Abstract
Calpastatin is special inhibitor of calpain that influence rate of muscle extension. It is a candidate marker for carcass and growth traits in livestock. In this study, a fragment of sheep calpastatin gene was analysed to investigate if mutations in this sequence might be responsible for carcass and growth traits. Carcass traits included fat and longissimus muscle (LM) thickness pre and post finishing and LM area after finishing. Growth traits included birth and weaning weight, 6 and 9 months weight, pre and post finishing weight and weight gain during different age intervals and finishing. Calpastatin polymorphism in Afshari sheep breed from Zanjan province in Iran was evaluated with RFLP method. The present study was conducted to evaluate the Calpastatin polymorphism of one Iranian sheep breed (Afshari), using RFLP method. The out coming result revealed 74% and 26% frequency for M and N alleles, respectively, which differ by having a number of restriction sites present in one allele but not in the other. Association of the polymorphisms with growth traits, carcass traits, and polymorphisms were analysed using a general linear model procedure. No significant associations were observed between these polymorphisms and growth and carcass traits.
Introduction
In some nutrient studies, were evaluated the influence of enzyme supplementation on growth performance and carcass characteristics (Ríos Rincón et al. Citation2010; Mota et al. Citation2011).
In our study we focused on Growth and carcass traits, which are under the control of multiple genes, are economically important traits in livestock. Selection of animals with higher growth rate and better carcass composition is of great significance to breeders and consumers. Current technologies enable scientists to improve the accuracy and efficiency of traditional selection methods by applying genetic markers through marker-assisted selection. Therefore, genetic polymorphisms that are significantly associated with certain traits of interest are very useful (Zhao et al. Citation2004).
Rate of muscle protein degradation play a very important role to rate and extend muscle mass. Differences in rate of muscle growth in domestic animals often are due to differences in rate of muscle protein degradation with little or no change in rate of protein synthesis. It was originally proposed that the calpain system was initiating metabolic turnover of the myofibrillar proteins and that it affected muscle protein degradation (Goll et al. Citation1998).
The calpain system presently is known to be constituted of three well-characterised proteins include two Ca2 +-dependent proteolytic enzymes, µ-calpain and m-calpain. Calpastatin is third member of calpain family, a multiheaded inhibitor capable of inhibiting more than one calpain molecule. Calpastatin is variable component of the calpain system and skeletal muscle calpastatin activity is highly related to rate of muscle protein turnover and post-mortem tenderisation. All these factors are main concerns for livestock industries to improve rate of growth and meat quality (Goll et al. Citation2003; Amanda et al. Citation2004).
Differences in calpastatin genotypes were found for growth traits in Angus bulls (Chung and Davis Citation2001). A significant difference was found between calpastatin genotype and meat tenderness in beef cattle (Casas et al. Citation2006; Schenkel et al. Citation2006). In transgenic mice over expressing calpastatin, autolysis of µ-calpain was slower than normal mice, and there was no obvious effect of the transgenic on mouse size or weight (Kent et al. Citation2004).
Afshari sheep contained one of the largest populations within Iranian sheep breeds that provide the major part of meet requirement. The current study was designed to screen calpastatin polymorphism and to analyse the association of these polymorphism with growth and carcass traits in Afshari lambs.
Materials and method
Animals and sampling
Fifty-one Afshari male lambs were selected from sheep flock in education farm of Department of animal Sciences, Zanjan University, Iran.
Ration formulated according to NRC (Citation1985). Lambs fed ad libitum access to (Alfalfa hay), corn silage and 1 Kg/day/lamb concentrate (Barely 64% DM, Cotton seed meal 20% DM, Wheat bran 14% DM, Vitamin and Mineral premix 1% DM, Sodium Bicarbonate 1% DM). Concentrate nutrient contains CP 15.41% DM, NDF 15% DM and ME 2.97 MCal/KgDM of concentrate. Salt and fresh water was available all time.
Ultrasonography measurement
Ultrasonography measurement was taken using a sunovet 600 real time mode scanner with a 6 MHz transducer head. In this study, ultrasonography measurement of subcutaneous fat over the ongissimus muscle (LM) at the C site (45 mm from the midline between 12th and 13th rib), LM muscle depth, area and live weight were recorded pre and post finishing lambs. For increase the accuracy and rate of measurement, the ultrasonography machine was connected to DVD recorder and using with media player and scion image (version 4.0.3.2) soft wares to take suitable photo and measurement particular positions.
Polymorphism detection
Blood samples were taken from 51 pure Afshari breed lambs. Genomic DNA was extracted from whole blood by slight modification of salting out method (Boom et al. Citation1990) and genotyped using PCR-RFLP. Exon and intron regions from a part of the first inhibitor repetitive domain of the ovine calpastatin gene were chosen for amplification. Two primers of Cast2F (5′TGGGGCCCAATGACGCCATCG-ATC3′) and Cast2R (5′GGTGATCAGAAGTGCTGCTCCACC3′), which have been designed based on calpastatin gene of bovine (GenBank accession NO: L14450) were used to amplify a fragment of about 600 bp. PCR thermal condition was as follows: 95°c 3 min, then 35 cycle of 95°c 1 min, 62°c 1 min, 72°c 1.5 min, a 10 min extra extension in the last cycle.
PCR master mix was consisted of 50–100 ng ovine genomic DNA, 10 pmol each primer, 200 µM dNTPs, 1.5 mM MgCL2, PCR buffer 1X, and 1 unit Taq polymerase per 25-µL reaction.
PCR amplified products electrophoresed in a 1.5% agarose gel and fragments were visualised by a uv transilluminator. PCR products were digested with MspI to determine the mutation. The reaction mixture, which included 8 µl of digested PCR product with 2 µl loading buffer loaded on 7% non-denaturing polyacrylamide gel. The samples were run in 1×TBE (Tris-EDTA) buffer at 150 V for 14 h, followed by using silver staining method to visualise the gels. Selected PCR products from each three genotypes (MM, MN and NN) were purified and sequenced by using Cast2F and Cast2R primers on AB1 sequencing machine at Kowsar Laboratory.
Sequence data were compared with other calpastatin gene with blast search. Gene runner version 3.05 (Hastings Software Inc.,), ClustalW (Thompson et al. Citation1994), ClustalX (Thompson et al. Citation1997) programs have been used for determination and alignment of nucleic acid and amino acid sequences.
Statistical analysis
Associations of the animal genotypes with growth traits and calpastatin gene pholymorphism were determined by analysis of quantitative traits, which included birth weight; weaning weight; 6 and 9 months weight; pre and post finishing weight; and weight gain during the period between different ages and carcass traits included back fat thickness (mm) pre and post finishing and LM area (mm2) post finishing were analysed by ANOVA using GLM procedures of SAS (SAS Inst., Inc., Cary, NC). Fixed effects of genotype, and birth type were included as independent variables in the linear model. Mother age was treated as a covariate for birth and weaning weight in the model.
Table 1. Alleles and genotype frequency of calpastatin gene in Afshari sheep papulation.
Results and discussion
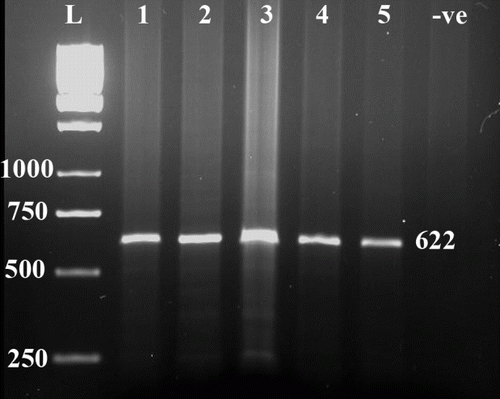
PCR amplification of genomic DNA in 51 Afshari sheep breed by Cast2F and Cast2R, followed by electrophores on 1.5% agarose gel have shown a band of about 600 bp (). Digestion of this PCR product by MspI on acrylamide gel revealed two fragments, one close to 300 bp and the other one, which stands higher than 300 bp band of DNA ladder. shows the alleles (M and N) and genotype (MM, MN and NN) frequency of calpastatin gene in Afshari sheep population. Sequencing of the three selected specimens corresponding to three genotypes of MM, MN and NN confirmed the correct size of 622 bp for the amplified fragment. Furthermore, restriction site analysis of these sequences revealed the presence of one site for MspI enzyme, representing two fragments of 336 and 286 bp on acrylamide gel () for allele N. An alignment of each genotypes sequence in Afshari sheep with the sequences deposited in GenBank revealed their 99% similarity with the bovine calpastatin gene (AY834770) in nucleic acid level. Similarly, this percentage between the sequences of the three genotypes was 99%. However, in amino acid level, no difference was detected within three genotypes.
Figure 1. L: one kb marker (SMO311), -ve: negative control, 1 to 5: PCR amplification of a partial calpastatin gene in Afshari sheep.
Carcass and growth traits estimate with corresponding means and SD for each genotype in 51 animals has shown in . No significant associations with growth and carcass traits were observed for the identified polymorphism. NN genotype had the highest eye muscle area, thickness, the lowest fat thickness, while genotype MM had the lowest weight and the highest fat thickness.
Palmer et al. (Citation1998) reported that Cast2F and Cast2R primers amplified a fragment of 622 bp in Dorset Down sheep, which is in the same size of amplified PCR product in current study carried out on Iranian Afshari sheep. In addition, restriction enzyme digestion of this fragment by MspI followed by electrophoresis revealed two bands of 286 and 336 bp on acrylamide gel (), that is corresponding to the expected sizes of these fragments reported by Palmer et al. (Citation1998).
Figure 2. Restriction digestion pattern for calpastatin gene (Cast1) with MspI. The 622 bp fragment was termed the M allele and the 336 and 286 bp fragments were termed the N allele. Assigned genotypes label each lane. L is 100 bp marker.

In spite of the role of calpastatin protein in muscle growth and the reported research works (Chung and Davis Citation2001; Kuryl et al. Citation2003), we may conclude that calpastatin gene has major effect in increasing the muscle mass. However, Kent et al. (Citation2004) reported that in transgenic mice, introduction of calpastatin gene to their genome, has not led to significant difference in weight between transgenic and control mice. The callipyge phenotype in sheep is inherited as an autosomal, dominant gene that maps to the ovine chromosome 18 (Cockett et al. Citation1994). Skeletal muscle mass in callipyge lambs are increasing in the pelvic limbs and back muscle. but muscle mass of fore limbs in callipyge lambs are similar to the normal sheep muscles (Jackson et al. Citation1997; Koohmaraie and Shackelford Citation1995). Calpastatin activity from callipyge lambs are 68–120% higher than in the same muscle from normal lambs and in the fore limbs muscles (Koohmaraie and Shackelford Citation1995).
Table 2. Phenotypic records for carcass and growth traits with corresponding means and SD for each genotype.
Although it seems that in current study, growth and carcass traits have not been affected from this fragment polymorphism, and there was not any correlation among those, perhaps due to the following reasons: (1)The polymorphisms we detected were in intron region, while the exons of calpastatin gene that we analysed are highly conserved. Although/Despite, silent mutation cause diseases (Montera et al. Citation2001) and introns are involved in controlling gene expression (Matthews et al. Citation1997), but introns have not direct role in amino acid sequence and the deduced protein structures (Matthews et al. Citation1997); (2)We selected the partial sequence of 12th and 13th exons and 12th intron that code sub-domain C and terminal sub-domain in bovine calpastatin protein (Raynaud et al. Citation2005), While it seems these partial sequences have not been the appropriate parts for detection of polymorphism in sheep breed. Moreover, Furthermore, Amanda et al (Citation2004) reported that sub-domain C and A have no or less inhibitor activity of calpain. Accordingly, we may suggest that further studies should be carried out on other positions of calpastatin gene especially in exon of sub-domain b B that has central role to inhibit calpain (Amanda et al. Citation2004). On the other hand, calpastatin promoter and domain L were considered as polymorphic regions that are related to carcass and growth traits; (3) Despite having calpastatin polymorphism we could not found the significance correlation between genetic variation with growth and carcass traits.
Conclusion
The partial segment of 12th and 13th exons were sequenced and the polymorphisms in 12th intron were identified. No significant associations were observed between these polymorphisms and growth and carcass traits.
Acknowledgements
We wish to thank colleagues for their technical assistance, H. Mehri, F. Emanikhah, and Dr S. Zakeri. We also wish to thank all staffs and students in Animal science department in the Zanjan University and also Malaria Research Group (MVRG) of biotechnology department in Pasteur Institute of Iran.
References
- Amanda , W , Thompson , VF and Goll , DE . 2004 . Interaction of calpastatin with calpain: a review . Biological chemistry , 385 : 465 – 472 .
- Boom , R , Sol , CJ , Salmans , MM , Jansoen , CL , Wertheim-van Dillen , PM and Noorda , JVD . 1990 . Rapid and simple method for purification of nucleic acids . Journal of Clinical Microbiology , 28 : 495 – 503 .
- Casas , E , White , SN , Wheeler , TL , Shackelford , SD , Koohmaraie , M , Riley , DG , Chase , JrCC , Johnson , DD and Smith , TPL . 2006 . Effects of calpastatin and µ-calpain markers in beef cattle on tenderness traits . Journal of Animal Science , 84 : 520 – 525 . doi: 10.2527/jas.2006-200
- Chung HY , Davis EM. 2001 . Effect of calpain and calpastatin genotypes on growth of angus bulls . Ohio State University-Research and Reviews: Beef and sheep [Internet]; [cited 2010]. Available from: http://ohioline.osu.edu/sc181/sc181_5.html
- Cockett NF , Jackson SP , Shay TL , Nielsen D , Moore SS , Stelle MR , Barendse W , Green RD , Georges M. 1994 . Chromosomal localization of the callipyge gene in sheep (Ovis aries) using bovine DNA markers . Proceedings of the National Academy of sciences of the USA . 91 : 3019 – 3023 .
- Goll , DE , Thompson , VF , Taylor , RG and Ouali , A . 1998 . The calpain system and skeletal muscle growth . Canadian Journal of Animal Science , 78 : 503 – 512 . doi: 10.4141/A98-081
- Goll , EG , Thompson , VF , Li , H , Wei , W and Cong , J . 2003 . The calpain system . Physiological Reviews , 83 : 731 – 801 .
- Jackson , SP , Miller , MF and Green , RD . 1997 . Phenotypic characterization of Rambouillet sheep expressing the callipyge gene: III. Muscle weight distribution . Journal of Animal Science , 75 : 133 – 138 .
- Kent , MP , Spencer , MJ and Koohmaraie , M . 2004 . Postmortem proteolysis is reduced in transgenic mice overexpressing calpastatin . Journal of Animal Science , 82 : 794 – 801 .
- Koohmaraie , M and Shackelford , SD . 1995 . A muscle hypertrophy condition in lamb (callipyge): characterization of effects on muscle growth and meat quality traits . Journal of Animal Science , 73 : 3596 – 3607 .
- Kuryl , J , Kapelanski , W , Pierzchala , M , Grajewska , S and Bocian , M . 2003 . Preliminary observations on the effect of calpastatin gene (CAST) polymorphism on carcass traits in pigs . Animal Science Papers and reports , 2 : 87 – 95 .
- Matthews , HR , Freedland , R and Miesfeld , RL . 1997 . Biochemistry a short course , New York : Wiley-liss, Inc .
- Montera , M , Piaggio , F , Marchese , C , Gismondi , V , Stella , A , Resta , N , Veresco , L , Guanti , G and Mareni , C . 2001 . A silent mutation in exon 14 of the APC gene is associated with exon skipping in a FAP family . Journal of Medical Genetics , 38 : 863 – 867 . doi: 10.1136/jmg.38.12.863
- Mota , N , Mendoza , GD , Plata , FX , Martínez , JA , Lee , H , Rojo , R and Crosby , MM . 2011 . Effect of exogenous glucoamylase enzymes and reduction of grain level on lamb performance . Journal of Applied Animal Research , 39 : 129 – 131 . doi: 10.1080/09712119.2011.565188
- NRC . 1985 . Nutrient Requirements of Sheep . Washington , DC : National Academy Press .
- Palmer , R , Roberts , N , Hickford , JGH and Bickerstaffe , R . 1998 . Rapid communication: PCR-RFLP for MspI and NcoI in the ovine calpastatin gene . Journal of Animal Science , 76 : 1499 – 1500 .
- Raynaud , P , Jayat-vignoles , C , Laforet , ML , Leveziel , H and Amarger , V . 2005 . Four promoters direct expression of the calpastatin gene . Archives of Biochemistry and Biophysics , 437 : 69 – 77 . doi: 10.1016/j.abb.2005.02.026
- Ríos Rincón , FGR , Barreras-Serrano , A , Estrada-Angulo , A , Obregón , JF , Plascencia-Jorquera , A , Portillo-Loera , JJ and Zinn , RA . 2010 . Effect of level of dietary zilpaterol hydrochloride (β2-agonist) on performance, carcass characteristics and visceral organ mass in hairy lambs fed all-concentrate diets . Journal of Applied Animal Research , 38 : 33 – 38 . doi: 10.1080/09712119.2010.9707150
- Schenkel , FS , Miller , SP , Jiang , Z , Mandell , IB , Ye , X , Li , H and Wilton , JW . 2006 . Association of a single nucleotide polymorphism in the calpastatin gene . Journal of Animal Science , 84 : 291 – 299 .
- Thompson , JD , Gibson , TJ , Plewniak , F , Jeanmougin , F and Higgins , DG . 1997 . The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools . Nucleic Acids Research , 24 : 4876 – 4882 . doi: 10.1093/nar/25.24.4876
- Thompson , JD , Higgins , DG and Gibson , TJ . 1994 . CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice . Nucleic Acids Research , 22 : 4673 – 4680 . doi: 10.1093/nar/22.22.4673
- Zhao , Q , Davis , ME and Hines , HC . 2004 . Associations of polymorphisms in the Pit-1 gene with growth and carcass traits in angus beef cattle . Journal of Animal Science , 82 : 2229 – 2233 .