpG sites (indicated by italics in ) in the target region. Six clones were analysed to ensure that an accurate methylation profile was obtained, because when small amounts of bisulphite-treated DNA were amplified, clones derived from a single PCR may not be fully representative of the original sample.
Figure 3. The schematic diagram of HOXC-8 and the region amplified by Bisulphite-PCR. (A) The schematic diagram of the HOXC-8 gene of Mongolian sheep. (B) A 293 bp region in exon-1 of the HOXC-8 gene amplified by Bisulphite-PCRs. The CpG sites in exon-1 of the HOXC-8 gene of Mongolian sheep (GenBank Accession No. EU817489) are marked in italics and numbered. The primer is underlined.
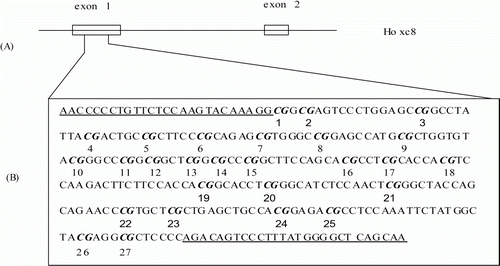
The bisulphite-modified genomic DNA of three multi-vertebrae and three normal Mongolian sheep were subjected to PCR, and the results were analysed by BiQ-Analyzer software (http://biq-analyzer.bioinf.mpi-inf.mpg.de). In the 293 bp region, there were six, three and six methylated Cs in the CpG Island of normal Mongolian sheep and 23, 20 and 21 in multi-vertebrae Mongolian sheep ().
Figure 4. Methylation profiles of CpG dinucleotides of exon-1 of the HOXC-8 gene in normal Mongolian sheep (N) and multi-vertebrae Mongolian sheep (T). Open and closed circles indicate non-methylated and methylated CpG sites, respectively.
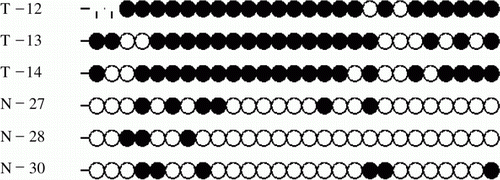
The ratios of methylated to non-methylated CpG dinucleotides of normal and multi-vertebrae Mongolian sheep were 22.2, 11.1 and 22.2% (in the normal sheep samples) and 85.2, 74.1 and 77.8% (in the multi-vertebrae sheep samples). A t-test showed that the means of the two experimental groups were 18.50±0.064 and 79.03±0.056% (P<0.01), respectively (). These results indicated that the methylation level of multi-vertebrae Mongolian sheep in this region is significantly higher than that of normal Mongolian sheep.
Discussion
In a mating experiment, Zhang (Citation2000) found that the heredity of multi-vertebrae characteristics significantly contradicted the classical Mendelian pattern. However, the pattern of inheritance of multi-vertebrae characters was similar to genomic imprinting. Genomic imprinting is an epigenetic mechanism that produces functional differences between the paternal and maternal genomes and plays an essential role in mammalian development and growth. Thus, non-Mendelian characteristics are inherited, for example, by incomplete penetrance or incomplete dominance, and a similar situation may occur in the genetics of the multi-vertebral column in Mongolian sheep. The sequences of exon-1 and exon-2 of the HOXC-8 gene from normal and multi-vertebrae Mongolia sheep were deposited in GenBank (Accession numbers GU479925 and FJ905472). Sequences from both types of sheep were aligned using CLC Free Workbench software, and no differences were observed between them, either in exon-1 or in exon-2 (Chen et al. Citation2011). Thus, the hypothesis that the multi-vertebrae character is related to mutations HOXC-8 mutation was not supported. However, the present study showed that the DNA methylation level in exon-1 of the HOXC-8 gene of the normal Mongolian sheep was much lower than that of the 14-vertebrae Mongolian sheep. Hypermethylation of HOXC-8 exon-1 probably affects gene expression and may be responsible for the increase in the number of thoracic vertebrae. Indeed, a previous study reported that extra vertebrae developed in mice in which the HOXC-8 gene had been knocked out, indicating an involvement of HOXC-8 in the control of the development of thoracic vertebrae (Griffiths et al. Citation1996). The extent of DNA methylation is usually inversely proportional to the amount of gene expression.
BSP is particularly useful when the methylation status of the gene of interest had not been previously characterised, because it provides a good overview of the methylation pattern. In addition, to determining the role of methylation in specific regulatory mechanisms, it is important to know the methylation status of individual CpG dinucleotides. In this study, DNA was embedded in agarose to avoid DNA loss during the incubation steps while maintaining a good bisulphite conversion rate.
In our previous study (Chen et al. Citation2009), the methylated cytosine concentrations of HOXC-8 exon-1 in the multi-vertebrae Mongolian sheep and the normal Mongolian sheep were analysed by methylated DNA immunoprecipitation real-time quantitative PCR technology. The difference between the corrected methylated cytosine concentrations of the samples in multi-vertebrae and normal Mongolia sheep was significant (P<0.05). The results of BSP presented here agree with those of methylated DNA immunoprecipitation real-time quantitative PCR. Further study is required to test the DNA methylation status of the HOXC-8 promoter and exon-1 in a large sample size, and to analyse the gene and protein expressions of HOXC-8 to clarify the genetic mechanism of the multi-vertebrae phenotype.
Acknowledgements
This work was supported by the 11th Five-Year Plan National Science and Technology Support Program of China (No. 2006BDA13B08) and National Natural Science Foundation of China (No. 30960245).
References
- Chen , Q , Zhao , J , Zhang , LL and Ma , YH . 2009 . Methylation analysis to Hoxc-8 exon-1 of multi-vertebrae Mongolia sheep . Chin J Anim Sci. , 45 ( 23 ) : 10 – 14 .
- Chen , Q , Zhao , J , Zhang , LL and Ma , YH . 2011 . Sequence analysis of Hoxc8 exon-1 and exon-2 of multi-vertebrae Mongolia sheep . J Tropical Org. , 2 ( 2 ) : 157 – 163 .
- Frommer , M , McDonald , LE , Millar , DS , Collis , CM , Watt , F , Grigg , GW , Molloy , PL and Paul , CL . 1992 . A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands . Proc Natl Acad Sci. , 89 ( 5 ) : 1827 – 1831 .
- Gopalakrishnan , S , Van Emburgn , BO and Robertson , KD . 2008 . DNA methylation in development and human disease . Mutat Res. , 647 ( 1/2 ) : 30 – 38 .
- Griffiths , AJF , Miller , JH , Suzuki , DT , Lewontin , RC and Gelbart , WM . 1996 . An introduction to genetic analysis , 6th ed , 772 New York , NY : W.H. Freeman and Company Press .
- Han , W , Cauchi , S , Herman , JG and Spivack , SD . 2006 . DNA methylation mapping by tag-modified bisulfite genomic sequencing . Anal Biochem. , 355 ( 1 ) : 50 – 61 .
- Hayatsu , H . 2008 . The bisulfite genomic sequencing used in the analysis of epigenetic states, a technique in the emerging environmental genotoxicology research . Mutat Res. , 659 : 77 – 82 .
- McLachlan , JA , Burow , M , Chiang , TC and Li , SF . 2001 . Gene imprinting in developmental toxicology: a possible interface between physiology and pathology . Toxicol Lett. , 120 : 161 – 164 .
- Olek , A , Oswald , J and Walter , J . 1996 . A modified and improved method for bisulphite based cytosine methylation analysis . Nucleic Acids Res. , 24 ( 24 ) : 5064 – 5066 .
- Zhang LL. 2000 . Studies on developmental and population genetics in Mongolia sheep [Doctoral thesis] . [Zhang, Y.] . Beijing : China Agricultural University . 36 – 46
- Zhang , LL , Ju , LH and Yang , LJ . 2000 . Researches on characters of parental imprinting genetics of vertebra number in Mongolia sheep . J Inner Mongolia Inst Agric Anim Husbandry. , 21 ( 2 ) : 1 – 6 .