Abstract
The objective of this study was to reveal the sequence characteristic and expression pattern of lipoprotein lipase (LPL) gene in high altitude yaks (Bos grunniens). A full-length cDNA of LPL was cloned from yak liver by RT-PCR. The cDNA obtained was 1591 nucleotide (nt) long with a 1437 nt open reading frame encoding 478 amino acids. Yak LPL shares 99.5% nucleotide sequence similarity and 100% amino acid sequence similarity with cattle. Quantitative real-time PCR analysis showed that yak heart and adipose tissue contained significantly higher LPL mRNA level than other tissues assayed. Yak calves contained lower level of LPL mRNA in longissimus dorsi than adult yaks (p < 0.01). Besides, LPL mRNA level in longissimus dorsi was markedly lower in adult yaks that in adult cattle living at low altitude region.
Introduction
Lipoprotein lipase (LPL, EC 3.1.1.34) is the rate-limiting enzyme involved in the clearance of triglyceride-rich chylomicrons and very-low-density lipoprotein from the blood, providing free fatty acids to adipose tissue for storage and to skeletal muscle tissue for oxidation (Brown and Rader Citation2007). LPL is highly expressed in adipose tissues, cardiac and skeletal muscle, kidney and mammary glands while lower levels are observed in liver and brain (Zhang et al. Citation2001). It has been reported that LPL activity in tissues is affected by many factors, such as weaning, nutrient states, hormonal levels and cold acclimation (Bégin-Heick and Heick Citation1977; Quig et al. Citation1983; Richelsen Citation1999; Hocquette et al. Citation2001). Due to the significance of LPL in fatty acid utilisation, tissue LPL activity may have important effects on energy metabolism and repartition of adipose tissue, and this may affect intramuscular fat content and meat sensory quality in meat animals. Saez et al. (Citation2009) reported that LPL protein content in pectorails major muscle was higher in duck breeds which had higher fat content.
Yak (Bos grunniens) inhabits steppes of the Himalayan highlands and is the sole cattle species adapted to the hypoxic environment of the Qinghai-Tibetan plateau. It was reported that permanent exposure or acclimation to severe hypoxia decreased intramyocellular lipid stores (for review see Hoppeler et al. Citation2003). Yaks have lower intramuscular fat content than cattle (Cai and Wiener Citation1995). Because of the high similarity (98–99%) in many protein sequences assayed between yak and cattle (Zheng et al. Citation2008), yak is considered a suitable animal model to study the influences of hypoxia on production performance. Considering the functions of LPL in lipid metabolism and adipose tissue repartition, we thus cloned yak LPL gene and assayed the expression profiles of LPL mRNA and compared with cattle, in order to highlight the molecular basis of meat quality in yaks.
Materials and methods
Experimental yaks and sampling
The experimental Jiulong yaks, a yak breed in China, were reared at Jiulong County of Sichuan Province, including male yaks at ages of 0.5 yr (n=5), 3.5–5.5 yr (n=9) and 9.5–11.5 yr (n=4). The experimental yaks grazed on the same natural pasture at an altitude of 3500 m without feed supplementation. Within 30 min after slaughter, longissimus muscle sample was taken from each yak at the position between the last thoracic spine and the third lumbar spine of right carcass. Heart, liver, kidney and adipose tissues were also taken from each yak. Longissimus muscles of Chinese Yellow cattle (4.8±0.9 yr, n=8) living at low altitude were taken for comparison. All of the samples were promptly frozen and stored at –80°C until analysis. This experiment was conducted according to the guidelines of Chinese government for the use of experimental animals including animal welfare and conditions.
cDNA cloning of yak LPL gene
The PCR primers were designed based on bovine LPL mRNA sequence in GenBank (NM_001075120.1) as follows: F: 5′-CTTGGGTTCAGCGGGTCT-3′, R: 5′ –AGCTGGGGTTAATACTCCGG-3′. Total RNA was extracted from yak liver using TRIzol Reagent (Invitrogen, New Zealand). cDNA was synthesised by reverse transcription from qualified total RNA as described in the manufacturer's protocol (Fermentas Life Science, ON, Canada). The above primers were used to amplify the entire open reading frame of yak LPL cDNA. The PCR conditions were as follows: 95°C, 4 min, then 38 cycles of 35 s at 95°C, 35 s at 54°C, 1.5 min at 72°C; 7 min at 72°C. The PCR product was purified and cloned into pMD 18-T Vector [TaKaRa Biotechnology (Dalian) Co., Ltd.]. Three positive clones were sequenced from both strands.
Analysis of LPL mRNA level in tissues of yak and cattle
Quantitative real-time PCR assay was developed for the quantitation of LPL mRNA level using β-actin as internal control. Total RNA and cDNA were prepared from tissues of yaks (n=18) and longissimus dorsi of Chinese Yellow cattle (n=8) as mentioned above. The primers were designed according to LPL mRNA sequences of yak (HM627756) and cattle (NM_001075120). LPL-F: 5′ TTATGAACTGGATGGCGGATGA 3′, LPL-R: 5′ TGGTTGGAAAGTGCCTCCGTTA 3′, fragment size 311bp; The primers for β-actin-F: 5′ CACAGCCGAGCGGGAAAT 3′, β-actin-R: 5′ CCGTGTTGGCGTAGAGGT 3′, fragment size 287bp. The amplification mixture contained 1 µL of RT reaction mix, 10 µL of SYBR® Premix Ex Taq TM (2×) (TaKaRa Biotechnology (Dalian) Co., Ltd.), 0.5 µL of 10 µmol/L each of primers and add ddH2O to 20 µL. mRNA and ddH2O were used as templates to determine the specificity of quantitative real-time PCR. Reactions were run on a thermo cycler iQ5 (Bio-Rad Laboratories), The PCR conditions were as follows: one cycle of 1 min at 95°C; 45 cycles of 30 s at 95°C, 30 s at 55°C, 30 s at 72°C. Each sample was run in duplicate. The threshold cycle was analysed using the 2–ΔΔCt method (Livak and Schmittgen Citation2001).
Statistical analysis
Data were analysed using Statistical Package for the Social Science (SPSS 17.0). Values were expressed as Mean±SE. The developmental pattern difference of LPL was assayed by one-way ANOVA, and the significance of LPL expression between yak and cattle was evaluated using independent-sample t-test and significance level was set at p<0.05.
Results and discussion
Yak LPL cDNA sequence
A 1591bp fragment was obtained by RT-PCR from yak liver. The CDS of this sequence (HM627756) is 1437bp in length (), with seven nucleotide differences compared to bovine LPL (NM_001075120) and sharing 99.5% similarity. All of the differences belong to T-C or A-G transition. This sequence encodes 478 amino acids fully identical to that of cattle. This result is consistent with many genes assayed in yaks (Zheng et al. Citation2008), and also provides evidence for the close phylogenetic relationship between yak and cattle.
Tissue expression profiles of LPL gene in yak
In adult yaks (3.5–5.5 yr; n=6), the highest level of LPL mRNA was observed in heart among tissues examined (p < 0.01), and adipose tissue also contained high level of LPL mRNA (). Longissimus dorsi of 0.5 yr yaks contained significantly lower level of LPL mRNA than those of 3.5–5.5 yr and 9.5–11.5 yr yaks ().
Figure 2. Relative abundance of LPL mRNA in tissues of adult yaks. Different capitalised and small letters indicate p < 0.01 and p < 0.05, respectively. Adult yaks (3.5–5.5 yr; n=6) were used for the measurement.
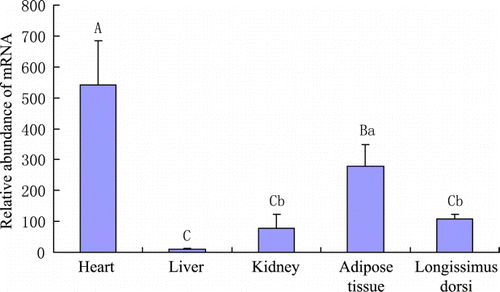
Figure 3. Relative abundance of LPL mRNA in longissimus muscles of yaks at different ages. Different capitalised letters indicate p < 0.01.
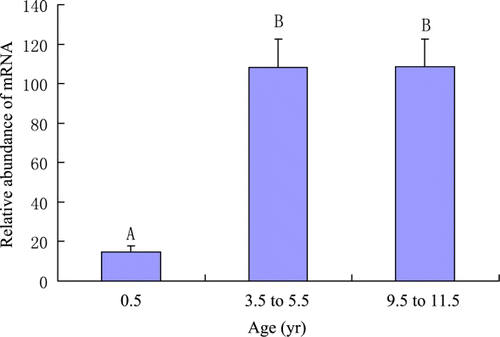
The expression pattern of LPL in yak tissues is generally similar to those in other animals (Hocquette et al. Citation1998; Zhang et al. Citation2001). It is discovered that the levels of LPL transcripts are positively related to LPL activity in bovine muscles and adipose tissues (Hocquette et al. Citation1998). Thus we suggest that the mRNA level of LPL may represent the actual activity of LPL in yak tissues. LPL activity in tissues is affected by many factors (Hocquette et al. Citation2001). Ruge et al. (Citation2004) reported that in some species LPL is subject to nutritional regulation by a posttranscriptional mechanism. The expression profile of LPL mRNA in longissimus dorsi of yaks at different ages is in a similar pattern as peroxisome proliferator-activated receptor gamma (PPARγ, our unpublished data). LPL is a downstream gene of PPARγ. The PPARγ/RXR complex would bind to the promoter region of LPL gene and increases its expression (Kota et al. Citation2004). The induction of LPL by PPARγ occurs mainly in the mature adipocytes in order to increase local free fatty acids (Rangwala and Lazar Citation2004).
LPL mRNA abundance comparison between yak and bovine muscles
Comparison of LPL mRNA abundance in longissimus muscles showed that adult yaks (3.5–5.5 yr) had significantly lower level of LPL mRNA compared to adult Chinese Yellow cattle (4.8±0.9 yr) as shown in . This is consistent with the lower intramuscular fat content in yaks, which might be related to hypoxia adaptation (Hoppeler et al. Citation2003). LPL is involved in the supply of free fatty acids to adipose tissue for storage and to skeletal muscle tissue for oxidation (Brown and Rader Citation2007). There is report that LPL protein is positively correlated with muscle fat deposition (Saez et al. Citation2009). Thereby we suppose that the lower LPL mRNA level of yak muscle compared to cattle might be responsible for yak's lower intramuscular fat content, which is a characteristic of acclimation to severe hypoxia.
Figure 4. Relative abundance of LPL mRNA in longissimus muscles of adult yaks and cattle. **p < 0.01.
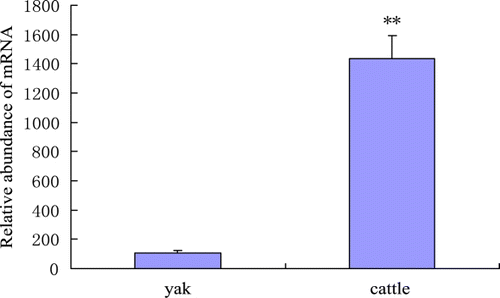
The oxidative metabolism decreases in the rank order of fibre type I, IIa, IIx, IIb (Lefaucheur et al. Citation2002; Lefaucheur et al. Citation2004; Moreno-Sánchez et al. Citation2008). In a previous study we found that myosin heavy chain I (MyHC I) in longissimus dorsi were significant lower (p < 0.01) in yak compared with Chinese Yellow cattle (Lin et al. Citation2011), suggesting that yak longissimus muscle is less oxidative than that of cattle. It was reported that LPL activity was higher in bovine heart and oxidative muscles than in mixed or glycolytic muscles (Hocquette et al. Citation2001). Thus the lower level of LPL, less oxidative muscle fibre type and lower intramuscular fat content of yak longissimus muscle relative to those of cattle seem to have cause-to-effect relationship, and are possibly important characteristics of yak muscle. Based on these comparisons, we suggest that the relatively lower expression of LPL mRNA in longissimus dorsi of adult yaks compared to cattle might contribute to the fat profile of yak muscle and LPL is a potential candidate gene for meat traits.
Acknowledgements
This work was supported by the National Basic Research Program of China (2007CB116204) and Animal Science Discipline Program of Southwest University for Nationalities (No. 2011XWD-S0905).
References
- Bégin-Heick , N and Heick , HM . 1977 . Increased lipoprotein lipase activity of skeletal muscle in cold-acclimated rats . Canadian Journal of Biochemistry , 55 ( 12 ) : 1241 – 1243 . doi: 10.1139/o77-186
- Brown , RJ and Rader , DJ . 2007 . Lipases as modulators of atherosclerosis in murine models . Current Drug Targets , 8 ( 12 ) : 1307 – 1319 . doi: 10.2174/138945007783220614
- Cai , L and Wiener , G . 1995 . The yak , 99 – 101 . Bangkok : The regional office for Asia and the Pacific of the food and agriculture organization of the United Nations .
- Hocquette , JF , Graulet , B and Olivecrona , T . 1998 . Lipoprotein lipase activity and mRNA levels in bovine tissues. Comparative Biochemistry and Physiology . Biochemistry and Molecular Biology , 121 ( 2 ) : 201 – 212 . doi: 10.1016/S0305-0491(98)10090-1
- Hocquette , JF , Graulet , B , Vermorel , M and Bauchart , D . 2001 . Weaning affects lipoprotein lipase activity and gene expression in adipose tissues and in masseter but not in other muscles of the calf . The British Journal of Nutrtion , 86 ( 4 ) : 433 – 441 . doi: 10.1079/BJN2001432
- Hoppeler , H , Vogt , M , Weibel , ER and Flück , M . 2003 . Response of skeletal muscle mitochondria to hypoxia . Experimental Physiology , 88 : 109 – 119 . doi: 10.1113/eph8802513
- Kota , BP , Huang , TH and Roufogalis , BD . 2004 . An overview on biological mechanisms of PPARs . Pharmacological Research , 51 ( 2 ) : 85 – 94 . doi: 10.1016/j.phrs.2004.07.012
- Lefaucheur , L , Milan , D , Ecolan , P and Le Callennec , C . 2004 . Myosin heavy chain composition of different skeletal muscles in Large White and Meishan pigs . Journal of Animal Science , 82 : 1931 – 1941 .
- Lefaucheur , L , Patrick , E , Plantard , L and Gueguen , N . 2002 . New insights into muscle fiber types in the pig . The Journal of Histochemistry and Cytochemistry , 50 : 719 – 730 . doi: 10.1177/002215540205000513
- Livak , KJ and Schmittgen , TD . 2001 . Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method . Methods , 25 : 402 – 408 . doi: 10.1006/meth.2001.1262
- Lin , YQ , Wang , GS , Feng , J , Huang , JQ , Xu , YO , Jin , SY , Li , YP , Jiang , ZR and Zheng , YC . 2011 . Comparison of enzyme activities and gene expression profiling between yak and bovine skeletal muscles . Livestock Science , 135 : 93 – 97 . doi: 10.1016/j.livsci.2010.06.134
- Moreno-Sánchez , N , Díaz , C , Carabaño , MJ , Rueda , J and Rivero , J-LL . 2008 . A comprehensive characterization of the fibre composition and properties of a limb (Flexor digitorum superficialis, membri thoraci) and a trunk (Psoas major) muscle in cattle . BMC Cell Biology , 9 : 67 – 81 . doi: 10.1186/1471-2121-9-67
- Quig , DW , Layman , DK , Bechtel , PJ and Hackler , LR . 1983 . The influence of starvation and refeeding on the lipoprotein lipase activity of skeletal muscle and adipose tissue of lean and obese Zucker rats . The Journal of Nutrition , 113 : 1150 – 1156 .
- Rangwala , SM and Lazar , MA . 2004 . Peroxisome proliferator-activated receptor ( in diabetes and metabolism . Trends in Pharmacological Sciences , 25 ( 6 ) : 331 – 336 . doi: 10.1016/j.tips.2004.03.012
- Richelsen , B . 1999 . Effect of growth hormone on adipose tissue and skeletal muscle lipoprotein lipase activity in humans . Journal of Endocrinological Investigation , 22 ( 5 Suppl ) : S10 – S15 .
- Ruge , T , Neuger , L , Sukonina , V , Wu , G , Barath , S , Gupta , J , Frankel , B , Christophersen , B , Nordstoga , K Olivecrona , T . 2004 . Lipoprotein lipase in the kidney: activity varies widely among animal species. American Journal of Physiology . Renal Physiology , 287 : F1131 – F1139 . doi: 10.1152/ajprenal.00089.2004
- Saez , G , Davail , S , Gentès , G , Hocquette , JF , Jourdan , T , Degrace , P and Baéza , E . 2009 . Gene expression and protein content in relation to intramuscular fat content in Muscovy and Pekin ducks . Poultry Science , 88 ( 11 ) : 2382 – 2391 . doi: 10.3382/ps.2009-00208
- Zhang , Y , Repa , JY , Gauthier , K and Mangelsdorf , DJ . 2001 . Regulation of lipoprotein lipase by the oxysterol receptors, LXRα and LXRβ . The Journal of Biological Chemistry , 276 ( 46 ) : 43018 – 43024 . doi: 10.1074/jbc.M107823200
- Zheng , YC , Si , XH , He , QH , Jin , SY and Hong , J . 2008 . Yak lactate dehydgogenase A: purification, properties, and cDNA cloning and sequencing . Bioscience, Biotechnology, and Biochemistry , 72 : 2448 – 2451 . doi: 10.1271/bbb.80208

